Abstract
Previous studies have reached conflicting conclusions regarding the proportion of Merkel cell carcinomas (MCCs) that contain the Merkel cell polyomavirus (MCPyV) and the clinical significance of tumor viral status. To address these controversies, we detected MCPyV large T antigen using immunohistochemistry with two distinct antibodies and MCPyV DNA using quantitative PCR. Tumors were called MCPyV-positive if two or more of these three assays indicated presence of this virus. A total of 53 of 282 (19%) MCC tumors in this cohort were virus-negative using this multimodal system. Immunohistochemistry with the CM2B4 antibody had the best overall performance (sensitivity = 0.882, specificity = 0.943) compared with the multimodal classification. Multivariate analysis including age, sex, and immunosuppression showed that, relative to MCC patients with virus-positive tumors, virus-negative MCC patients had significantly increased risk of disease progression (hazard ratio = 1.77, 95% confidence interval = 1.20–2.62) and death from MCC (hazard ratio = 1.85, 95% confidence interval = 1.19–2.89). We confirm that approximately 20% of MCCs are not driven by MCPyV and that such virus-negative MCCs, which can be quite reliably identified by immunohistochemistry using the CM2B4 antibody alone, represent a more aggressive subtype that warrants closer clinical follow-up.
INTRODUCTION
Merkel cell carcinoma (MCC) is a rare and aggressive neuroendocrine skin cancer with a disease-associated mortality of greater than 40% (Lemos et al., 2010). The incidence of this malignancy has quadrupled over the last 20 years, likely because of the increasing prevalence of risk factors including advanced age, cumulative UV exposure, and systemic immune suppression (Fitzgerald et al., 2015; Lemos and Nghiem, 2007). The discovery that HIV-positive patients have a greater than 10-fold risk of developing MCC suggested that the immune system plays a critical role in the development of this cancer, which in turn led to the search for a potential infectious etiology (Engels et al., 2002). In 2008, the Moore-Chang group found that most MCCs were associated with the newly discovered, highly prevalent Merkel cell polyomavirus (MCPyV) (Feng et al., 2008). After the discovery of MCPyV, viral large and small T antigens have been shown to be constitutively expressed in MCC and capable of driving oncogenesis through a variety of mechanisms, including binding and inactivation of the retinoblastoma tumor suppressor (Church and Nghiem, 2015; DeCaprio and Garcea, 2013; Houben et al., 2010).
The initial report identified MCPyV in eight of 10 (80%) MCCs (Feng, 2008). Subsequent studies using PCR and/or immunohistochemistry (IHC) targeting MCPyV large T antigen have at times provided disparate estimates of viral positivity in MCC, ranging from as low as 24% in some series to 100% in another (Garneski et al., 2009; Rodig et al., 2012). With wide variation in methods and populations, the remaining 10 of 12 major studies to address this question to date have estimated anywhere from 46% to 89% MCPyV positivity, with the aggregate estimate of all studies being 76% (453 of 595 unique MCCs) (Andres et al., 2010; Feng et al., 2008; Foulongne et al., 2008; Garneski et al., 2009; Harms et al., 2013; Kassem et al., 2008; Katano et al., 2009; Leroux-Kozal et al., 2015; Nardi et al., 2012; Rodig et al., 2012; Schrama et al., 2011; Sihto et al., 2009). At this time, there is currently no accepted standard method for determining MCC viral status, neither is there consensus on which single assay might be most appropriate for routine clinical application.
Beyond the question of the prevalence of MCPyV in MCC, there are also conflicting reports as to whether tumor viral status has prognostic significance. One study of 114 MCC patients found that virus-negative patients had significantly worse overall survival compared with their virus-positive counterparts in a multivariate model (13.0% vs. 45.0% 5-year survival) (Sihto et al., 2009). A later study among 60 MCC patients found that virus-negative patients had significantly worse overall and recurrence-free survival in a univariate model but no such association when tumor stage and nodal involvement were taken into account (Nardi et al., 2012). The largest published cohort to date to address this question, however, found no significant survival difference on the basis of viral status among 127 MCC patients (Schrama et al., 2011).
Because of the lack of a standard clinical test for MCPyV and the unclear prognostic significance of tumor viral status, MCCs are not currently routinely analyzed for the presence of MCPyV. To identify the prevalence of MCPyV among 282 tumors and determine the best clinical test for viral detection, we used optimized quantitative PCR (qPCR) and IHC. We then assessed the impact of tumor viral status on clinical outcomes including progression-free survival, MCC-specific survival, and overall survival.
RESULTS
Multimodal approach for MCPyV viral status determination
All 282 MCC tumors were analyzed by qPCR for MCPyV DNA with the LT4 primer set normalized against a control gene, thyroid peroxidase (TPO), previously shown to have stable copy number in MCC (Figure 1 and Materials and Methods section). All samples were also stained with both CM2B4 and Ab3 antibodies, which target overlapping portions of the MCPyV large T antigen. Using the combination of qPCR, CM2B4, and Ab3 data and requiring a given tumor to have at least two modalities in agreement for viral classification, we determined 229 of 282 tumors (81.2%) to be MCPyV-positive and 53 of 282 tumors (18.8%) to be MCPyV-negative (Table 1). All three tests agreed on tumor viral status (positive or negative) in 167 (59.2%) of the 282 samples, and the remaining tumors were determined to be positive on two of three assays (see Supplementary Figure S1 online).
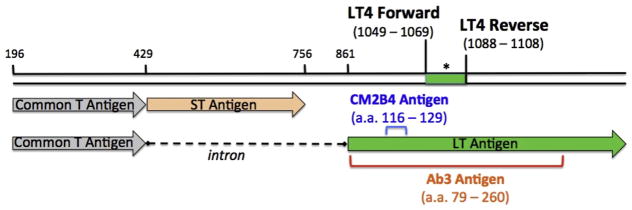
Figure 1. Detection of MCPyV DNA and oncoprotein in MCC tumors.
Schematic diagram of the MCPyV small and large Tantigens. Numbers correspond to the nucleotide position in the MCPyV genome. The LT4 primer set targets a 60-base pair segment of the persistently expressed portion of the truncated large T antigen. The asterisk indicates the TaqMan probe location (see Materials and Methods for details). The colored brackets in the lower portion of the figure show amino acid positions of antigens used to generate the large T antibodies: CM2B4 (blue) and Ab3 (red). a. a., amino acid; MCC, Merkel cell carcinoma; MCPyV, Merkel cell polyomavirus.
Table 1.
Individual performance of the three assays (qPCR, CM2B4, and Ab3) in classifying intratumoral MCPyV in Merkel cell carcinoma compared with the multimodal approach1
| Virus-Positive, n* | Virus-Negative, n* | Total, n | Assay Sensitivity | Assay Specificity | |
|---|---|---|---|---|---|
| qPCR-pos | 189 | 10 | 199 | ||
| qPCR-neg | 40 | 43 | 83 | ||
| Total | 229 | 53 | 282 | 0.825 | 0.811 |
| CM2B4-pos | 202 | 3 | 205 | ||
| CM2B4-neg | 27 | 50 | 77 | ||
| Total | 229 | 53 | 282 | 0.882 | 0.943 |
| Ab3-pos | 225 | 29 | 254 | ||
| Ab3-neg | 4 | 24 | 28 | ||
| Total | 229 | 53 | 282 | 0.983 | 0.453 |
Abbreviations: neg, negative; pos, positive.
Tumor viral status as determined by the multimodal approach (at least two of the three assay results in agreement).
As a means of validation, we cross-referenced this classification with available data from a serologic assay that detects circulating antibodies against the MCPyV T antigens (Paulson et al., 2010). In general, circulating T antigen antibodies would not be expected to be present in patients who do not harbor MCPyV in their tumors and would be present in most patients whose tumors were virus-positive. A total of 58 of the 229 patients classified as having MCPyV-positive tumors had serologic data available; 37 of these 58 (63.8%) patients had detectable T antigen antibodies at the time of diagnosis. In addition, 17 of the 53 patients classified as having MCPyV-negative tumors had serologic data available. None of these 17 patients had detectable T antigen antibodies at the time of diagnosis. Thus, results from the oncoprotein antibody assay further support the viral status determinations made with the multimodal approach.
Detection of MCPyV DNA by qPCR
A total of 199 of 282 (70.6%) MCC tumors produced an LT4/TPO ratio of 0.01 or greater (range = 0.01–504.5) and were called virus-positive by qPCR (Table 1). The remaining 83 (29.4%) samples (LT4/TPO ratios between 0 and 0.009) were called virus-negative by qPCR. Most samples classified as PCR-negative did in fact have low-level LT4 positivity, although with LT4/TPO ratio below the cutoff of 0.01, suggesting that fewer than 1% of cells were positive for viral DNA and that the signal represented background infection that can be seen in normal tissues. On the basis of this classification, qPCR was 82.5% sensitive and 81.1% specific for identifying intratumoral MCPyV compared the multimodal approach described earlier (Table 1 and Materials and Methods section).
Three of 16 (18.8%) non-MCC skin cancers (basal cell carcinoma, squamous cell carcinoma, melanoma) were positive for MCPyV by PCR (LT4/TPO ratio range = 0.0277–1.858; data not shown). All three of these tumors had Ab3 and CM2B4 Allred scores less than or equal to 2 and were therefore considered to have amplification that was false-positive or indicative of background MCPyV infection.
Detection of MCPyV large T antigen by IHC
Using the CM2B4 antibody, we found that 205 of 282 (72.7%) MCC tumors were called virus-positive because they had an Allred score greater than 2 (Table 1). The remaining 77 (27.3%) samples were virus-negative by CM2B4. On the basis of this classification, CM2B4 was 88.2% sensitive and 94.3% specific for identifying intratumoral MCPyV versus the multimodal approach (Table 1 and Materials and Methods section). None of 16 control tissues (non-MCC skin cancers) were virus-positive by CM2B4 (data not shown).
Using the Ab3 monoclonal antibody, 254 of 282 (90.1%) MCCs were called virus-positive because they had an average Allred score greater than 2 (Table 1). The remaining 28 (9.9%) samples were virus-negative by Ab3. On the basis of this classification, Ab3 was 98.3% sensitive and 45.3% specific for identifying intratumoral MCPyV versus the multimodal approach (Table 1 and Materials and Methods section). Three of 16 (18.8%) control tissues (non-MCC skin cancers) were virus-positive by Ab3 (data not shown). All three of these tumor samples had LT4/TPO ratios less than 0.01 and CM2B4 Allred scores less than or equal to 2 and were therefore considered to have false-positive staining.
In some cases, staining of keratinocytes and adnexal and vascular structures by Ab3 was observed. Similarly, low-level staining of tonsillar tissue by CM2B4 was occasionally observed. In each case, this was considered nonspecific and did not affect the interpretation of tumor tissue staining.
MCC cohort and survival analysis
From the 281 patients with available clinical data (Figure 2), 1,211 person-years of follow-up time were contributed. The median age at diagnosis was 71 years for both the virus-negative and virus-positive groups (Table 2). The proportion of male and white patients and the presence of systemic immunosuppression were similar between groups. Virus-negative tumors tended to be smaller than virus-positive tumors at presentation (median = 1.1 cm vs. 1.9 cm, respectively). Despite this trend, patients with MCPyV-negative tumors were more likely to present with advanced (nodal or distant metastatic) disease than patients with virus-positive tumors (66.7% vs. 48.3%). Patients with virus-negative tumors were less likely to undergo surgical excision (74.4% vs. 93.0%). This is likely due to the increased tendency to present with stage III or IV disease; however, administration of radiation and chemotherapy were similar between the groups.
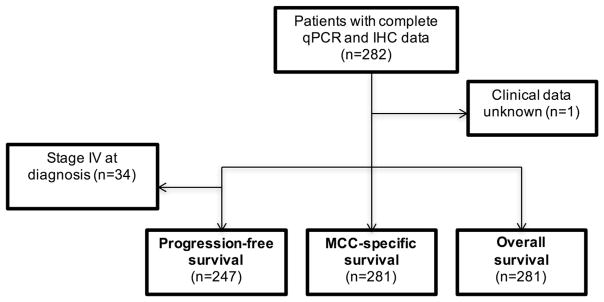
Figure 2. Flowchart of survival analysis inclusion criteria.
All patients with clinical follow-up data were included in survival analysis. For progression-free survival, patients who presented with metastatic disease were excluded from analysis. IHC, immunohistochemistry; MCC, Merkel cell carcinoma; qPCR, quantitative PCR.
Table 2.
Baseline characteristics of MCC patients, by tumor MCPyV status, Fred Hutchinson Cancer Research Center, 2004–2012
| Characteristics | Virus-Negative n = 53 | Virus-Positive n = 229 | ||
|---|---|---|---|---|
| Patient characteristics | Median | SD | Median | SD |
| Median age, years | 71 | 11.2 | 71 | 12.6 |
| n | % | n | % | |
| Sex | ||||
| Female | 18 | 34.0 | 87 | 38.0 |
| Male | 35 | 66.0 | 142 | 62.0 |
| Race | ||||
| White | 51 | 98.1 | 221 | 96.9 |
| Other | 1 | 1.9 | 7 | 3.1 |
| Immunosuppression | ||||
| Absent | 51 | 96.2 | 211 | 92.1 |
| Present | 2 | 3.8 | 18 | 7.9 |
| Tumor characteristics | Median | SD | Median | SD |
| Primary tumor size, cm | 1.1 | 1.7 | 1.9 | 1.7 |
| n | % | n | % | |
| Site of primary tumor | ||||
| Head and neck | 22 | 41.5 | 74 | 32.3 |
| Trunk | 6 | 11.3 | 10 | 4.4 |
| Buttock/genitalia | 2 | 3.7 | 18 | 7.9 |
| Extremity | 10 | 18.8 | 100 | 43.7 |
| Unknown primary | 13 | 24.5 | 27 | 11.8 |
| Stage at Diagnosis | ||||
| Local | 14 | 33.3 | 93 | 51.7 |
| I | 10 | 23.8 | 65 | 36.1 |
| II | 4 | 9.5 | 28 | 15.6 |
| Advanced | 28 | 66.7 | 87 | 48.3 |
| III | 20 | 47.6 | 61 | 33.9 |
| IV | 8 | 19.1 | 26 | 14.4 |
| Treatment | ||||
| Surgical excision | ||||
| Yes | 29 | 74.4 | 187 | 93.0 |
| No | 10 | 25.6 | 14 | 7.0 |
| Radiation | ||||
| Yes | 29 | 61.7 | 138 | 64.2 |
| No | 18 | 38.3 | 77 | 35.8 |
| Chemotherapy | ||||
| Yes | 10 | 19.2 | 30 | 13.3 |
| No | 42 | 80.8 | 195 | 86.7 |
Abbreviations: MCC, Merkel cell carcinoma; MCPyV, Merkel cell polyomavirus; SD, standard deviation.
A total of 88 of 202 (43.6%) virus-positive and 30 of 45 (66.7%) virus-negative MCC patients with stage I–III disease at presentation experienced progression over the assessed time interval (P = 0.0052) (Figure 3a). The median time to progression for the entire cohort was 5.1 years (not reached among the virus-positive patients, and 1.2 years among the virus-negative patients). In univariate analysis, virus-negative patients had a 1.80-fold higher risk of progression (95% confidence interval [CI] = 1.22–2.64, P = 0.003) (Table 3). Sex and immune status, but not age or stage at presentation, were significantly associated with progression. In multivariate analysis including age, sex, and immune status, virus-negative patients were 1.77 times more likely to progress (95% CI = 1.20–2.62, P = 0.004). After disease stage at presentation was accounted for, the hazard ratio fell to 1.55 (95% CI = 0.96–2.49, P = 0.073), suggesting that stage accounted for 0.22 (28.5%) of the increased risk of progression among virus-negative patients.
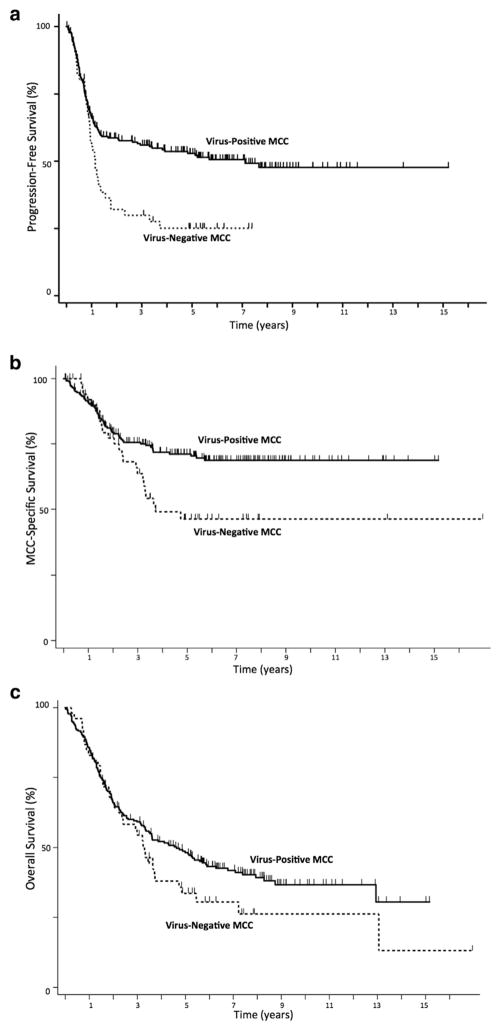
Figure 3. Kaplan-Meier curves of survival among MCC patients whose tumors were MCPyV-positive or -negative.
(a) Patients with virus-negative tumors had significantly decreased progression-free survival (P = 0.0052). (b) Patients with virus-negative tumors had significantly decreased MCC-specific survival (P = 0.015). (c) Patients with virus-negative tumors had a nonsignificant trend for poorer overall survival (P = 0.14). MCC, Merkel cell carcinoma.
Table 3.
Univariate and multivariate analysis of progression-free survival, MCC-specific survival, and overall survival on the basis of MCPyV tumor viral status
| Risk Factor | Hazard Ratio | 95% Confidence Interval | P-Value |
|---|---|---|---|
| Progression-Free Survival | |||
| Univariate analysis | |||
| Tumor viral status | |||
| MCPyV-positive | Reference | Reference | — |
| MCPyV-negative | 1.80 | 1.22–2.64 | 0.003 |
| Multivariate analysis excluding stage | |||
| Tumor viral status | |||
| MCPyV-positive | Reference | Reference | — |
| MCPyV-negative | 1.77 | 1.20–2.62 | 0.004 |
| Age | 1.01 | 0.99–1.02 | 0.27 |
| Sex | |||
| Female | Reference | Reference | — |
| Male | 1.73 | 1.14–2.62 | 0.010 |
| Immunosuppression | |||
| Absent | Reference | Reference | — |
| Present | 2.14 | 1.15–4.01 | 0.017 |
| Multivariate analysis including stage | |||
| Tumor viral status | |||
| MCPyV-positive | Reference | Reference | — |
| MCPyV-negative | 1.55 | 0.96–2.49 | 0.073 |
| Age | 1.02 | 1.00–1.03 | 0.063 |
| Sex | |||
| Female | Reference | Reference | — |
| Male | 1.76 | 1.07–2.91 | 0.027 |
| Immunosuppression | |||
| Absent | Reference | Reference | — |
| Present | 2.28 | 1.12–4.63 | 0.022 |
| Stage | |||
| I | Reference | Reference | — |
| II | 1.61 | 0.91–2.86 | 0.10 |
| III | 1.42 | 0.87–2.32 | 0.16 |
| IV | Omitted | Omitted | — |
| MCC-Specific Survival | |||
| Univariate analysis | |||
| Tumor viral status | |||
| MCPyV-positive | Reference | Reference | — |
| MCPyV-negative | 1.79 | 1.14–2.80 | 0.011 |
| Multivariate analysis excluding stage | |||
| Tumor viral status | |||
| MCPyV-positive | Reference | Reference | — |
| MCPyV-negative | 1.85 | 1.19–2.89 | 0.006 |
| Age | 1.00 | 0.99–1.02 | 0.57 |
| Sex | |||
| Female | Reference | Reference | — |
| Male | 1.84 | 1.13–2.99 | 0.014 |
| Immunosuppression | |||
| Absent | Reference | Reference | — |
| Present | 2.64 | 1.29–5.41 | 0.008 |
| Multivariate analysis including stage | |||
| Tumor viral status | |||
| MCPyV-positive | Reference | Reference | — |
| MCPyV-negative | 1.50 | 0.88–2.58 | 0.14 |
| Age | 1.00 | 0.98–1.02 | 0.90 |
| Sex | |||
| Female | Reference | Reference | — |
| Male | 1.93 | 1.11–3.33 | 0.019 |
| Immunosuppression | |||
| Absent | Reference | Reference | — |
| Present | 3.94 | 2.14–7.26 | <0.001 |
| Stage | |||
| I | Reference | Reference | — |
| II | 1.79 | 0.77–4.17 | 0.18 |
| III | 2.92 | 1.52–5.63 | 0.001 |
| IV | 5.29 | 2.21–12.65 | <0.001 |
| Overall Survival | |||
| Univariate analysis | |||
| Tumor viral status | |||
| MCPyV-positive | Reference | Reference | — |
| MCPyV-negative | 1.32 | 0.93–1.87 | 0.12 |
| Multivariate analysis excluding stage | |||
| Tumor viral status | |||
| MCPyV-positive | Reference | Reference | — |
| MCPyV-negative | 1.25 | 0.90–1.73 | 0.180 |
| Age | 1.05 | 1.03–1.06 | <0.001 |
| Sex | |||
| Female | Reference | Reference | — |
| Male | 1.52 | 1.11–2.06 | 0.008 |
| Immunosuppression | |||
| Absent | Reference | Reference | — |
| Present | 2.39 | 1.29–4.44 | 0.006 |
| Multivariate analysis including stage | |||
| Tumor Viral Status | |||
| MCPyV Positive | Reference | Reference | — |
| MCPyV Negative | 1.30 | 0.86–1.96 | 0.21 |
| Age | 1.04 | 1.02–1.06 | <0.001 |
| Sex | |||
| Female | Reference | Reference | — |
| Male | 1.47 | 1.01–2.14 | 0.044 |
| Immunosuppression | |||
| Absent | Reference | Reference | — |
| Present | 3.25 | 1.81–5.81 | <0.001 |
| Stage | |||
| I | Reference | Reference | — |
| II | 1.60 | 0.91–2.81 | 0.10 |
| III | 2.03 | 1.30–3.17 | 0.002 |
| IV | 3.38 | 1.91–5.97 | <0.001 |
Abbreviations: MCC, Merkel cell carcinoma; MCPyV, Merkel cell polyomavirus.
A total of 60 of 228 (26.3%) virus-positive and 24 of 53 (45.3%) virus-negative MCC patients died from MCC over the assessed time interval (P = 0.015) (Figure 3b). Median time to death from MCC was not reached in the virus-positive patients, and it was 3.7 years among the virus-negative patients. In univariate analysis, virus-negative patients had a 1.79-fold higher risk of death from MCC (95% CI = 1.14–2.85; P = .011) (Table 3). Sex, immune status, and stage at presentation, but not age, were significantly associated with MCC-specific survival. In multivariate analysis including age, sex, and immune status, virus-negative patients were 1.85 times more likely to die from MCC (95% CI = 1.19–2.89; P = 0.006). After disease stage at presentation was accounted for, the hazard ratio fell to 1.50 (95% CI = 0.88–2.58; P = 0.14), suggesting stage accounted for 0.35 (41.2%) of the increased risk of MCC-specific death among virus-negative cases.
A total of 132 of 228 (57.9%) virus-positive and 37 of 53 (69.8%) virus-negative MCC patients died over the assessed time interval (P = 0.14) (Figure 3c). Median overall survival time was 3.7 years in this cohort (4.6 years for virus-positive and 3.3 years for virus-negative patients). Age, sex, immune status, and stage at presentation were all statistically significantly associated with overall survival (Table 3). Tumor viral status was not statistically significantly associated with overall survival in univariate or multivariate analyses.
Survival analysis using CM2B4 Allred score alone to classify MCPyV tumor viral status yielded almost identical results to those described above (see Supplementary Figure S2 and Supplementary Table S1 online). The use of a higher LT4/TPO ratio of 0.1 as a cutoff to determine viral status as in Goh et al. (2015) also yielded similar results (data not shown). Among patients whose tumors were identified as virus-positive, neither viral copy number nor intensity of IHC staining correlated with risk of progression or death from MCC (data not shown).
DISCUSSION
With 282 unique MCC tumors, to the best of our knowledge, this is the largest study that addresses the prevalence and prognostic significance of MCPyV in MCC. We assessed the presence of MCPyV with qPCR using a primer set (LT4) with superior performance to previously reported primers, as well as with IHC using both CM2B4 and Ab3 antibodies, which each target the MCPyV large T antigen. Recognizing from these data and the prior literature that no standard criteria exists for determining MCPyV tumor viral status, we required tumors to show detectable MCPyV by at least two of these three modalities to be classified as MCPyV-positive. This approach meant that the presence of viral DNA alone, in the absence of detectable oncoprotein expression in tumor cells, was insufficient to classify a tumor as virus-positive. This requirement is relevant because MCPyV is prevalent in normal skin, and a signal from adjacent stromal MCPyV should not be interpreted as causal for tumorigenesis in the absence of viral protein expression in the tumor. Using this multimodal approach, we found that 19% (approximately one fifth) of MCCs are not driven by the virus. This finding is highly consistent with the aggregate result from prior studies (76% MCPyV positivity in MCC tumors, as discussed), although we note that individual rates of viral positivity in MCC cohorts have varied widely between 24% and 100% (Andres et al., 2010; Feng et al., 2008; Foulongne et al., 2008; Garneski et al., 2009; Harms et al., 2013; Kassem et al., 2008; Katano et al., 2009; Leroux-Kozal et al., 2015; Nardi et al., 2012; Rodig et al., 2012; Schrama et al., 2011; Sihto et al., 2009).
Patients with virus-negative tumors were more likely to recur after treatment and to die from MCC, even in multivariate models including prognostic factors known to have significant impact on MCC survival. Because advanced stage at presentation is likely to be a causal mechanism between tumor viral status and poorer outcome in MCC, we included this variable in multivariate analyses in an attempt to understand its role as a mediator. When stage at presentation was added to the analysis, the associations with viral status were no longer statistically significant, suggesting that advanced stage associated with virus-negative patients partially but not entirely accounts for the poorer clinical outcomes seen in the cohort. The lack of statistical significance in overall survival is likely due to the many competing causes of death in this population with a median age at diagnosis of 71 years. Taken together, these analyses support the conclusion that virus-negative tumors may be more clinically aggressive even though they tended to be smaller in diameter at presentation.
Plausible biological mechanisms to explain this finding have been previously described. MCPyV-negative tumors have been shown to have a higher number of chromosomal aberrations (Goh et al., 2016; Paulson et al., 2009), a greater nucleotide mutation burden (Goh et al., 2016; Harms et al., 2015), and a higher number of mutations in known oncogenic pathways including PI3K/pAKT, P53, and RB1 (Goh et al., 2016; Nardi et al., 2012). Virally driven MCCs may also be more immunogenic because of their constitutive expression of oncoproteins that may serve as targets for cytotoxic tumor-infiltrating lymphocytes.
This study has important limitations. Several samples were determined to be virus-positive despite having lower-than-cutoff qPCR values, thus raising the question of whether the IHC assay results were false-positive. In these 40 (of 229) such cases, we considered this to be more likely a deficiency of the qPCR assay than a false-positive result for both IHC assays. Importantly, 39 of those 40 “PCR-negative” samples had detectible MCPyV DNA at a level of at least one copy per 1,000 cells. To be consistent with the Allred IHC system (one cell positive in 100 tumor cells), we opted to keep a viral copy number/cell threshold of 0.01 (equivalent to one DNA viral copy per 100 cells; see Materials and Methods section). Possible sources of lower-than-cutoff qPCR values include dilution of tumor tissue by surrounding stromal tissue, mutations in the region of MCPyV targeted by the LT4 primer, and variable DNA quality extracted from archival tumor tissue embedded in paraffin blocks of various ages and processing techniques. We opted not to test the samples with alternative primers, because more strongly positive results would only affirm the virus-positive call, whereas further negative results would be subject to the same sources of potential error outlined above and would not resolve the question. Regarding the clinical significance of tumor viral status, this retrospective cohort study suffers from possible biases. Specifically, although we included known confounders of survival in MCC (e.g., age, sex, immunosuppression), it is possible that there is confounding from other characteristics not captured by our database (e.g., smoking, medical comorbidities, etc). Lastly, despite its relatively large size, this study may have been underpowered to detect significant differences in clinical outcomes when including clinical stage in survival models.
Despite these limitations, both the multimodal classification system and CM2B4 IHC alone reliably identified a cohort with worse outcomes from their cancer. The observed higher risk of recurrence, progression, and death from MCC in patients with virus-negative tumors suggests that it may be clinically indicated to determine tumor viral status at the time of diagnosis, because the results may affect prognosis and optimal initial management. To this end, we believe that the CM2B4 antibody test may be well suited for routine clinical use because of the following factors: its improved sensitivity and specificity profile versus PCR with the LT4 primer and versus IHC with Ab3, its ability to identify essentially the same virus-negative cohort as the multimodal approach that used all three assays (see Results, Supplementary Figure S2, and Supplementary Table S1), and its commercial availability and the ease with which it could be included in the workflow of clinical laboratories accustomed to IHC. Immune therapy with PD1 pathway blockade has recently been shown to be effective in treating both virus-negative and virus-positive MCC tumors and can thus be considered regardless of tumor viral status (Nghiem et al., 2016). However, with the knowledge that virus-negative tumors may be more aggressive, clinicians may consider larger initial surgical margins, larger radiotherapy fields, and the use of regional nodal therapy even in the absence of documented nodal metastasis. Closer clinical follow-up and more frequent radiologic surveillance may be justified for patients with virus-negative MCC tumors because of their higher risk of recurrence and the fact that serologic monitoring (Paulson et al., 2010) is not feasible for this patient population.
MATERIALS AND METHODS
Patient selection and study approval
A total of 1,078 patients with pathologically verified MCC diagnosed between 1980 and 2015 were enrolled in an ongoing, institutional review board-approved (FHCRC-6585) repository of clinical data and specimens based in Seattle between 2004 and 2015. MCC patients gave their written informed consent for participating in research to examine their clinical data, leftover tissue, and blood. At the time of enrollment, patient data including age, sex, presence of systemic immunosuppression (e.g., chronic lymphocytic leukemia, non-Hodgkin lymphoma, HIV, solid organ transplantation), tumor characteristics, American Joint Committee on Cancer clinical stage at diagnosis, and therapies received were ascertained. Follow-up data were obtained at intervals varying from 3–12 months. Patients were censored at the last follow-up date if they could no longer be reached or chose to no longer participate.
For the purposes of this study, all patients who had pathology specimens available for qPCR and IHC analysis and had clinical follow up information, 282 of 1,078 enrolled patients, were included. Each patient contributed one tumor specimen. Primary tumors were used for evaluation whenever possible (242 of 282 patients). In the remaining patients presenting with stage III or IV disease but with no known primary cutaneous tumor (i.e., unknown primary, 40 of 282 patients), metastatic tumor tissue was used instead.
IHC
IHC staining was performed at Fred Hutchinson Cancer Research Center Experimental Histopathology Laboratory. Formaldehyde-fixed paraffin-embedded (FFPE) tumor blocks were sectioned and stained with hematoxylin and eosin. IHC was performed using 4-μm–thick tissue sections in large batches of samples to minimize staining variation between runs. Mouse monoclonal antibodies Ab3 (Rodig et al., 2012) (a generous gift from James DeCaprio, Dana-Farber Cancer Institute) and CM2B4 (Shuda et al., 2009) (SC-136172; Santa Cruz Biotechnology, Santa Cruz, CA) were applied at the following final concentrations: 2.4 μg/ml and 2.0–4.0 μg/ml, respectively, after signal optimization for each batch. Slides were stained using a DAKO Autostainer (Agilent Technologies, Santa Clara, CA) platform. Images were acquired using a Leica DFC290 Microscope (Leica, Wetzlar, Germany) and accompanying Image-Scope Viewer software (Leica). Slides were scored on the Allred scoring system, which combines intensity of staining and proportion of cells stained into a single score of 0–8 points (Allred et al., 1998). Samples were scored by either one or two independent pathologists. In cases for which two pathologists reviewed the samples, the mean Allred score was used. An Allred score of less than or equal to 2 (<1% of cells with weak staining) was considered negative for presence of the viral antigen. Samples of known MCPyV viral status (negative, low positive, and high positive) were used for verification of consistent antibody performance over time.
DNA extraction and quantification
DNA was extracted from samples that were originally obtained at the time of diagnosis. Extractions were performed on three or four 4-μm tumor tissue curls from paraffin-embedded tumor using the QIAamp DNA-FFPE Tissue Kit (Qiagen, Valencia, CA) following the manufacturer’s instructions. Cell lines that had previously been established to be virus-positive (MKL-1) and virus-negative (UISO) (Houben, 2010), as well as peripheral blood mononuclear cells, were used as controls and extracted in parallel with the tumor tissue. DNA was also extracted from basal cell carcinomas (n = 12), squamous cell carcinomas (n = 2), and malignant melanomas (n = 2). DNA concentration and quality control for impurities were determined with a NanoDrop spectrophotometer (ThermoFisher Scientific, Waltham, MA).
qPCR
qPCR was performed at the University of Washington Molecular Virology Laboratory. Sequences of MCPyV were aligned using Sequencher program (Gene Codes Corporation, Ann Arbor, MI). Multiple real-time TaqMan primer/probe sets were then selected from the conserved regions of different genes (Large T, Small T, and VP 2) using the Primer Express program (Life Technologies, Waltham, MA). After comparing the robustness of amplification of the newly selected primer sets and previously published primer sets (LT2, LT3, SET 6, SET 7, SET 9) (Rodig et al., 2012), LT4 and LT3 primers were selected for sensitivity testing on 157 unique tumor samples. The LT4 primer set displayed a higher rate of amplification of target viral DNA across the tumor set and was thus selected for use in this study. Specifically, in the subset of tumors analyzed with both LT3 and LT4 primer sets, there was no case of a tumor with results that were negative by LT4 and positive by LT3. Sequences for the LT4 primer set are as follows: forward primer: TTCCTCTGGGTATGGGTCCTT, TaqMan probe: Fam-TCAGCGTCCCAGGCT-MGB, Reverse primer: GGTCCTCTGGACTGGGAGTCT.
Primers that target the TPO gene were also multiplexed into each PCR reaction. TPO is located on chromosome 2, which is known to have a stable genome copy number in Merkel cell carcinoma, and was thus used as a genomic control for each sample as described previously (Van Gele et al., 1998). MCPyV copy number was calculated relative to the number of copies of TPO; therefore, the LT4/TPO ratio is a surrogate marker for viral copy number per cell. A ratio of LT4 to TPO that was less than 0.01 (i.e., one copy of viral DNA per 100 cells) was considered negative for MCPyV DNA. We chose this threshold primarily because it corresponds to the Allred scoring system cutoff of less than or equal to 2, which similarly corresponds to positive staining of 1% of cells. The use of a lower cutoff also helps to maintain a conservative estimate of tumors that are virus-negative.
TaqMan Exogenous Internal Positive Control Reagents (Thermo-Fisher Scientific, Waltham, MA) were spiked into all the PCR reactions to monitor PCR inhibition (Limaye et al., 2001). Negative results were accepted only if the TaqMan Exogenous Internal Positive Control Reagents were detected within the acceptable range. Each 30 μl PCR reaction contained 10 μl of DNA, 830 nmol/L of each primer, 100 nmol/L of probe, exogenous internal control, 0.03 units of uracil N-glycosylase (an enzyme that eliminates carryover PCR products) (Longo et al., 1990), 15 μl of QuantiTect Multiplex PCR master mix (Qiagen). The thermocycling conditions were as follows: one cycle of 50 °C for 2 minutes then 95 °C for 15 minutes, followed by 45 cycles of 94 °C for 1 minute and 60 °C for 1 minute.
Viral status determination
Tumor viral status was determined by the consensus of the three tests (DNA qPCR, IHC using Ab3, and IHC using CM2B4). For example, if a sample was negative for presence of the virus by qPCR and CM2B4, the sample was deemed virus-negative, even if the third test (Ab3) result was positive. We refer to this classification method as the “multimodal approach.” Individual test performance characteristics (e.g., sensitivity and specificity) were estimated using standard 2 × 2 contingency tables compared with the multimodal approach described.
Statistical analysis
Statistical analyses were performed with STATA Systems software version 13.1 (StataCorp, College Station, TX). Chi-square tests were used to compare categorical variables. Student t tests were used to compare continuous variables. All hypothesis tests were two-tailed, and P-values less than 0.05 were considered statistically significant.
Progression-free survival was defined as the duration from the date of diagnosis to the date of first progression (new disease, or increase in existing disease) after initial treatment. MCC-specific survival was defined as the duration from the date of diagnosis to the date of death due to MCC. Overall survival was defined as the duration from the date of diagnosis to the date of death due to any cause. Standard Kaplan-Meier survival curves and Cox proportional hazard models were generated for each clinical outcome. With respect to the latter, we chose to fit three different models: first, a univariate model using MCPyV tumor viral status alone; second, a multivariate model including characteristics known to be associated with survival in MCC (e.g., sex, age, and immunosuppression) but excluding stage; and third, a multivariate model including stage. We considered the second model to best estimate the independent relationship between MCPyV tumor viral status and each outcome, because it includes the effect of tumor stage as a potential causal mechanism between viral status and outcome. We fit the third model to quantify the effect of stage as a mediator of progression and survival, rather than as a confounder.
Acknowledgments
We thank James DeCaprio at the Dana Farber Cancer Institute for the generous gift of the Ab3 monoclonal antibody. We thank Julie Randolph-Habecker and Kim Melton of the FHCRC experimental histopathology core facility for performing immunohistochemistry. We acknowledge support from the following National Institutes of Health grants, RO1CA176841, R01CA162522, K24CA139052, and TL1TR000422, as well as the Colin Johnston Fund and the Janet Canning Fund.
Abbreviations
- CI
confidence interval
- IHC
immunohistochemistry
- MCC
Merkel cell carcinoma
- MCPyV
Merkel cell polyomavirus
- qPCR
quantitative PCR
- TPO
thyroid peroxidase
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at http://dx.doi.org/10.1016/j.jid.2016.10.028.
CONFLICT OF INTEREST
Maryam M. Asgari received funding from Valeant and Pfizer to her research institute, Massachusetts General Hospital, but it was not related to this project.
References
- Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–68. [PubMed] [Google Scholar]
- Andres C, Belloni B, Puchta U, Sander CA, Flaig MJ. Prevalence of MCPyV in Merkel cell carcinoma and non-MCC tumors. J Cutan Pathol. 2010;37:28–34. doi: 10.1111/j.1600-0560.2009.01352.x. [DOI] [PubMed] [Google Scholar]
- Church CD, Nghiem P. How does the Merkel polyomavirus lead to a lethal cancer? Many answers, many questions, and a new mouse model. J Invest Dermatol. 2015;135:1221–4. doi: 10.1038/jid.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio JA, Garcea RL. A cornucopia of human polyomaviruses. Nat Rev Microbiol. 2013;11:264–76. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels EA, Frisch M, Goedert JJ, Biggar RJ, Miller RW. Merkel cell carcinoma and HIV infection. Lancet. 2002;359(9305):497–8. doi: 10.1016/S0140-6736(02)07668-7. [DOI] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319(5866):1096–100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald TL, Dennis S, Kachare SD, Vohra NA, Wong JH, Zervos EE. Dramatic increase in the incidence and mortality from Merkel cell carcinoma in the United States. Am Surg. 2015;81:802–6. doi: 10.1177/000313481508100819. [DOI] [PubMed] [Google Scholar]
- Foulongne V, Kluger N, Dereure O, Brieu N, Guillot B, Segondy M. Merkel cell polyomavirus and Merkel cell carcinoma, France. Emerg Infect Dis. 2008;14:1491–3. doi: 10.3201/eid1409.080651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneski KM, Warcola AH, Feng Q, Kiviat NB, Leonard JH, Nghiem P. Merkel cell polyomavirus is more frequently present in North American than Australian Merkel cell carcinoma tumors. J Invest Dermatol. 2009;129:246–8. doi: 10.1038/jid.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G, Walradt T, Markarov V, Blom A, Riaz N, Doumani R, et al. Mutational landscape of MCPyV-positive and MCPyV-negative merkel cell carcinomas with implications for immunotherapy. Oncotarget. 2016;7:3403–15. doi: 10.18632/oncotarget.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms PW, Patel RM, Verhaegen ME, Giordano TJ, Nash KT, Johnson CN, et al. Distinct gene expression profiles of viral- and nonviral-associated merkel cell carcinoma revealed by transcriptome analysis. J Invest Dermatol. 2013;133:936–45. doi: 10.1038/jid.2012.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms PW, Vats P, Verhaegen ME, Robinson DR, Wu YM, Dhanasekaran SM, et al. The distinctive mutational spectra of polyomavirus-negative Merkel cell carcinoma. Cancer Res. 2015;75:3720–7. doi: 10.1158/0008-5472.CAN-15-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, et al. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84:7064–72. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem A, Schopflin A, Diaz C, Weyers W, Stickeler E, Werner M, et al. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res. 2008;68:5009–13. doi: 10.1158/0008-5472.CAN-08-0949. [DOI] [PubMed] [Google Scholar]
- Katano H, Ito H, Suzuki Y, Nakamura T, Sato Y, Tsuji T, et al. Detection of Merkel cell polyomavirus in Merkel cell carcinoma and Kapos’s sarcoma. J Med Virol. 2009;81:1951–8. doi: 10.1002/jmv.21608. [DOI] [PubMed] [Google Scholar]
- Lemos B, Nghiem P. Merkel cell carcinoma: more deaths but still no pathway to blame. J Invest Dermatol. 2007;127:2100–3. doi: 10.1038/sj.jid.5700925. [DOI] [PubMed] [Google Scholar]
- Lemos BD, Storer BE, Iyer JG, Phillips JL, Bichakjian CK, Fang LC, et al. Pathologic nodal evaluation improves prognostic accuracy in Merkel cell carcinoma: analysis of 5823 cases as the basis of the first consensus staging system. J Am Acad Dermatol. 2010;63:751–61. doi: 10.1016/j.jaad.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux-Kozal V, Leveque N, Brodard V, Lesage C, Dudez O, Makeieff M, et al. Merkel cell carcinoma: histopathologic and prognostic features according to the immunohistochemical expression of Merkel cell polyomavirus large T antigen correlated with viral load. Hum Pathol. 2015;46:443–53. doi: 10.1016/j.humpath.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Limaye AP, Jerome KR, Kuhr CS, Ferrenberg J, Huang ML, Davis CL, et al. Quantitation of BK virus load in serum for the diagnosis of BK virus-associated nephropathy in renal transplant recipients. J Infect Dis. 2001;183:1669–72. doi: 10.1086/320711. [DOI] [PubMed] [Google Scholar]
- Longo MC, Berninger MS, Hartley JL. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–8. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- Nardi V, Song Y, Santamaria-Barria JA, Cosper AK, Lam Q, Faber AC, et al. Activation of PI3K signaling in Merkel cell carcinoma. Clin Cancer Res. 2012;18:1227–36. doi: 10.1158/1078-0432.CCR-11-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem PT, Bhatia S, Lipson EJ, Kudchadkar RR, Miller NJ, Annamalai L, et al. PD-1 blockade with pembrolizumab in advanced Merkel-cell carcinoma. N Engl J Med. 2016;374:2542–52. doi: 10.1056/NEJMoa1603702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson KG, Carter JJ, Johnson LG, Cahill KW, Iyer JG, Schrama D, et al. Antibodies to merkel cell polyomavirus T antigen oncoproteins reflect tumor burden in merkel cell carcinoma patients. Cancer Res. 2010;70:8388–97. doi: 10.1158/0008-5472.CAN-10-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson KG, Lemos BD, Feng B, Jaimes N, Penas PF, Bi X, et al. Array-CGH reveals recurrent genomic changes in Merkel cell carcinoma including amplification of L-Myc. J Invest Dermatol. 2009;129:1547–55. doi: 10.1038/jid.2008.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodig SJ, Cheng J, Wardzala J, DoRosario A, Scanlon JJ, Laga AC, et al. Improved detection suggests all Merkel cell carcinomas harbor Merkel polyomavirus. J Clin Invest. 2012;122:4645–53. doi: 10.1172/JCI64116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrama D, Peitsch WK, Zapatka M, Kneitz H, Houben R, Eib S, et al. Merkel cell polyomavirus status is not associated with clinical course of Merkel cell carcinoma. J Invest Dermatol. 2011;131:1631–8. doi: 10.1038/jid.2011.115. [DOI] [PubMed] [Google Scholar]
- Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernandez-Figueras MT, et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int J Cancer. 2009;125:1243–9. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sihto H, Kukko H, Koljonen V, Sankila R, Bohling T, Joensuu H. Clinical factors associated with Merkel cell polyomavirus infection in Merkel cell carcinoma. J Natl Cancer Inst. 2009;101:938–45. doi: 10.1093/jnci/djp139. [DOI] [PubMed] [Google Scholar]
- Van Gele M, Speleman F, Vandesompele J, Van Roy N, Leonard JH. Characteristic pattern of chromosomal gains and losses in Merkel cell carcinoma detected by comparative genomic hybridization. Cancer Res. 1998;58:1503–8. [PubMed] [Google Scholar]