Abstract
The Universal Protein Resource (UniProt) is a freely available comprehensive resource for protein sequence and annotation data. UniProt is a collaboration between the European Bioinformatics Institute (EMBL-EBI), the SIB Swiss Institute of Bioinformatics and the Protein Information Resource (PIR). Across the three institutes more than 100 people are involved through different tasks such as expert curation, software development and support.
This chapter introduces the functionality and data provided by UniProt. It describes example use cases for which you might come to UniProt and the methods to help you achieve your goals.
Keywords: UniProt, protein data, protein tools
1. Introduction
The Universal Protein Resource (UniProt) is a freely available comprehensive resource for protein sequence and annotation data1. UniProt provides a number of datasets, the main ones being the UniProt Knowledgebase (UniProtKB), Proteomes, UniProt Reference Clusters (UniRef) and the UniProt Archive (UniParc). An overview of these datasets can be seen in Fig 1. UniProtKB is the central hub for all functional information on proteins2. It consists of two sections, the reviewed (Swiss-Prot) section contains expertly annotated entries and the unreviewed (TrEMBL) section contains computationally analyzed and annotated entries. UniRef provides clusters of UniProtKB sequences (including isoforms) based on sequence identity at resolutions of 100% identity, 90% identity and 50% identity. This helps compress sequence redundancy and speed up sequence similarity searches. UniParc is the sequence archive of all publicly available protein sequences, including those not part of the UniProtKB set. UniProt also provides the Proteomes dataset for species with completely sequenced genomes. A proteome is the set of proteins thought to be expressed by an organism. In addition to these core protein datasets, UniProt provides supporting datasets for Literature Citations, Taxonomy, Keywords, Subcellular locations, Cross-referenced databases and Human diseases. UniProt also provides Automatic annotation rules for UniRule (expertly curated rules) and SAAS (statistical automatic annotation system).
Fig. 1.
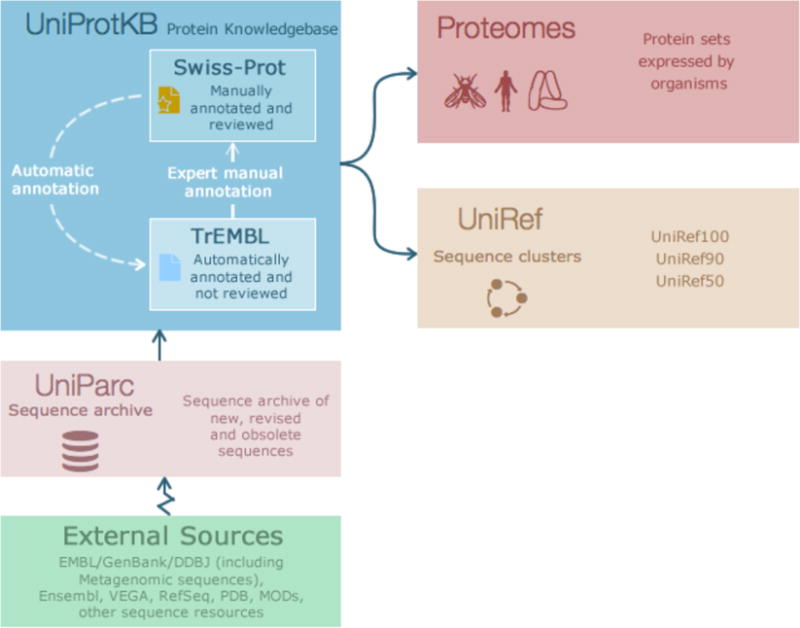
Overview of key UniProt datasets
UniProt also provides three tools embedded into workflow through datasets and on their own dedicated pages. These are the BLAST sequence search tool, the Align multiple sequence alignment tool and the Retrieve/ID mapping tool which allows you to use a list of UniProtKB accessions to download a batch of UniProtKB entries or map the accessions to an external database and vice versa3. The tools are available through their own dedicate pages on the UniProt website at www.uniprot.org. They are also integrated into search results and entry pages from where you can access them while in the process of exploring data.
Understanding protein function is critical to research in many areas of science such as biology, medicine and biotechnology. As the number of completely sequenced genomes continues to increase, huge efforts are being made in the research community to understand as much as possible about the proteins encoded by these genomes. This work is generating large amounts of data, which are spread across multiple locations including scientific literature and many biological databases. Keeping up with all of this information is a daunting task for most researchers and UniProt supports this by providing a comprehensive body of protein information. Here we describe the key use cases supported by UniProt for researchers to be able to achieve their goals at a single site.
2. Methods
2.1. Searching and exploring protein data
The UniProt website provides an intuitive interface to help you find your protein of interest and explore protein data. You can use the search bar in the UniProt banner at the top of all pages to search the various UniProt datasets. Here we consider searching for ‘insulin’ as an example.
Go to http://www.uniprot.org/. You will see is a drop-down list to the left of the search bar that allows you to select a dataset, see Fig 2. You can search all UniProt datasets by selecting them from this drop-down. If you’re looking for protein information about function, subcellular location, interactions, etc., use the default selection of ‘UniProtKB’ and enter your search term in the search box (for example ‘insulin’) and click on the search button.
In order to make your search more specific, you can use the advanced search function. Click on the ‘advanced’ link towards the right of the search box. Click on the dropdown in the advanced search panel to define the type of query you’re making. For example, you can select ‘Protein name’ from the first dropdown to correspond to the query ‘insulin’. You can also add additional parameters like ‘Organism’, ‘Protein existence’, etc. as shown in Fig 3. To add more than two rows of parameters, click on the ‘+’ icon and to delete a row of parameters, click on the bin icon. When you have entered all your parameters, click on the search button.
Once you have submitted your search, you will arrive at the relevant results page, for example the UniProtKB results page in Fig 4. The results page offers a panel of filters on the left to help you refine your search. To the right of the filters is the main results table. Above the results table is a row of action buttons. You can select entries via checkboxes and then directly run a BLAST search, a multiple sequence alignment, download them in a number of available formats or add them to your basket for later use (see Note 1). You can also edit the columns you’re seeing to see more or less information by using the Columns button.
When you have found your exact protein of interest in your dataset, you can click on the entry accession link highlighted in blue font to view the full protein entry page, as shown in Fig 5. When viewing a UniProtKB entry, the menu bar on the left-hand side of the screen lists the entry sections, allowing you to move easily between sections. The entry provides all annotated data for the protein, its sequence(s) and cross-references to over 150 relevant databases. You can also use action buttons on this page to run a BLAST search on the entry, align all isoforms (if any), view or download the entry in different formats and add it to your basket for later.
Fig. 2.
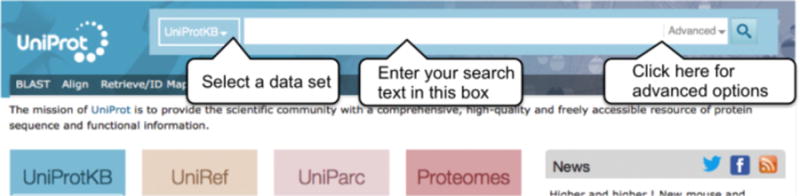
UniProt header search bar
Fig. 3.
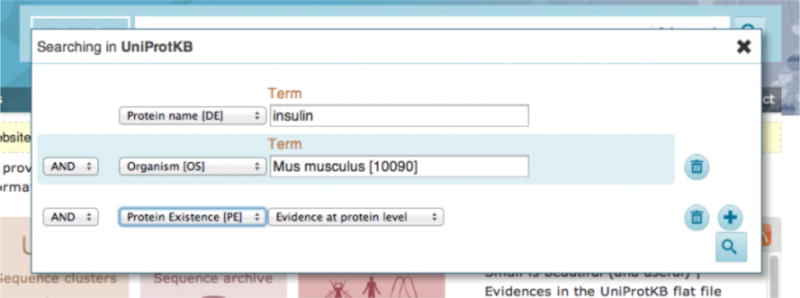
Advanced search
Fig. 4.
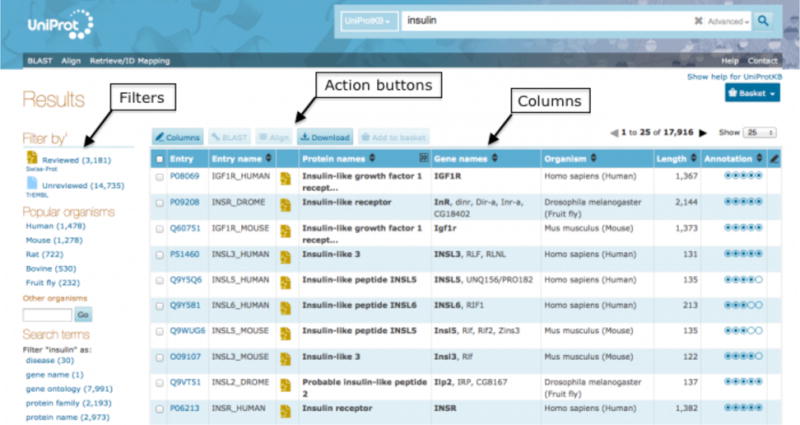
UniProtKB search results page
Fig. 5.
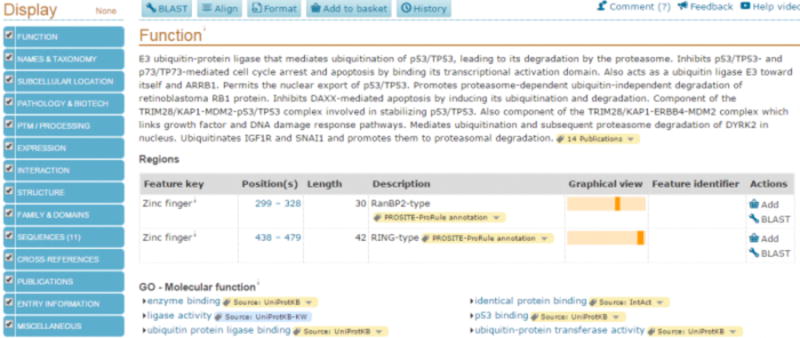
UniProtKB protein entry
2.2. Finding the Proteome (complete protein set) for an organism
A proteome is the full set of proteins thought to be expressed by an organism and the UniProt websites provides proteomes datasets for species with completely sequenced genomes.
Click on the dropdown to the left of the search bar and select ‘Proteomes’.
Enter your query directly into the search box, for example ‘Homo Sapiens’, or click on the ‘advanced’ button to the right of the search box and build a query using the parameters provided. This can help find exact results for the organism or taxonomy level you would like to specify. Click on the search button or hit enter to get to your results page.
You will be presented with a table of results for your proteomes search, as shown in Fig 6.
Click on the proteome identifier to go to the detailed proteome page where you will see a summary of the organism, information about the genome assembly, proteins arranged by the chromosome or plasmid that they belong to, links to the protein entries in UniProtKB and publications related to the proteome as shown in Fig 7.
Fig. 6.
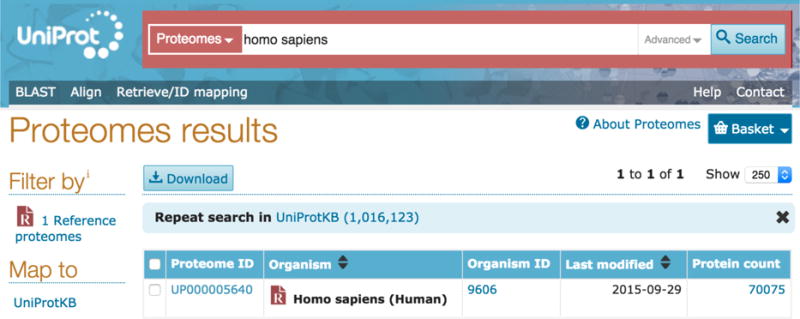
Proteomes results page
Fig. 7.
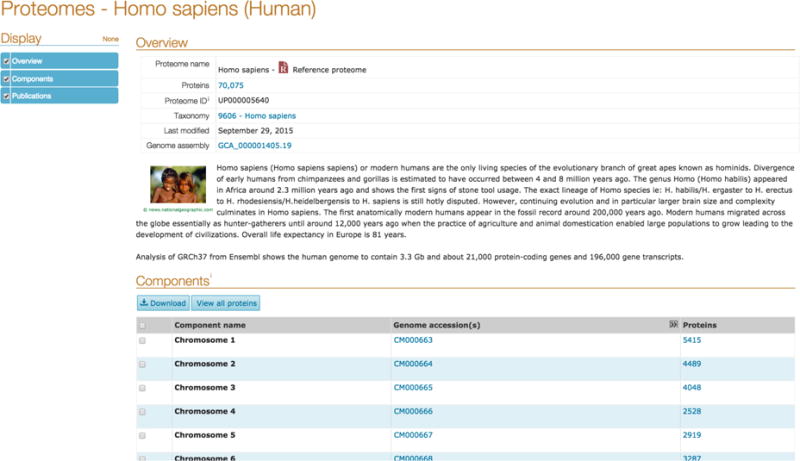
Human Proteome entry
2.3. Finding all proteins involved in a disease
Studying the involvement of proteins in diseases is important to help identify drug targets and better understand disease mechanisms in the human body. The best way to look for all proteins involved in a disease is to begin your search with the Human diseases dataset provided by UniProt4. Here we consider this use case with the disease Breast Cancer as an example.
Go to http://www.uniprot.org/ and click on the drop-down to the left of the search bar. Select ‘Human diseases’ under the ‘Supporting data’ section in the dropdown. Type your query into the search box, for example ‘Breast cancer’, and hit the search button.
You will arrive at a results page. The results table presents results that match your query such as is ‘Breast cancer, lobular’, ‘Breast cancer’ and so on. You will see a definition of the disease and a link to UniProtKB to see all proteins linked to this disease. For example, in case of the result ‘Breast cancer’, there are 12 linked UniProtKB entries as shown in Fig 8.
To view all linked proteins, simply click on the UniProtKB link under the disease definition. You can also click on the disease name to view the detailed definition for the disease, its synonyms and cross-references to related resources (like MIM, MeSH, etc.).
Clicking on the UniProtKB link will bring you to a UniProtKB results page for all proteins linked to this disease. Each of these UniProtKB protein entries provides information about various biological aspects such as function, taxonomy, subcellular location, pathology and biotech, etc. The ‘Pathology & Biotech’ section of UniProtKB entries lists all diseases that the protein is involved in, the supporting evidence and a list of natural variants linked to the disease.
Fig. 8.
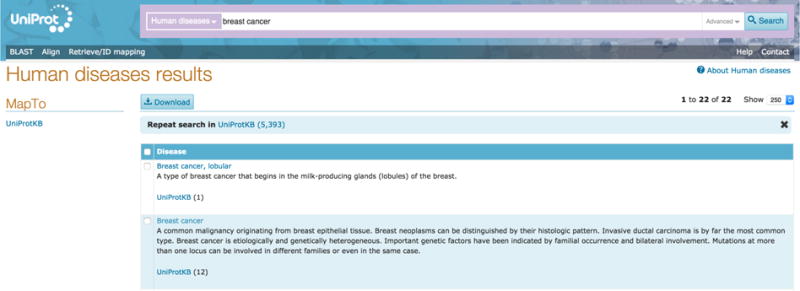
Human diseases results page
2.4. Identifying your sequence using the BLAST search
If you have a protein sequence you would like to identify, you can use the Basic Local Alignment Search Tool to find closely matching sequences from UniProt that can help you understand evolutionary relationships and make functional inferences based on sequence identity. The UniProt website provides a form to submit your own sequences or any UniProtKB protein accession, UniParc sequence archive accession number or UniRef cluster accession to the BLAST tool, using the NCBI BLAST algorithm5. It supports an integrated workflow that allows you to submit protein entries to BLAST from a search results page, the UniProt basket and also a protein entry page.
Click on the BLAST link in the header of the UniProt website. This will bring you to the form submission page for BLAST.
Enter a protein or nucleotide sequence or a UniProtKB, UniParc or UniRef cluster identifier or accession in the input field, for example P00750, as shown in Fig 9.
You have a number of optional settings that you can change or leave as default. The options include ‘Target database’, ‘E-Threshold, ‘Matrix’, ‘Filtering’, ‘Gapped’ (yes or no) and number of Hits you’d like to get from the tool. For example, if you would like to find sequence matches only from a particular taxonomic level like ‘mammals’ instead of from all of UniProtKB, you can select this from the ‘Target database’ dropdown. You can also use the ‘Target database’ dropdown to search against UniRef clusters instead of UniProtKB. UniRef clusters consist of UniProtKB sequences clustered based on identity at 100%, 90% and 50%. Searching against clusters hence speeds up BLAST searches.
Click on the Run BLAST button to execute your query. You will see a ‘Job status: RUNNING’ page while your query is being run. This page provides details of your query sequence and settings.
You will arrive at the BLAST results page once your query has been executed, as shown in Fig 10. On the left hand side, this page provides filters, mapping links to map your results to other datasets like UniProtKB and alternative views of the results by taxonomy tree, text or XML versions. The upper half of the page provides an overview which you can expand to see all results by clicking on the ‘Show all 250’ link. In Fig 10, the overview shows the UniProtKB entry accession number, the protein names and species, a diagrammatic view of your matches that is colour coded by identify and the actual identity percentage. The lower half of the page shows your alignments in detail with each one represented diagrammatically in related to the query sequence. You can click on the graphic to view the raw sequence alignment in detail. The page also provides a Job identifier that you can use to retrieve your results page for up to 7 days.
You can also submit a UniProtKB, UniParc or UniRef entry to the BLAST tool from a search results page by selecting the checkbox for that entry and clicking on the ‘BLAST’ button above the results page. Alternatively you can click on the checkbox and then click on the ‘Add to Basket’ button above the search results table to build a collection of entries in your basket and submit one of them to BLAST at a later point.
When on a UniProtKB entry page, a UniRef cluster page or a UniParc sequence archive page, you can simply click on the BLAST button near the top of the entry to submit the sequence to the BLAST tool. In case of a UniRef cluster entry with multiple sequences in the page, you can choose one by ticking on the checkbox to the left of it and then click on the ‘BLAST’ button to submit to the tool.
Fig. 9.
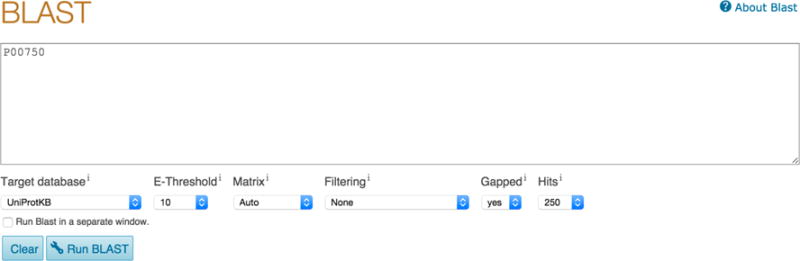
BLAST input page
Fig. 10.

BLAST results page
2.5. Multiple sequence alignment
Aligning multiple sequences can help understand evolutionary relationships and identify areas of conservation between your sequences that can have structural or functional associations. UniProt provides a multiple sequence alignment tool ‘Align’ which uses the Clustal Omega algorithm to align sequences6. For the most meaningful results, you should try and align sequences that are likely to be related so that you can explore evolutionary, structural and functional relationships. You can access the tool through its own form submission page or directly through search results pages and protein entry pages. Integrating the tool into the data exploration workflow offers you a flexible way to find and analyse your data.
Click on the ‘Align’ link in the header of the UniProt website. You will see a form submission page with an input box.
If you have two or more sequences that you would like to align to find areas to conservation and divergence, you can submit the sequences in FASTA format or accessions into the input box on this page. Click on the ‘Align’ button to execute your query.
You will see a ‘Job status: RUNNING’ page while your query is being executed.
Once completed, you will be presented the Alignment results page. The results page presents the alignment information, an evolutionary relationship tree for your sequences and the results information able at the bottom. On the left hand side, you have Highlight options that allow you to select checkboxes to visually highlight sequence areas corresponding to annotations like active sites, domains, glycosylation, etc. and amino acid properties like hydrophobicity as shown in Fig 11. This allows you to have a quick visual overview of important annotations or amino acid properties and their conservation or distribution in the sequences you have aligned. The results information on the page provides a job identifier that you can use to access your results for up to 7 days.
You can also submit UniProt entries to the Align tool from a search results page by selecting the checkboxes for entries and clicking on the ‘Align’ button above the results page. Alternatively you can click on the checkboxes and then click on the ‘Add to Basket’ button above the search results table to build a collection of entries in your basket and submit them to the Align tool at a later point.
When on a UniProtKB protein entry page with multiple sequences (i.e. isoforms), you can click on the ‘Align’ button towards the top of the entry page to align the isoforms. When on a UniRef cluster entry page, you can click on checkboxes to select constituent entries that you would like to align and then click on the ‘Align’ page towards the top of the page to submit them to the Align tool.
Fig. 11.
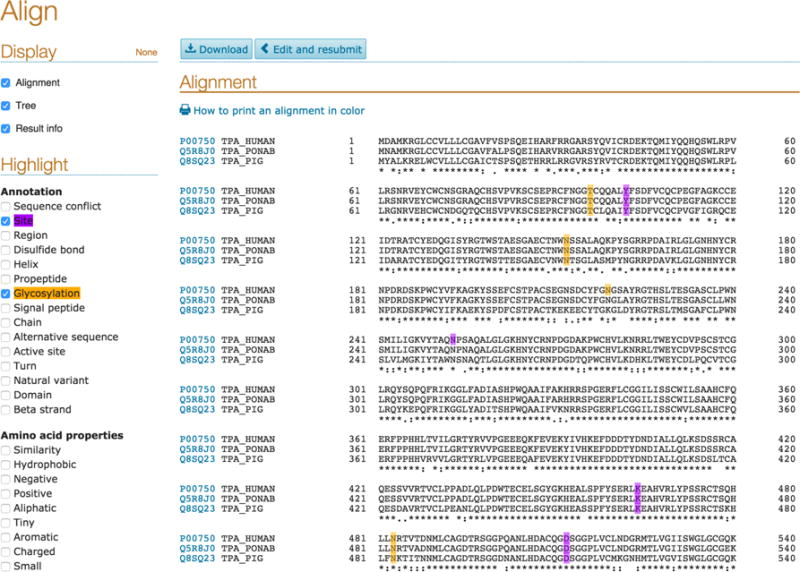
Align results page
2.6. ID mapping
If you have a list of UniProtKB accessions that you need to map or convert to identifiers from another database, for example if you have a list of UniProt accessions from a mass spectrometry experiment that you would like to map to other databases (for example, Ensembl, PDB, InterPro, etc.), you can use the Retrieve/ID mapping tool on the UniProt website. You can also map identifiers from external databases to UniProt using this tool3.
Click on the ‘Retrieve/ID mapping’ link the UniProt header. This will bring you to a form submission page with an input box, ‘from and to’ database options and a ‘Go’ button.
To convert UniProtKB accessions to an external database, for example Ensembl, paste your list of UniProtKB accessions in the input box or upload them as a file. Now click on the ‘From’ dropdown and select UniProtKB and click on the ‘To’ dropdown to select your target database (in this case Ensembl). The tool allows you to convert or map your accessions from UniProt to over 100 external databases that UniProt is cross-referenced to and vice versa (e.g. Ensembl, PDB, Refseq, etc.). You will get a results page showing a table of your input IDs and the mapped IDs from your target database as shown in Fig 12.
To convert external database identifiers to UniProtKB accessions or identifiers, for example Ensembl to UniProtKB, select the external database from the ‘From’ dropdown and UniProtKB from the ‘To’ dropdown. You will get a results page with your mapped UniProt entries and the default columns of data that you can customise, as shown in Fig 13. You can select entries using checkboxes to BLAST them, align them, download them or add them to your basket. You are also presented with filters o the left hand side of the page to help narrow down your results.
UniProt also provides the flexibility of submitting UniProt accessions to the ID mapping tool from your basket. Just add entries to your basket as you explore data and then you can use checkboxes to select them inside your basket and click on the ‘map IDs’ tool to arrive on the Retrieve/ID mapping tool with your input pre-filled in the input box.
Fig. 12.
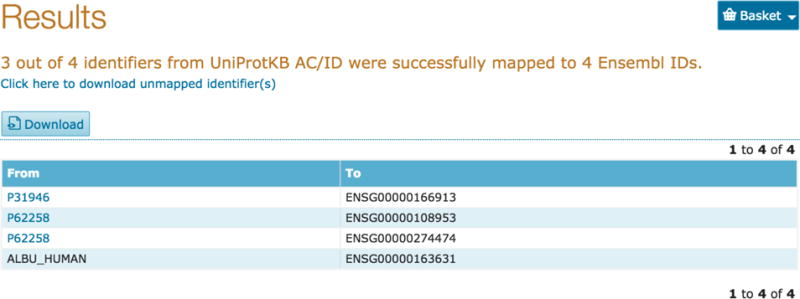
Retrieve/ID mapping results page from UniProt to external IDs
Fig. 13.

Retrieve/ID mapping results page from external database to UniProt ACs
2.7 Retrieving UniProt entries for a list of identifiers
If you have a list of UniProt accessions and would like to retrieve information for them from the UniProt website in a single step, you can use the Retrieve/ID mapping tool.
Click on the ‘Retrieve/ID mapping’ link the UniProt header. This will be you to a form submission page with an input box, ‘from and to’ database options and a ‘Go’ button.
To retrieve UniProtKB information corresponding to UniProtKB accessions or identifiers, paste your list of UniProtKB accessions in the input box or upload them as a file. You can leave the ‘From’ dropdown and the ‘To’ dropdown selections as the default UniProtKB since you’re not converting or mapping identifiers between different databases.
You will get a results page with your requested UniProt entries and the default columns of data that you can customise, as shown in Fig 14. You can select entries using checkboxes to BLAST them, align them, download them or add them to your basket. You are also presented with filters on the left hand side of the page to help narrow down your results.
Fig. 14.
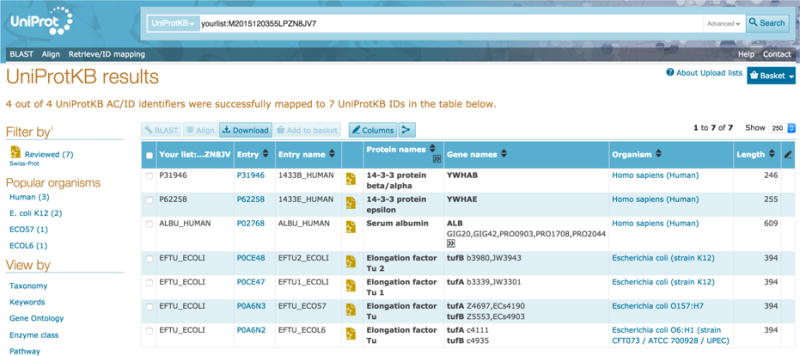
Retrieve/ID mapping results page for batch UniProtKB entry retrieval
Acknowledgments
UniProt has been prepared by Alex Bateman, Maria Jesus Martin, Claire O’Donovan, Michele Magrane, Emanuele Alpi, Ricardo Antunes, Benoit Bely, Mark Bingley, Carlos Bonilla, Ramona Britto, Borisas Bursteinas, Hema Bye-A-Jee, Andrew Cowley, Alan Da Silva, Maurizio De Giorgi, Tunca Dogan, Francesco Fazzini, Leyla Garcia Castro, Luis Figueira, Penelope Garmiri, George Georghiou, Daniel Gonzalez, Emma Hatton-Ellis, Weizhong Li, Wudong Liu, Rodrigo Lopez, Jie Luo, Yvonne Lussi, Alistair MacDougall, Andrew Nightingale, Barbara Palka, Klemens Pichler, Diego Poggioli, Sangya Pundir, Luis Pureza, Guoying Qi, Steven Rosanoff, Rabie Saidi, Tony Sawford, Aleksandra Shypitsyna, Elena Speretta, Edward Turner, Nidhi Tyagi, Vladimir Volynkin, Tony Wardell, Kate Warner, Xavier Watkins, Rossana Zaru and Hermann Zellner at the European Bioinformatics Institute; Ioannis Xenarios, Lydie Bougueleret, Alan Bridge, Sylvain Poux, Nicole Redaschi, Lucila Aimo, Ghislaine Argoud-Puy, Andrea Auchincloss, Kristian Axelsen, Parit Bansal, Delphine Baratin, Marie-Claude Blatter, Brigitte Boeckmann, Jerven Bolleman, Emmanuel Boutet, Lionel Breuza, Cristina Casal-Casas, Edouard de Castro, Elisabeth Coudert, Beatrice Cuche, Mikael Doche, Dolnide Dornevil, Severine Duvaud, Anne Estreicher, Livia Famiglietti, Marc Feuermann, Elisabeth Gasteiger, Sebastien Gehant, Vivienne Gerritsen, Arnaud Gos, Nadine Gruaz-Gumowski, Ursula Hinz, Chantal Hulo, Florence Jungo, Guillaume Keller, Vicente Lara, Philippe Lemercier, Damien Lieberherr, Thierry Lombardot, Xavier Martin, Patrick Masson, Anne Morgat, Teresa Neto, Nevila Nouspikel, Salvo Paesano, Ivo Pedruzzi, Sandrine Pilbout, Monica Pozzato, Manuela Pruess, Catherine Rivoire, Bernd Roechert, Michel Schneider, Christian Sigrist, Karin Sonesson, Sylvie Staehli, Andre Stutz, Shyamala Sundaram, Michael Tognolli, Laure Verbregue and Anne-Lise Veuthey at the SIB Swiss Institute of Bioinformatics; Cathy H. Wu, Cecilia N. Arighi, Leslie Arminski, Chuming Chen, Yongxing Chen, John S. Garavelli, Hongzhan Huang, Kati Laiho, Peter McGarvey, Darren A. Natale, Karen Ross, C. R. Vinayaka, Qinghua Wang, Yuqi Wang, Lai-Su Yeh and Jian Zhang at the Protein Information Resource.
Footnotes
UniProt provides a basket functionality to help you store your UniProt entries of interest and then analyse then, download them or view them at a later point. The basket saves entries from UniProtKB, UniParc and UniRef. You can add entries to the basket from search results pages of these three datasets or from their individual entry pages through the ‘Add to basket’ button. The basket lets you select entries using checkboxes and submit them to the BLAST, Align and ID mapping tools. You can also download your entries in formats lie List, Text, FASTA, Tab-separated, Excel, GFF and XML. It also provides a ‘Clear’ button and a ‘Full view’ button which shows you your saved entries in a full results screen where you can use filters and add or remove columns to the results table. The basket keeps your saved entries until you clear your browser cookies.
References
- 1.The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–212. doi: 10.1093/nar/gku989. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Magrane M, The UniProt Consortium UniProt Knowledgebase: a hub of integrated protein data. Database: The Journal of Biological Databases and Curation 2011. 2011 doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang H, McGarvey PB, Suzek BE, Mazumder R, Zhang J, Chen Y, Wu CH. A comprehensive protein-centric ID mapping service for molecular data integration. Bioinformatics. 2011;27(8):1190–1191. doi: 10.1093/bioinformatics/btr101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magrane M, Pundir S. UniProt: Exploring protein sequence and fucntional information. 2015 http://www.ebi.ac.uk/training/online/course/uniprot-exploring-protein-sequence-and-functional. Accessed 17 December 2015.
- 5.Ladunga I. Finding homologs in amino acid sequences using network BLAST searches. In: Baxevanis Andreas D., editor. Current protocols in bioinformatics / editoral board. 2009. Chapter 3: Unit 3.4. [DOI] [PubMed] [Google Scholar]
- 6.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]


