Abstract
Duchenne muscular dystrophy is caused by mutations in DMD which disrupt the reading frame. Therapeutic strategies that restore DMD’s reading frame, such as exon skipping and CRISPR/Cas9, need to be tested in the context of the human DMD sequence in vivo. We have developed a novel dystrophic mouse model by using CRISPR/Cas9 to delete exon 45 in the human DMD gene in hDMD mice, which places DMD out-of-frame. We have utilized this model to demonstrate that our clinically-relevant CRISPR/Cas9 platform, which targets deletion of human DMD exons 45–55, can be directly applied in vivo to restore dystrophin.
Keywords: Duchenne muscular dystrophy, gene editing, CRISPR, mice, animal models
INTRODUCTION
Duchenne muscular dystrophy (DMD) is a progressive muscle degenerative disease, affecting about 1 in 5000 male births [1]. It is most often caused by out-of-frame mutations in the DMD gene, which prevent expression of dystrophin protein. Dystrophin normally stabilizes and assembles the dystrophin glycoprotein complex (DGC) at the muscle fiber sarcolemma to connect the actin cytoskeleton to the extracellular matrix. Lack of dystrophin and the DGC leads to muscle fiber damage and progressive degeneration, resulting in fibro-adipogenic accumulation in the muscle. Potential therapeutic strategies, such as exon skipping, or clustered, regularly interspaced, short palindromic repeats (CRISPR) and -associated protein (Cas) 9 (CRISPR/Cas9), aim to restore the DMD reading frame, which will lead to the milder, allelic disease Becker muscular dystrophy [2]. Exon skipping therapies use antisense oligonucleotides to target a specific sequence in the dystrophin mRNA and alter the splicing pattern to reframe the transcript [3]. CRISPR/Cas9-based therapies use guide RNAs (gRNAs) and the Cas9 nuclease to target specific sequences of genomic DNA for deletion or editing [4–6]. Currently, there are no mouse models that allow for testing of such therapies on the human DMD sequence in a dystrophic context in vivo.
A humanized mouse (hDMD) was previously generated in which the entire human DMD sequence was integrated into mouse chromosome 5 [7]. However, this mouse expresses the wildtype human DMD transcript and does not present a dystrophic phenotype, even if crossed to the mdx mouse which lacks murine dystrophin, since human dystrophin can functionally replace the mouse protein. Since the CRISPR/Cas9 system has shown much promise in quickly and easily generating novel mouse models, we used CRISPR/Cas9 to make an out-of-frame mutation by deleting exon 45 of the human DMD gene in hDMD mouse zygotes (hereafter referred to as hDMD del45 mice). We have crossed this mouse to both the mdx and mdxD2 backgrounds, both of which have a premature stop codon in the mouse Dmd gene. MdxD2 mice are on the DBA2 background and have a more severe phenotype than mdx mice on the C57BL/10 background, primarily due to two modifier alleles, Ltbp4 and Anxa6 [8–10]. Here we describe the initial characterization of the hDMD del45 model. We show that muscles of hDMD del45 mdxD2 mice contain a mutated human DMD gene lacking exon 45 and are dystrophic. We also show proof-of-principle in vivo application of our CRISPR/Cas9 gene editing platform which targets human DMD exons 45–55 for deletion to restore the reading frame for up to 60% of Duchenne patients [11].
MATERIALS AND METHODS
Mice
All animal work was conducted under protocols approved by the UCLA Animal Research Committee in the Office of Animal Research Oversight. hDMD (Tg(DMD)72Thoen/J, 018900), C57BL/10 mdx (001801), and mdxD2 (D1.B10-Dmdmdx/J, 013141) mice were obtained from Jackson Laboratories.
Generation of hDMD del45 mice
CRISPR/Cas9 injection into hDMD zygotes was performed by the University of California, Davis, Mouse Biology Program. 100 ng/µl mRNA Cas9 was mixed with 20 ng/µl of each of 44C1, 44C2, 45C2, 45C3 gRNAs (sequences in Supplemental Table 2) produced via in vitro transcription. 2–4pL (0.2–0.4pg Cas9 and 0.04–0.08pg each gRNA) was injected into the nucleus with excess positive flow into the cytoplasm. Pups were PCR screened for the deletion and the sequence confirmed via Sanger sequencing.
In vivo electroporation
hDMD del45 mdx and hDMD del45 mdxD2 mice were electroporated as described [12] with 20µg of px333 plasmid DNA (Addgene 64073, Andrea Ventura [13]) containing CRISPR guides 44C4 and 55C3 (from [11]) or pmaxGFP as a control forhDMDdel45 mdxD2mice. In brief, 5µl hyaluronidase was injected into the flexor digitorum brevis (FDB) muscle and 1hr later the DNA was injected and electroporated 20 times for 20 ms at 1 Hz.
hDMD del45 mdx mice were harvested 22 or 33 days later and genomic DNA was extracted by digesting the muscles with proteinase K then using the Quick-gDNA™ Miniprep Kit (Zymo Research). PCR for an exon 45–55 deletion was performed as described using Accuprime Taq HiFi (Thermo Fisher Scientific) or Herculase II Fusion Polymerase (Agilent Genomics) [11]. Sequencing of blunt cloned PCR products from Zero Blunt® TOPO® (Life Technologies) was done by Laragen Inc.
hDMD del45 mdxD2 mice were harvested 24 days post-electroporation. The interosseous (IO) and FDB were flash frozen and samples of 10µm cryosections taken throughout the whole muscle. Intervening sections as well as the lumbricalis were used for genomic DNA extraction and PCR as above.
Please see expanded Materials and Methods in the Supplementary Data.
RESULTS
We sought to generate a mouse model that expresses a mutated human DMD gene in mice lacking murine dystrophin. To this end, we designed gRNAs targeting DMD introns 44 and 45 to cause deletion of exon 45 via non-homologous end joining (NHEJ) of the human DMD gene in hDMD mice (Fig. 1A). The gRNAs were screened individually and in pairs in HEK293FT cells, and their ability to cause Cas9 cutting (Supplemental Figure 1A) or an exon 45 deletion (Supplemental Figure 1B), respectively, was assessed. Two pairs that had the highest deletion efficacy were chosen and in combination, microinjected into hDMD zygotes. 15 pups were screened and one female heterozygous pup and one male compound heterozygous pup were obtained (data not shown), hereafter referred to as hDMD del45 mice. Upon crossing, both founder mice were shown to have germline transmission of the exon 45 deletion (Fig. 1B), which was confirmed by sequencing of the intron 44/45 rejoining site (Fig. 1C).
Fig. 1.
Generation and characterization of hDMD del45 mice. A) Cartoon showing the region of the human DMD gene targeted for mutation in hDMD mice. Guide RNAs (represented as lightning bolts) flank exon 45 to cause an exon 45 deletion by NHEJ, which puts the gene out-of-frame. The Dp140 isoform promoter is shown (arrow). B) Genotyping PCR of genomic DNA from pups (labeled #1–6) from the founder mice F#4 and M#14 crossed to mdxD2 mice showing germline transmission of the deletion. The red arrow highlights the undeleted band seen in wildtype (wt) hDMD mice and the purple arrow depicts the exon 45 deletion seen in the pups. Primers are depicted as arrows in A. Since two pairs of gRNAs were injected together, different deletion sizes occurred due to the various target sites. C) Sequencing of the introns 44 and 45 rejoining site in two pups shows an exon 45 deletion. D) Western blot of whole muscle extracts probed with human dystrophin. Ponceau stain is shown to demonstrate loading. The gastrocnemius (gastroc), tibialis anterior (TA), diaphragm (dia), and heart from an hDMD del45/mdxD2 mouse (second backcross, 6wks old) show lack of dystrophin compared to an hDMD (wt)/mdxD2 mouse (third backcross, 6.5wks old) gastroc muscle. E) Immunohistochemistry of muscle sections stained with anti-laminin (white) and human dystrophin (red). Laminin was used to delineate muscle fibers. hDMD del45/mdxD2 muscles (second backcross, 6wks old) show a lack of dystrophin staining with a few faint revertant fibers (*and inset at higher contrast). Scale bar 50µm. F) Hematoxylin and eosin (H&E) staining of muscle sections show dystrophic pathology in muscles of hDMD del45/mdxD2 mice (second backcross, 6wks old). Scale bar 100µm. Similar analyses from another hDMD del45/mdxD2 mouse is shown in Supplemental Figure 2 (representative of n = 4 mice analyzed).
hDMD del45 mice were crossed to mdxD2 and then mdx mice to generate fully dystrophic models. As a control, wildtype hDMD mice were also crossed to mdx and mdxD2 mice, so these mice expressed human dystrophin but lacked the murine equivalent. The resulting hDMD del45 mdxD2 mice and hDMD del45 mdx mice (from the first or second cross) lacked human dystrophin by western blot (Fig. 1D, Supplemental Figures 2A and 3A) and immunostaining (Fig. 1E, Supplemental Figures 2B and 3B). Occasionally, a few revertant dystrophin+ fibers were seen (see asterisks in Fig. 1E) but these events were infrequent. The muscle pathology of hDMD del45 mdxD2 mice was dystrophic, with features of fibrosis, inflammation, and calcium deposits, compared to the hDMD mdxD2 mice which express the wildtype DMD gene that rescues the dystrophic phenotype (Fig. 1F and Supplemental Figure 2C).
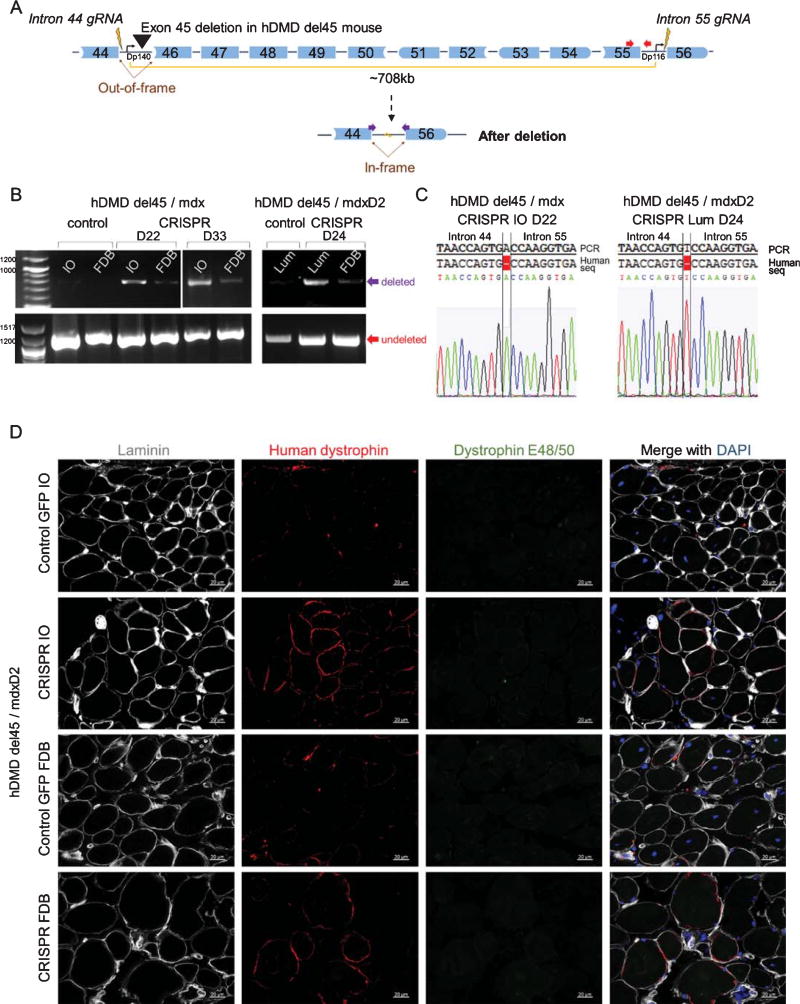
To demonstrate the in vivo utility of this mouse, we applied our therapeutic CRISPR/Cas9 gene editing platform that targets human DMD exons 45–55 for deletion. Treatment with the CRISPR/Cas9 platform will restore the reading frame for out-of-frame mutations in this region, such as an exon 45 deletion (Fig. 2A). We electroporated a plasmid containing one gRNA targeted to intron 44 and one to intron 55 [11] in tandem with Cas9 into the FDB muscle of 12 week old hDMD del45 mdx mice and 18.5 week old hDMD del45 mdxD2 mice. After 22–33 days, an exon 45–55 deletion in the FDB, IO, and/or lumbricalis (Lum) muscles was observed by genomic DNA PCR (Fig. 2B) and sequencing of the intron 44/55 rejoining site (Fig. 2C) in the electroporated mice. Dystrophin restoration was observed by immunostaining in the IO and FDB of the electroporated hDMD del45 mdxD2 mice (Fig. 2D). Thus, our novel humanized dystrophic hDMD del45 mouse model demonstrates proof-of-principle that a human targeted CRISPR/Cas9 platform is functional in vivo. This mouse model will be a useful resource for testing different CRISPR/Cas9 strategies and other therapies that target the human DMD gene.
Fig. 2.
CRISPR/Cas9 deletion ofDMDexons 45–55 to restore the reading frame in hDMD del45 mdx and mdxD2 mice. A) Cartoon showing our therapeutically relevant CRISPR/Cas9 platform that was electroporated into the FDB of hDMD del45 mdx and hDMD del45 mdxD2 mice. This platform consists of guide RNAs (represented as lightning bolts) in introns 44 and 55 that cause deletion of exons 45–55 through NHEJ to restore the reading frame for the out-of-frame exon 45 deletion in hDMD del45 mice. The Dp140 and Dp116 isoform promoters are shown (arrows). B) Genomic DNA PCR using primers for the undeleted allele (red, 1201bp, bottom) or the deletion (purple, 788bp, top). An exon 45–55 deletion was observed in the FDB, IO and/or Lum muscles 22, 24, and 33 days (D22, D24, and D33) after in vivo electroporation of the CRISPR/Cas9 platform compared to the control mice. C) Sequencing of the intron 44/55 rejoining site from the IO and Lum muscle of the CRISPR electroporated mice, which showed a 1bp insertion. D) Immunohistochemistry of hDMD del45/mdxD2 IO and FDB muscles stained with anti-laminin (grey), human dystrophin (red), and dystrophin targeting exons 48/50 (E48/50, green). Laminin was used to delineate muscle fibers. Dystrophin E48/50 was used to exclude revertant fibers from the analysis, since after CRISPR/Cas9 application exons 45–55 will be deleted. Dystrophin positive fibers can be seen in the CRISPR electroporated sample compared to the GFP electroporated control. Scale bar 20 µm.
DISCUSSION
Here we describe a novel humanized mouse model of Duchenne muscular dystrophy. We have created an out-of-frame human DMD exon 45 deletion in the hDMD mouse (referred to as hDMD del45) and have crossed it to mdx and mdxD2 backgrounds so that the mice completely lack dystrophin. We have validated that this model lacks human dystrophin and presents with a dystrophic muscle pathology in multiple muscles across the body. Lastly, we have shown one utility of this mouse by applying our CRISPR/Cas9 platform which targets human DMD exons 45–55 for deletion. After delivering the CRISPR/Cas9 platform via electroporation in vivo we have demonstrated a restored DMDreading frame and dystrophin expression in the hDMD del45 mdx and mdxD2 muscles.
This mouse has a wide range of uses. The model will be useful for testing a variety of antisense oligonucleotides against a mutated human sequence in vivo since the reading frame can be restored using an exon 44 or 46 single skip, a 46/47 double skip, or a 43/44/46 or 46/47/48 triple skip. For CRISPR/Cas9-based therapies, the model can be used for testing various human targeted gRNAs around this region. Additionally, the hDMD del45 model is a relevant negative control for western blotting of human dystrophin protein from the hDMD wildtype mouse. Lastly, it presents a unique model in which to study endogenous skipping of the human DMD gene which is a phenomenon known to occur at a low level in human patients with an exon 45 deletion [14]. The ability to study the human protein and/or test potential therapeutics in a dystrophic context in vivo on the human DMD gene rather than just in human cells offers obvious advantages and will allow for faster translation of human specific therapies.
Supplementary Material
Acknowledgments
We would like to thank Diana Becerra, Joana Capote, Natalia Ermolova, Brandon Lippold, Chino Kumagai-Cresse, and Jane Wen for technical assistance. We are grateful for the technical support, products, and/or services provided to our research by the Mouse Biology Program at the University of California Davis and the Center for DMD at UCLA Muscle Phenotyping and Imaging Core. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1144087 (CSY). Additional funding was provided by a pilot and feasibility seed grant from the Center for DMD at UCLA (NIH NIAMS 5P30AR05723UCLA and CTSI Grant #UL1TR000124), the Eli & Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA, the California Institute of Regenerative Medicine, Wellstone Cooperative Muscular Dystrophy Center (U54AR052646-Sweeney-PI, Spencer-Co-I) and NIAMS of the NIH (R01AR064327-ADP).
Footnotes
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
The supplementary material is available in the electronic version of this article: http://dx.doi.org/10.3233/JND-170218.
References
- 1.Mendell JR, Shilling C, Leslie ND, Flanigan KM, Al-Dahhak R, Gastier-Foster J, et al. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann Neurol. 2012;71(3):304–13. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- 2.Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2(1):90–5. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 3.Aartsma-Rus A, van Ommen G-JB. Antisense-mediated exon skipping: A versatile tool with therapeutic and research applications. RNA. 2007;13(10):1609–24. doi: 10.1261/rna.653607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–7. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L, Ran AF, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–22. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.’t Hoen PAC, de Meijer EJ, Boer JM, Vossen RHAM, Turk R, Maatman RGHJ, et al. Generation and characterization of transgenic mice with the full-length human DMD gene. J Biol Chem. 2008;283(9):5899–907. doi: 10.1074/jbc.M709410200. [DOI] [PubMed] [Google Scholar]
- 8.Heydemann A, Ceco E, Lim JE, Hadhazy M, Ryder P, Moran JL, et al. Latent TGF-β – binding protein 4 modifies muscular dystrophy in mice. JCI. 2009;119(12):3703–12. doi: 10.1172/JCI39845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coley WD, Bogdanik L, Vila MC, Yu Q, van der Meulen JH, Rayavarapu S, et al. Effect of genetic background on the dystrophic phenotype in mdx mice. Hum Mol Genet. 2016;25(1):130–45. doi: 10.1093/hmg/ddv460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swaggart KA, Demonbreun AR, Vo AH, Swanson KE, Kim EY, Fahrenbach JP, et al. Annexin A6 modifies muscular dystrophy by mediating sarcolemmal repair. PNAS. 2014;111(16):6004–9. doi: 10.1073/pnas.1324242111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, et al. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell Stem Cell. 2016;18(4):533–40. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiFranco M, Quinonez M, Capote J, Vergara J. DNA transfection of mammalian skeletal muscles using in vivo electroporation. JoVE. 2009;(32):1–7. doi: 10.3791/1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han Y-C, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423–7. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thanh LT, Nguyen TM, Helliwell TR, Morris GE. Characterization of revertant muscle fibers in Duchenne muscular dystrophy, using exon-specific monoclonal antibodies against dystrophin. Am J Hum Genet. 1995;56(3):725–31. [PMC free article] [PubMed] [Google Scholar]
- 15.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Man N, Morris GE. Use of epitope libraries to identify exon-specific monoclonal antibodies for characterization of altered dystrophins in muscular dystrophy. Am J Hum Genet. 1993;52:1057–66. [PMC free article] [PubMed] [Google Scholar]
- 17.Kendall GC, Mokhonova EI, Moran M, Sejbuk NE, Wang DW, Silva O, et al. Dantrolene enhances antisense-mediated exon skipping in human and mouse models of Duchenne muscular dystrophy. Sci Transl Med. 2012;4:164ra160. doi: 10.1126/scitranslmed.3005054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.