Abstract
Energy production is inevitably linked to the generation of toxic metabolites, such as reactive oxygen and carbonyl species, known as major contributors to ageing and degenerative diseases. It remains unclear how cells can adapt to elevated energy flux accompanied by accumulating harmful by-products without taking any damage. Therefore, effects of a sudden rise in glucose concentrations were studied in yeast cells. This revealed a feedback mechanism initiated by the reactive dicarbonyl methylglyoxal, which is formed non-enzymatically during glycolysis. Low levels of methylglyoxal activate a multi-layered defence response against toxic metabolites composed of prevention, detoxification and damage remission. The latter is mediated by the protein quality control system and requires inducible Hsp70 and Btn2, the aggregase that sequesters misfolded proteins. This glycohormetic mechanism enables cells to pre-adapt to rising energy flux and directly links metabolic to proteotoxic stress. Further data suggest the existence of a similar response in endothelial cells.
Keywords: Methylglyoxal, Reactive metabolites, Glucose metabolism, Nutrient signalling, Protein quality control system, Heat shock proteins
Graphical abstract

Highlights
-
•
Low-dose MG induces tolerance towards toxic levels of MG and ROS in yeast cells.
-
•
This preconditioning effect is mediated via a multi-layered defence mechanism.
-
•
The hormetic defence is composed of prevention, detoxification and damage remission.
-
•
Low MG induces the PQS including protein sorting and handling via HSPs.
1. Introduction
Energy flux is essential for life, but at the same time, it leads unavoidably to the generation of highly reactive metabolites, such as reactive carbonyl (RCS) and reactive oxygen (ROS) species. RCS and ROS cause cellular damage through the production of advanced glycation endproducts (AGEs) and oxidative stress [1], [2] and are thought to contribute to ageing. This view is supported by the observation that long-lived mutants from different species produce decreased levels of ROS [3], [4], and lowering the burden of oxidative stress leads to lifespan extension [5], [6], [7], [8], [9], [10], [11], [12]. However, an increasing body of evidence indicates that ROS can also act as a secondary messenger to induce pathways that prolong lifespan in yeast and C. elegans [13], [14], [15], [16], [17] and may also be beneficial in mice [18], [19]. This effect has been described as mitohormesis and has been defined as a non-linear response to increased levels of ROS [20].
ROS are only one type of reactive metabolites that have been reported to result from increased energy flux, and they are formed at a rather late stage of energy production, namely during the concerted four-electron reduction of molecular oxygen catalysed by cytochrome C oxidase of complex IV of the respiratory chain. Therefore, it is unclear whether mitohormesis alone would be sufficient to protect cells from metabolic stress. In comparison to ROS, the role and effects of RCS have received less attention. RCS are formed endogenously mainly by carbohydrate metabolism but also during lipid peroxidation and autoxidation of reducing substrates [21]. Several of the most reactive RCS including methylglyoxal (MG) are produced non-enzymatically during glycolysis, mainly from intermediates such as glyceraldehyde 3-phosphate and dihydroxyacetone phosphate (triosephosphates) [22], [23]. The production of RCS, such as MG poses more of an immediate threat to the integrity of the cell as they are produced at three metabolic steps upstream of the step producing ROS. Given its early production in energy metabolism upon increasing glucose levels, RCS would be appropriately positioned to act as a potential gatekeeper in an immediate feedback loop, signalling that an increase in metabolic stress is to be expected even before oxidative stress levels have augmented. It is currently unknown whether RCS, such as MG, can exert a hormetic effect. However, hormesis dose-response relationships have been reported for various preconditioning agents and can be regarded as a measure of biological plasticity [24], [25]. Therefore it is probable, that cells can also respond to RCS or MG in a hormetic manner.
The major cellular detoxification system for MG is the glyoxalase system, which requires catalytic amounts of reduced glutathione (GSH) to convert MG in a two-step reaction to D-lactate [23], [26]. In the absence of glyoxalase 1 (Glo1), however, MG can also be degraded via aldoketo-reductases to acetol [27], [28]. As MG is very toxic, the capacity of corresponding detoxifying enzymes has to be tightly adapted to the energy flux. Indeed, Glo1 is highly efficient, with reaction kinetics close to the rate of diffusion [29]. However, when MG levels reach a saturating amount which overwhelms the intracellular GSH pool, Glo1 will become inactive [30]. Under such circumstances, intracellular MG would accumulate and could potentially cause downstream damage. To handle such a dynamic situation, it seems plausible that cells have evolved additional defence mechanisms.
Yeast is a model organism well suited for the study of MG metabolism, as it possesses all enzymes needed to deal with glucose metabolites similar to mammalian cells. It has been reported that during a shift from low to high glucose concentrations, MG levels increase and cause protein damage, visible through increased amounts of AGE modified proteins, such as argpyrimidine [31], [32]. When MG is added to the medium or during high osmotic stress, Glo1 is induced via the High Osmolarity Glycerol 1/Mitogen-activated protein kinase (Hog1) pathway [33], [34], [35]. Additionally, increased levels of MG can activate the yeast oxidative stress transcription factor Yap1 [36]. Furthermore, a genome wide unbiased screen for gene deletions that render yeast cells more sensitive towards MG identified gene deletions conferring increased sensitivity to MG [37]. These were enriched for protein, mRNA and DNA metabolic processes, but most hits were never followed up upon in a systematic way. An important question that remained open is whether any of these processes could be modulated in response to MG stress.
In this study, the hypothesis was investigated that cells are equipped with an elementary, evolutionary conserved pathway that is triggered by RCS and enables cells to handle increasing accumulation of toxic metabolites on different levels, to prevent further formation of toxic metabolites, to detoxify metabolites, and to repair metabolite-induced damage. If existing, this pathway should render cells more resistant to the physiological relevant reactive dicarbonyl MG as well as to ROS upon increasing glucose levels. Our results suggest that, when reaching a threshold level, MG can initiate a positive feedback mechanism in a hormetic manner, leading to activation of a multi-layered defence response.
2. Materials and methods
2.1. Chemicals and antibodies
Methylglyoxal solution (40% w/v; M0252), 2-Deoxy-D-glucose (D8375), Aminoguanidine hydrochloride (396494), Hydrogen peroxide solution ≥ 30% (95302), Doxycycline hyclate (D9891) as well as Paraformaldehyde (16005) were purchased from Sigma-Aldrich. D(+)-Glucose was from Merck. Aprotinin was from Applichem, Pepstatin was from Pepta Nova GmbH, Leupetin was from Peptide Institute, Inc., HygromycinB was from InvivoGen, Nourseothricin (cloNAT) was from Werner BioAgents. Zymolyase 20T was purchased from Amsbio. DAPI solution was from Thermo Fisher scientific. The anti-MG-H1 antibody was generated as described previously [28]. The anti-GFP antibody was purchased from Roche (Cat. No. 11 814 460 001). The anti-actin antibody was from Millipore (clone C4, Cat. No. MAB1501). As secondary antibodies horseradish-linked goat anti-rat (Cell Signalling Technology, # 7077S) goat anti-mouse (Cell Signalling Technology, # 7076S) were used.
2.2. Yeast strains, plasmids, growth conditions and standard methods
Yeast strains used in this study are derivatives of the BY4741 strain (Euroscarf). Single Knock-outs of Glo1, Btn2, Msn2, Msn4, Yap1, Cad1, Rim101 and all double and triple Knock-outs were created freshly using a gene replacement strategy [38] and confirmed by PCR. All additional knock-outs mentioned derived from the yeast deletion library (Invitrogen). The tetracycline regulatable Sln1 construct was from the “Yeast Tet promoters Hughes Collection (yTHC), Dharmacon”. The sln1-gene was repressed in the presence of doxycycline with a final concentration of 50 µg/ml in the growth medium for 12 h [39]. The plasmid for galactose-based overexpression of Ssa4 derived from a yeast ORF overexpression library [40]. The Heat shock reporter construct consists of a heat shock element (HSE) cloned in front of the Cyc1 minimal promoter sequence followed by 4 tandem repeats of sfGFP. To gain a stronger fluorescent signal, the reporter plasmid contains 2 copies of these expression cassettes in inverted repeats. For a stable expression pattern, the construct was integrated into the genome. All yeast cultures were grown in standard rich media (YPD; BD Difco 1% yeast extract, 2% peptone and 2% glucose) or plated on standard YPD-plates.
2.3. MG tolerance test in yeast cells
For preconditioning with low concentrations of MG or H2O2, cells growing in logarithmic growth phase were divided into two samples. One sample was left untreated (control), while MG or H2O2 was added to the second aliquot and the cells were further incubated at 30 °C for 45 min. After that, the two samples were adjusted to the same OD600 of 0,4, divided into aliquots and treated with increasing concentrations of MG for 60 min at 30 °C prior to harvesting the cells and resuspending them in fresh YPD such that each aliquot contained the same OD600. Subsequently, cells were serially diluted in 1:5 steps, spotted onto YPD plates and incubated for 48 h at 30 °C to determine cell survival.
2.4. Preparation of total/cytosolic yeast protein extracts
50 ml of a yeast culture in logarithmic growth phase at an OD600 ~ 0.4–0.6 were harvested and the pellet was transferred to 1.5 ml eppendorf tubes, resuspended in 100 µl of lysis buffer (10 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.1% NP40, 1 mM PMSF, protease inhibitors) and dripped in liquid nitrogen present in an 2 ml round bottom eppendorf tube that contained a 7 mm stainless steel ball. After boiling out of the liquid nitrogen, the tubes were closed and placed in an adaptor for 2 ml tubes into a Retsch mixer Mill MM 400 and agitated for 2 × 2 min at 30 Hz. The sample was cooled in liquid nitrogen in between the two rounds of agitation. The resulting powder of lysed cells was transferred into a 1.5 ml tube and resuspended into 500 µl of lysis buffer. Samples were centrifuged (2 × 5.000 rpm for 3 min at 4 °C) and supernatant was used for protein determination and further analysis. All protein concentrations were determined using the Bradford technique and BSA as calibration standard as described previously [41].
2.5. Western blotting
15 µg protein was incubated in 2x Laemmli buffer (Sigma) at 95 °C for 10 min and separated by a Mini-PROTEAN® TGX (Bio-Rad) precasted gel (4–20% acrylamide). Proteins were then transferred to a PVDF membrane and blocked with 2% dry milk (in PBS) or in the case of MG-modifications with a Pierce® protein-free blocking buffer (Thermo) at room temperature for 1 h. Membranes were then incubated overnight at 4 °C with antibodies against MG-H1 in protein-free blocking buffer (1:500 dilution), Actin in 2% dry milk containing PBS and 0.05% Tween20 (PBS-T) (1:1000 dilution) or GFP in PBS-T (1:1000 dilution). After 4 washing steps (10–15 min each) with PBS-T, membranes were incubated with secondary antibodies (anti-rat or anti-mouse, 1:2000 dilution) for 1 h at room temperature. Proteins were visualized on X-Ray films using ECL detection reagents (GE healthcare) with varying exposure time (0.5–5 min).
2.6. GLO1 enzyme activity assay
Activity of GLO1 was determined spectrophotometrically as described previously [41]. Briefly, the method monitors the initial rate of change in absorbance at 235 nm caused by the formation of S-D-lactoylglutathione through catalysis of GLO1. The assay mixture contained 2 mM MG and 2 mM GSH in sodium phosphate buffer (50 mM, pH 6.6, 37 °C) and was incubated for 15 min to guarantee the complete formation of HTA. After the addition of the cytosolic protein fraction (1 μg/µl) the change in absorbance was monitored for 15 min. The activity of GLO1 described in units (U), where 1 U is the amount of GLO1 which catalyzes the formation of 1 μmol of S-D-lactoylglutathione per minute [28].
2.7. RNA-isolation
Extraction of RNA was achieved using the RNEAsy Mini Kit (QIAGEN). For yeast cells, enzymatic lysis using zymolase (150 U for 3 × 107 cells) was performed. For RNA isolation from MCECs, the standard QIAGEN protocol for animal cells was followed. Afterwards, RNA was converted into cDNA with a High-Capacity cDNA Reverse Transcription Kit (Thermo).
2.8. RNA-Seq and data-analysis
Total RNA isolated from yeast cells after MG preconditioning and respective controls (n = 3) was analysed using an Illumina HiSeq. 2000. Differential expression analysis was carried out using DESeq. 2 [42]. For subsequent analyses, a statistical cut-off of padj < 0.005 and log2 fold change (FC) > +/−1 was applied. Functional enrichment analysis was performed with HOMER [43] using pathways from the KEGG database [44]. Pathways comprised in either metabolism, genetic information processing or cellular processes are shown in separate diagrams/tables. Terms of the same pathway are only listed ones, e.g. oxidative branch of pentose phosphate pathway is included in pentose phosphate pathway. HOMER was used for performing de novo motif analyses in the promoter region (−300, 50) of regulated genes.
2.9. Real-time PCR
qPCR was performed using DyNAmo ColorFlash SYBR Green qPCR Master Mix (Thermo) and a LightCycler® 480 Instrument II (Roche). Signals of amplified products were verified using melting curve analysis and mRNA levels were normalised to Beta-Actin. Relative expression levels were calculated using the Ct method described elsewhere [45]. Primer sequences used for analyzing mRNA content were: Hsp70 (PrimerBank ID: 387211a2), forward `5- GAGATCGACTCTCTGTTCGAGG -3` and reverse `5- GCCCGTTGAAGAAGTCCTG -3`; Beta-Actin (PrimerBank ID: 6671509a1), forward `5- GGCTGTATTCCCCTCCATCG -3` and reverse `5- CCAGTTGGTAACAATGCCATGT -3`.
2.10. Heat-shock element promoter assay
BY4741 wt strains were transformed with heat shock reporter constructs, grown to mid-log phase at 30 °C and subjected to either a mild heat shock at 38 °C or 2.5 mM MG for 45 min. Afterwards, one half of the cell culture was directly fixed in PFA (see PFA-fixation prior to fluorescence microscopy) or recovered for 30 min at 30 °C prior to PFA-fixation. GFP expression was analysed by flow cytometry at a low flow rate in BD FACSCanto, equipped with a 488 nm laser (BD Biosciences). For each sample 50,000 events were collected. Average GFP intensity was determined using BD FACSDiva Software Version 6.1.3.
2.11. Identification of proteins from MG-induced aggregates using HPLC-MSMS mass spectrometry
BY4741 cells were grown to an OD600 of 0,4 and left untreated (control) or were treated with 10 mM MG for 60 min. Cells were harvested by centrifugation and ≈ 100 OD600 units were resuspended in 1 ml 1 M sorbitol, 0,1 mM EDTA, 5 mM GdnHCl (inhibition of Hsp104 to avoid resolubilisation of aggregates), supplemented with 3 mg zymolyase T20 and further incubated for 30 min at 30 °C. After washing with lysis buffer (50 mM Tris-HCl, pH 7.6, 50 mM NaCl, 5 mM MgCl2, 1 mM ß-mercaptoethanol, 1 mM PMSF + peptidase inhibitors Leupeptin, Pepstatin, Aprotinin), cells were pelleted and resuspended in 400 µl of lysis buffer. Cells were lysed by freezing in liquid nitrogen, thawing and vortexing with acid washed glass beads (5 × 1 min). After removal of the debris (2 min, 500 × g), insoluble material was pelleted by a centrifugation for 30 min at 14,000 × g, resuspended in 100 µl of lysis buffer and the protein concentration of the pellet fraction was determined by Bradford. 30 µg/lane were loaded in Laemmli sample buffer onto an SDS gel and separated by SDS-PAGE. Gel pieces from different molecular weight ranges of the lanes with untreated and MG-treated cells were cut out in parallel and subjected to in-gel trypsin digestion. The resulting peptides were stably labelled through reductive methylation with H2CO or D2CO at the free amino groups [46]. This resulted in a molecular weight increase of 4 Da/aminogroup of the peptides labelled with D2CO as compared to the same peptide labelled with H2CO. The peptide samples from MG-treated and non-treated cells (control) were mixed and analysed by HPLC-MSMS mass spectrometry (nanoUPLC-LTQ-Orbitrap MS). Subsequently, the molar ratio of a peptide from the MG treated sample and the control can be determined by the ratio of the signal intensity from heavy and light peptides. The identity of the peptides was determined using the Swissprot database (selected for Saccharomyces cerevisiae (7798 entries)).
2.12. Fluorescence microscopy
Paraformaldehyde (PFA) fixation of yeast cells prior to microscopy was performed by adding equal volumes of 8% PFA in PBS to a yeast culture (final PFA concentration 4%) and incubation for 10 min at room temperature, followed by washing with PBS. Fluorescence microscopy was performed with an Olympus IX81 inverted microscope with a 100×/1.45 oil objective and narrow band-pass filters and a Hamamatsu ORCA-R2 camera in the Olympus Excellence Software. Unless indicated differently, z-stacks of cells with a step width of 0.2 µm were taken and the single layers were merged as maximum intensity projection into 1 image. Images were analysed with the ImageJ software and brightness and contrast were linearly adjusted. DAPI staining was performed where indicated by incubation of the PFA-fixed cells in 70% EtOH (in PBS) for 30 min on ice. Subsequently, cells were re-isolated by centrifugation and resuspended in low volumes of PBS. 200 ng/ml of DAPI was added directly prior to microscopic analysis.
2.13. Cell culture
Primary mouse cardiac endothelial cells immortalized with SV40 large T antigen were obtained from Biozol/CELLutions Biosystems Inc. (Catalogue No. CLU5109). Cells were grown in DMEM (gibco) with 1 g/ml glucose containing 5% FCS (Sigma), 1% penicillin (10,000 Units/ml) (gibco), 1% streptomycin (10 mg/ml) (gibco) and 1% amphotericin B (250 μg/ml) (gibco) at 37 °C in a saturated humidity atmosphere containing 95% air and 5% CO2. Cells were grown to 70% confluence for in vivo experiments and passaged at 90% confluence using 0.05% Trypsin-EDTA (gibco).
2.14. MTT assay
Viability assays were performed with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT-method). Cells were seeded into 96-well plates and grown to 70% confluency under basal growth conditions. Cells were then customized to DMEM medium with only 0.1% FCS for 24 h and afterwards exposed to increasing concentrations of MG (50–1000 μM) for 48 h. For viability determination 50 µl of MTT solution (2 mg/ml in PBS) was added to the medium and cells were incubated at 37 °C for 3–4 h. Reduced MTT was then solubilized in 200 µl DMSO and absorbance was measured at 590 nm (reference at 690 nm) using a FLUOstar OMEGA multiplate reader (BMG Labtech). Viability was calculated as a percentage of controls (untreated cells). Data were fitted by nonlinear regression using the Graphpad PRISM 6 software (GraphPad Software Inc.) and values of LD50 determined [28].
2.15. Measurement of methylglyoxal
Intracellular methylglyoxal in treated yeast was determined by derivatization with 1,2-diamino-4,5-dimethoxybenzene, according to the method described by McLellan et al. [47].
2.16. Measurement of advanced glycation endproducts (AGEs)
AGEs (MG-H1, G-H1, CEL, CML, Argpyrimidine, Fructosyl-lysine) as well as methionine sulphoxide and 3-nitrotyrosine were determined by stable isotope dilution analysis liquid chromatography with tandem mass spectrometric detection (LC-MS/MS) after exhaustive enzymatic hydrolysis [48].
2.17. ROS measurements
Yeast cells were pre-grown in YPD-medium to logarithmic growth at 30 °C and MG was added while incubating for 1 h shaking. Cells were harvested by centrifugation, resuspended in PBS and supplemented with Dihydroethidium (Invitrogen) to a concentration of 5 µM for 1 h at 30 °C. FACS analysis was performed immediately after the incubation with a FACS-Canto II (BD Bioscience) using 488 nm excitation and 585/42 nm emission wavelength. Only single cells, defined by forward- and side-scatter (width-parameter), were considered for data analysis.
2.18. Statistical methods
Data are either expressed as mean values ± standard deviation or as mean values ± standard error of the mean (SEM) and were analysed for significance using two-tailed unpaired t-test with Welch's correction. Differences were considered significant at p < 0.05.
3. Results
3.1. High glucose and low MG induce tolerance towards high MG and ROS
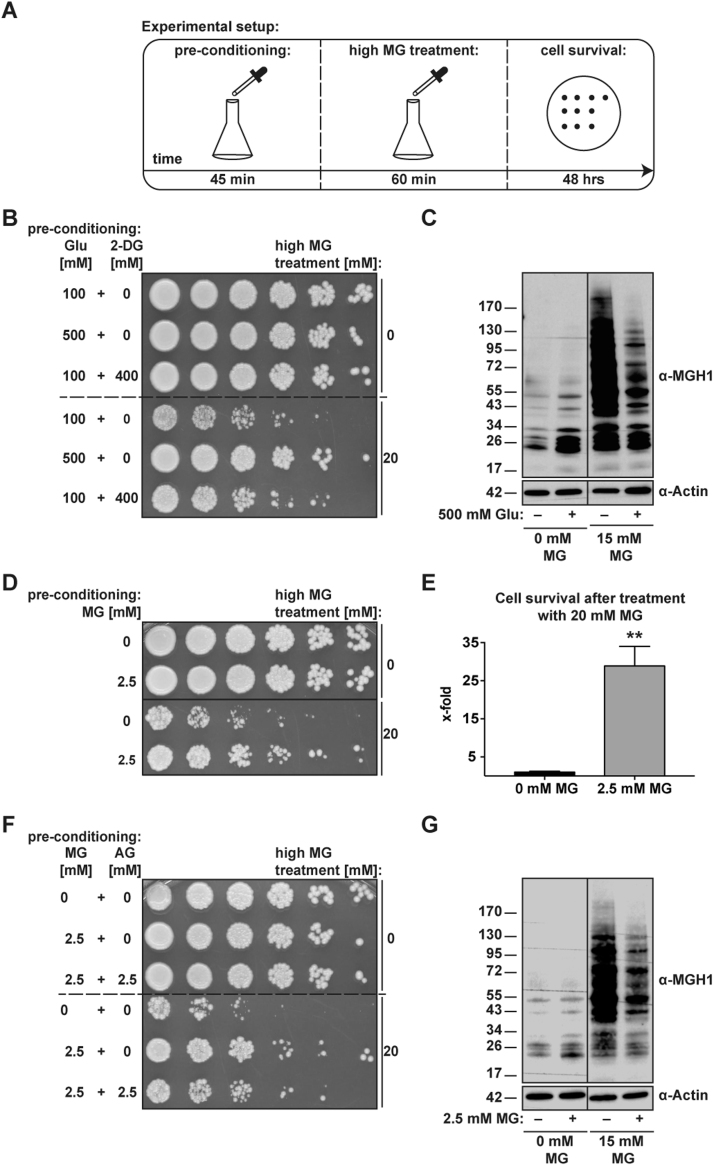
If cells were able to respond to metabolism by sensing rising levels of reactive metabolites to activate an immediate feedback loop, then short-term treatment with increased glucose concentrations should act protective during subsequent metabolic stress. Therefore, we challenged yeast cells with high levels of glucose prior to treatment with otherwise toxic concentrations of MG and ROS (Fig. 1). Preconditioning with high (500 mM, HG) but not standard (100 mM, NG) glucose concentrations led to an increased cell survival following incubation with toxic MG concentrations for 1 h (Fig. 1B). Consistently, pre-treatment with standard glucose (100 mM) plus non-fermentable 2-deoxy-D-glucose (400 mM) as a control had no effect on cell survival (Fig. 1B), demonstrating that the effect was dependent on glucose metabolism rather than on osmotic changes. Preconditioning with HG increased the amount of adduct of MG with arginine, hydroimidazolone (shortly MG-H1) (Fig. 1C, left panel), which confirmed that intracellular MG levels did indeed increase during the preconditioning with high glucose [31], [49]. At the same time, preconditioning prevented the accumulation of MG-H1 after high MG treatment (Fig. 1C, right panel), probably as a result of increased MG-detoxification or improved handling of MG-modified proteins. Together, these results suggest that increased glycolytic flux can trigger a defence response that prepares the cell for rising RCS.
Fig. 1.
Preconditioning with high glucose or low MG increases tolerance against MG and ROS. (A) Experimental setup of preconditioning experiment. In log-phase growing cells were treated for 45 min with high glucose (500 mM). Afterwards increasing MG (0, 15, 20, 25 mM) concentrations were added for 60 min, followed by direct MG removal and spotting onto YPD-plates in 1:5 dilution steps. Cell survival was recorded after 48 h of growth on YPD plates. (B) Cell survival of wild-type (wt) cells that were pre-treated with 100 mM glucose, 500 mM glucose or 100 mM glucose + 400 mM 2-deoxy-D-glucose and afterwards incubated with 0 or 20 mM MG. (C) Preconditioning with high glucose leads to increased intracellular MG-H1 concentrations shown by western blotting using whole cell lysates (left panel). Intracellular MG-H1 levels after treatment with 15 mM MG are ameliorated after preconditioning with 500 mM glucose (right panel). (D) Cell survival of wt cells after preconditioning with 2.5 mM MG and incubation with 0, and 20 mM MG versus respective controls. (E) Statistical analysis of surviving colonies after treatment with 20 mM MG with or without MG preconditioning. Values are shown as mean ± SEM (n = 3–4). (F) Simultaneous treatment with the MG scavenger aminoguanidine (AG) abolishes the protective effects of low MG pre-treatment. (G) Preconditioning with 2.5 mM MG results in an increase of intracellular MG-H1 concentrations in wild-type cells shown by western blotting using whole cell lysates (left panel). Cells that were pre-treated with low MG accumulate less MG-H1 after incubation with 15 mM MG as compared to control cells (right panel).
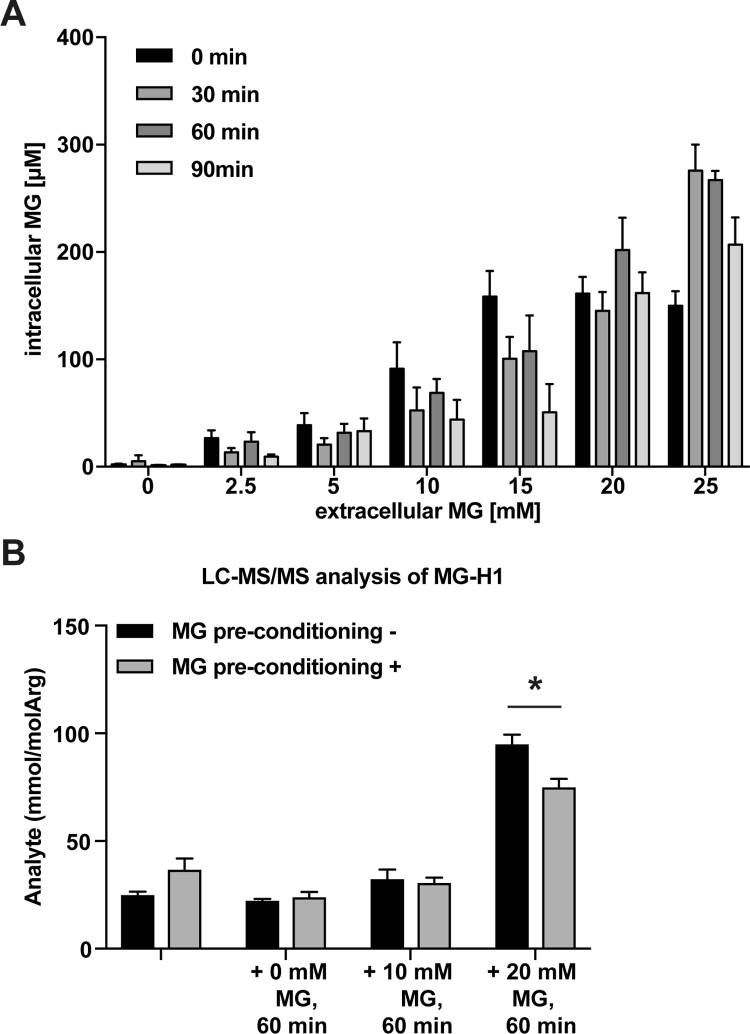
To establish whether the increased MG levels during HG treatment mediates this protective effect, cells were pre-conditioned with low (2.5 mM) levels of MG instead of high glucose before adding more toxic MG doses. It should be noted that the concentrations of MG added to the media were much higher than the resulting intracellular MG levels (Supplementary material Fig. S1A). Pre-treatment with low MG had the same effect as HG on cell survival (Fig. 1D and Supplementary material Fig. S2A), which we quantified to be 28-fold increased (Fig. 1E). Furthermore, it prevented from increased accumulation of MG-H1 (Fig. 1G and Supplementary material Fig. 1B). Similar data were obtained when MG-H1 was determined by LC-MS/MS, showing a decrease of MG-H1 accumulation in cells pre-conditioned and thereafter incubated with toxic MG concentrations (Supplementary material Fig. 1B). Simultaneous treatment with MG and aminoguanidine, a MG scavenger [50], diminished the protective effects of low MG treatment (Fig. 1F). Next, we measured ROS levels after preconditioning with low MG to reveal whether the protective effect observed here was mediated through ROS signalling (Supplementary material Fig. 2A), but we did not find any increase in ROS under these conditions. Interestingly, however, producing ROS by pre-treatment with low H2O2 concentrations also resulted in increased MG and ROS tolerance (Supplementary material Fig. 2B). Thus, both ROS and the dicarbonyl MG can induce a state of resistance towards both molecular species independently of each other.
3.2. Increased MG tolerance is inducible independently of Glo1
A likely explanation for the protective effect of preconditioning with low MG concentrations is that it induces Glo1 expression [33], [49]. This would also explain the lower amounts of accumulating MG-H1 upon subsequent treatment with high MG concentrations (Fig. 1G). In fact, Glo1 activity was up-regulated after MG preconditioning by approx. 1.5 times (Fig. 2A). Therefore, we reasoned that if the preconditioning effect is caused by boosting the capacity of the glyoxalase system, then MG tolerance should not be inducible any longer in cells lacking Glo1. As expected, ∆glo1 cells were more sensitive to MG (Fig. 2B) and accumulated more MG-H1 adduct (Fig. 2C) as compared to wild-type cells. Surprisingly, however, pre-treatment with low concentrations of MG (1.5 mM) still led to an increase in MG-tolerance (Fig. 2D) and lowered MG-H1 accumulation after high MG treatment (Fig. 2C), suggesting that increasing MG tolerance must involve additional defence strategies.
Fig. 2.
MG tolerance is not only mediated by Glo1 induction. (A) Glo1 enzyme activity in wt cells after MG preconditioning with 2.5 mM for 45 min and respective controls. Values are shown as mean ± SEM (n = 4). (B) Cell survival of wt versus Δglo1 cells after preconditioning with 2.5 mM MG and incubation with 0 or 15 mM MG. (C) Western blot analysis of MG-H1 concentrations in Δglo1 cells as compared to wt cells after incubation with 15 mM MG for 60 min with or without previous MG preconditioning. (D) Cell survival of Δglo1 cells after preconditioning with 1.5 mM MG and respective controls.
3.3. HOG1 pathway is part of a MG triggered feedback mechanism
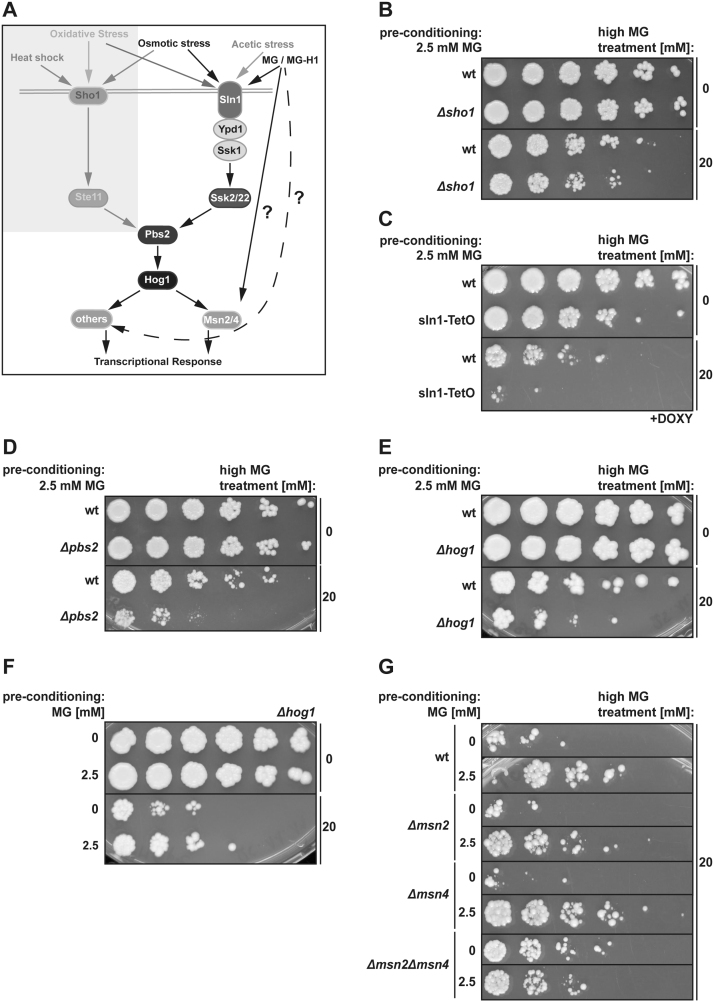
MG triggered Glo1 induction was previously described to occur through activation of the Hog1 pathway that activates the transcription factors Msn2/Msn4 (Fig. 3A) [33], [49]. As increased MG tolerance can be induced in Δglo1 cells, we questioned whether the additional defence pathways are also induced via Hog1 and Msn2/Msn4. First, we confirmed that MG activates the Hog1 signalling cascade via the Sln1 branch and not the Sho1 branch (Fig. 3A), by demonstrating that deleting Sho1 did not effect MG sensitivity (Fig. 3B), whereas reducing the levels of the essential Snl1 protein via the Doxycylin-inducible TetO-system [39] did (Fig. 3C). Moreover, loss of the downstream protein-kinases Pbs2 (Fig. 3D) and Hog1 (Fig. 3E) decreased MG-tolerance. Thus the MG-triggered Hog1 pathway plays a role in mediating the MG-induced feedback mechanism, as previously described [33], [35]. Unexpectedly, Δhog1 cells could still induce MG-tolerance (Fig. 3F) and loss of Msn2 or Msn4 had only minor effects on MG tolerance (Fig. 3G). However, simultaneous loss of both Msn2 and Msn4 resulted in cells that were even more resistant to MG as compared to wild type cells without preconditioning, suggesting that alternative MG defence pathways were constitutively activated in this strain (Fig. 3G). Consistently, preconditioning could not further increase MG tolerance in this strain (Fig. 3G). This clearly demonstrates that alternative pathways or mechanisms besides the involved Hog1/Msn2/Msn4 system (Fig. 3A) must exist that contribute to MG tolerance induction.
Fig. 3.
MG triggered defence response comprises HOG1 pathway. (A) Scheme of MG signal transduction. (B) Cell survival after MG preconditioning of wt yeast cells was compared to Δsho1 cells after incubation with 0 and 20 mM MG. (C) As Δsln1 cells are not viable, a conditional knockdown of Sln1 was induced via the TetO system and doxycycline (+DOXY) treatment. Cell survival of wt and Sln1-TetO cells under doxycycline treatment was compared after MG preconditioning and additional incubation with 0 or 20 Mm MG. (D) Δpbs2 and (E) Δhog1 cells were compared to wt cells after MG preconditioning and treatment with 0 or 20 mM MG via spotting assay for analysis of cell survival. (f) In order to test whether Δhog1 cells can still induce MG tolerance, cells without preconditioning were compared to cells pre-treated with 2.5 mM MG. Cell survival was analysed after additional incubation with 0 or 20 mM MG. (g) MG tolerance induction in wild-type, Δmsn2, Δmsn4 and Δmsn2Δmsn4 double-knockout cells after MG-preconditioning with 2.5 mM MG.
3.4. MG preconditioning leads to massive changes in gene expression
In order to identify alternative pathways induced by preconditioning with low concentrations of MG, RNA-Seq was carried out. Using DESeq. 2, approximately 2/3 of the yeast genes showed to be differentially expressed (padj < 0.005 and log2 FC > ±1) in low MG (2.5 mM) pre-stimulated cells (Fig. 4A), which was an unexpectedly high number, indicating that the cells undergo extensive transcriptional changes. Out of these, 1239 genes were ≥ 2-fold up-regulated and 1368 were ≥ 2-fold down-regulated (Tables S1 and S3; Figs. S3 and S4B).
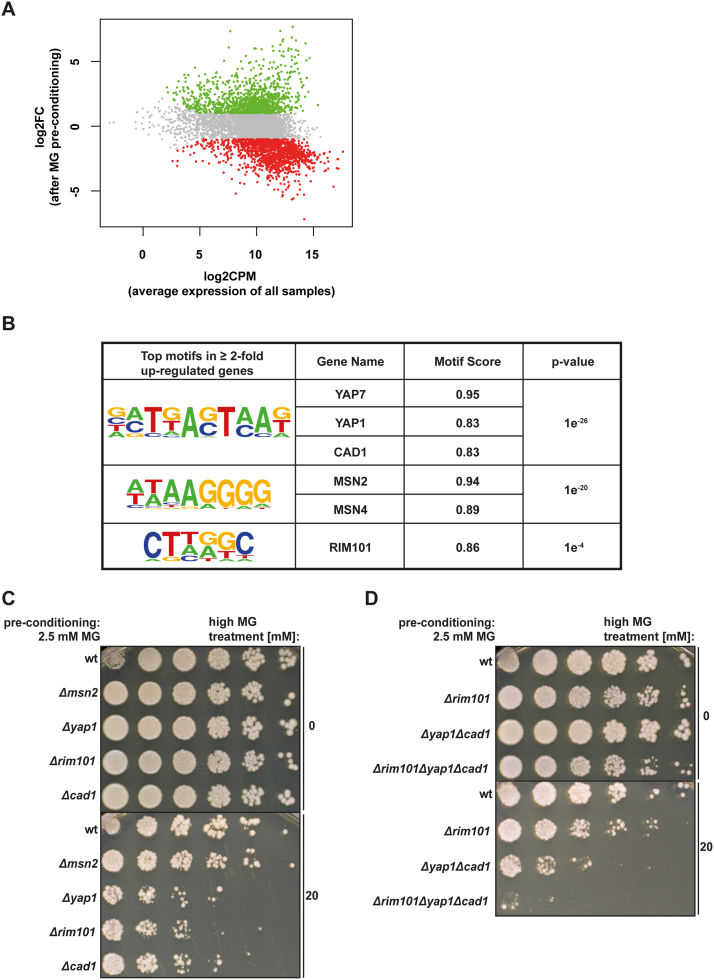
Fig. 4.
Transcription factors involved in transducing MG-triggered changes in gene expression. (A) MA plot of the average (normalised) expression of all the samples (both control and MG pre-conditioned) against the log2FC, showing whether the MG pre-conditioned samples are increased (green) or decreased (red) compared to control. (B) Top motifs resulting from de novo motif analysis in the promoters of ≥ 2-fold up-regulated genes. (C) Cell survival of wt cells compared to Δmsn2, Δyap1, Δrim101 and Δcad1 cells after MG preconditioning and incubation with 0 and 20 mM MG. (D) Cell survival of wt cells as compared to Δrim101, Δyap1Δcad1 double knockout or Δrim101Δyap1Δcad1 triple knockout cells after MG preconditioning and incubation with 0 and 20 mM MG. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To identify putative transcriptional regulators, de novo motif analysis was performed in the promoters of significantly regulated genes, and several highly enriched motifs were identified (Fig. 4B and Supplementary material Fig. 3). For genes up-regulated by preconditioning with low MG concentrations, the top-scoring motif (motif 1) showed the highest motif score for three transcription factors of the YAP family: Yap7, Yap1 and Cad1. Targets of Yap1 and Cad1 are well defined, whereas little is known about genes regulated by Yap7. Consistently, activation of Yap1 by MG treatment was previously described [36]. Motif 2 supported the previously described MG-triggered activation of Msn2/Msn4 via the Hog1 pathway [33], [49]. Finally, motif 3 showed a high motif score for the transcription factor Rim101, which is involved in pH regulation and yeast cell wall assembly [51].
To experimentally validate our motif analyses, the effect of genetic loss of Msn2, Yap1, Cad1 and Rim101 on MG-tolerance induction was further investigated. Without additional MG treatment, all knockout cells did not differ in cell survival as compared to wild-type cells after MG preconditioning (Fig. 4C). Loss of Msn2 only resulted in minor reduction of MG-tolerance induction after MG preconditioning as compared to controls (Figs. 3G and 4C) whereas Yap1, Cad1 and Rim101 knockout cells showed elevated MG sensitivity despite MG preconditioning. Double and triple knockouts of the candidate genes did not show any difference in cell survival as compared to wild-type cells after treatment with low MG (Fig. 4D). However, MG-tolerance was increasingly reduced from single to double and further to triple knockouts despite MG preconditioning, with the Δrim101Δyap1Δcad1 cells becoming highly sensitive to MG treatment (Fig. 4D). This implicates that these three transcription factors play a central role in mediating the MG-triggered defence mechanism.
3.5. MG triggered defence response involves metabolic changes and activation of the protein quality control system
To identify the MG-regulated pathways rendering the cells resistant, genes changed ≥ 2-fold were analysed using KEGG pathway analyses. The highest enrichment of the up-regulated genes was found in carbohydrate and sulfur/GSH metabolism, as well as in pathways involved in the protein quality control system (PQS), including protein folding, autophagy, ubiquitin-mediated proteolysis and protein processing in the ER (Fig. 5A). This implies that cells respond on three different levels to handle increased MG production: Level 1 shifts the carbohydrate metabolism towards the pentose phosphate pathway and increases the sulfur/GSH metabolism in order to balance the intracellular redox-state and prevent further RCS and ROS production. Level 2 leads to up-regulation of detoxifying enzymes like oxidoreductases, aldoketo-reductases and glyoxalases. Level 3, comprised of components of the protein quality control, takes care of the damage processing.
Fig. 5.
MG preconditioning leads to changes in metabolism and activates the protein quality control system (PQS). (A) KEGG pathway analysis was carried out with ≥ 2-fold up-regulated genes after MG preconditioning. Shown are logP values of significantly enriched terms of metabolism or cellular processes. (B) Table of heat-shock proteins significantly up-regulated by MG preconditioning. (C) Flow Cytometry-based GFP reporter assays for the heat-shock element was carried out in wt yeast cells immediately (45 min) and 30 min after MG-preconditioning (45 min + 30 min recovery) with 2.5 mM MG as well as heat-shock at 38 °C. (D) Protein levels analysed via western blotting of the two inducible forms of Hsp70, Ssa3 and Ssa4, after MG preconditioning and heat-shock at 38 °C. Ssa3 and Ssa4 were genomically tagged with GFP to be able to use an anti-GFP antibody.
Furthermore, an enrichment of pathways that deal with the maintenance of genetic information processing such as DNA repair was observed, however with much lower logP values (Supplementary material Table S2; Fig. 4A). Analysis of the ≥ 2-fold down-regulated genes revealed enrichment for genes involved in transcription and translation (Supplementary material Table S3; Fig. 4B), indicating that cells undergo growth reduction.
Analysis of the ≥ 2-fold up-regulated genes showed a high number of heat shock proteins (HSPs) being up-regulated by low-dose MG pre-treatment (Fig. 5B). Many of the genes encoding the up-regulated HSPs revealed to have promoter motifs for one or more of the identified transcriptional regulators (Fig. 5B). Surprisingly, motif analyses did not show a high score for heat-shock factor 1 (Hsf1), which is known to be a major transcription factor in the regulation of expression of HSPs under conditions of proteotoxic stress [52]. Therefore, the activation of the heat-shock response element (HSE) present in promoters of Hsf1 target genes was nonetheless tested for MG responsiveness using a GFP reporter assay. In line with data from the motif analyses, HSE activation was not triggered by MG preconditioning, but responded well to a 45 min heat shock at 38 °C (Fig. 5C). Consequently, upon MG preconditioning, genes encoding for HSPs are up-regulated independently of Hsf1.
Proteotoxic stress leads to increased expression of the two inducible forms of the cytosolic yeast Hsp70 family, Ssa3 and Ssa4. Both were strongly up-regulated on mRNA levels after MG preconditioning (Ssa3 7-fold and Ssa4 19-fold, Fig. 5B). To find out whether elevated mRNA levels also resulted in increased concentrations of the corresponding proteins, Ssa3 and Ssa4 levels were analysed after MG preconditioning and heat shock at 38 °C (Fig. 5D). Ssa4 was expressed at higher baseline levels than Ssa3 and also responded strongly to treatment with low MG concentrations (Fig. 5D). This finding supported the data from the mRNASeq and indicated that up-regulation of molecular chaperones on the mRNA levels during MG preconditioning resulted in up-regulation on the protein level, too.
3.6. Molecular chaperones and protein sorting to deposition sites are mediators of protective mechanisms during increased metabolic stress
Next, the physiological impact of up-regulation of molecular chaperones on development of MG tolerance was tested. Several of the chaperones listed in Fig. 5B were deleted, and possible changes in MG tolerance were investigated (Supplementary material Fig. 5A). A clear reduction in MG tolerance was observed in particular for Δssa3Δssa4 cells that lack both inducible cytosolic Hsp70 genes (Fig. 6A). This could be rescued by overexpression of Ssa4 (Fig. 6B). Therefore, protein misfolding and aggregation seem to be one aspect of MG toxicity.
Fig. 6.
Heat-shock proteins and protein sorting to deposition sites are mediators of MG induced defence response. (A) Cell survival of wt as compared to Δssa3Δssa4 cells after MG preconditioning and incubation with 0 or 20 mM of MG. (B) Same experiment as in (A) but, additionally, the effect of overexpression of Ssa4 via an inducible galactose promoter in Δssa3Δssa4 knockout cells was analysed. (C) Proteins enriched in MG-induced protein aggregates identified by mass spectrometry. (D) Fluorescence microscopy of wild-type cells expressing Hsp104-GFP after MG preconditioning and high MG treatment compared to cells without MG preconditioning. Statistical analysis shows mean values of foci per 100 cells ± SD (n = 4). (E) Fluorescence microscopy of Δbtn2 cells expressing the same Hsp104-GFP construct as wild-type cells shown in (D). Δbtn2 cells are unable to form foci with and without MG preconditioning and high MG treatment. Microscopy images represent the maximum projection of z-stacks with a step width of 0,2 µm. Scale bar = 2 µm.
To test whether MG stress results in significant protein aggregation in our conditions and whether preconditioning with low MG influences this process, a GFP tagged version of the disaggregase Hsp104 [38] was employed to visualize aggregates. Yeast cells expressing this construct were then subjected to MG pre-treatment followed by a higher dose of MG, and the protein aggregates were visualized by fluorescence microscopy [53]. After treatment with high MG (10 mM), pre-conditioned cells showed more visible fluorescent foci representing Hsp104-GFP bound aggregates as compared to untreated cells (Fig. 6C). This may appear counterintuitive, as one might expect more aggregates in cells without preconditioning. However, there was mainly one fluorescent focus per cell that was located almost exclusively at the site of the nucleus (Supplementary material Fig. 5B), suggesting that MG-modified proteins are sorted to the intranuclear aggregate deposition site (INQ) and this is the reason why distinct fluorescent foci become visible. INQ formation in response to heat-induced protein misfolding strictly depends on the protein aggregase Btn2 [54], [55], [56]. Interestingly, Btn2 was up-regulated after MG preconditioning by more than 8-fold (Supplementary material Table S1). To elucidate whether sorting of MG-induced aggregates to the INQ is Btn2 dependent, Hsp104-GFP localisation in Δbtn2 cells after preconditioning and a subsequent high dose of MG was analysed. In ∆btn2 cells, MG-induced foci formation was completely abolished (Fig. 6D). Interestingly, Δbtn2 cells were slightly more sensitive towards MG after preconditioning (Supplementary material Fig. 5C), indicating that Btn2 and protein sorting to INQ is necessary to achieve full cellular protection via the MG-triggered defence response.
Next, the protein aggregates induced by MG treatment were isolated from control and high MG treated wild-type yeast cells and analysed by mass spectrometry. Enrichment of Ssa4 and other heat shock proteins confirmed the recruitment of components of the protein quality control to MG-induced aggregates (Fig. 6E). As the aggregate isolation was not specific for Hsp104 or Btn2 binding, also aggregates outside the INQ were analysed, which explains enrichment of mitochondrial proteins (Fig. 6E).
3.7. MG triggered stress resistance and protein quality control is also relevant in mammals
To investigate whether there exists a MG-triggered tolerance induction in mammalian cells, mouse cardiac endothelial cells (MCECs) were treated for 24 h with low MG concentrations before a MG toxicity test was carried out over 48 h followed by an MTT assay (Fig. 7A). Pre-treatment with 100 nM or 500 nM MG for 24 h significantly increased the MG-specific EC50 value in MCECs. Furthermore, mRNA levels of mammalian Hsp70 following treatment with 100 nM MG for 24 h were significantly increased (Fig. 7B), confirming that also in mammals, MG tolerance involves the protein quality control system.
Fig. 7.
MG preconditioning is also relevant for mammalian cells. (A) Mouse cardiac endothelial cells (MCECs) were treated for 24 h with low concentrations of MG before a MG toxicity test was carried out over 48 h in order to determine the specific EC50 value. Data are shown as mean ± SEM (n = 4–5). (B) Changes in Hsp70 mRNA expression were analysed after MG preconditioning with 0,1 µM MG as compared to untreated controls using RT-PCR. Data are shown as mean ± SD (n = 4–5).
4. Discussion
Nutrition and metabolism inevitably result in the production of toxic metabolites. We show here that to ensure effective energy production without the accumulation of toxic metabolites, nature has evolved a mechanism, which enables cells to cope with the negative consequences from elevated energy flux. The data shown in this study establish a regulatory mechanism by which cells can adapt to an increasing metabolic rate as soon as reactive metabolites start to rise. This feedback mechanism initiates a transcriptional response that includes three different levels in order to handle the otherwise harmful effects of highly reactive molecules: i) prevention of enhanced production, ii) detoxification of reactive metabolites and iii) remission of damage caused when the first two levels of defence are overwhelmed.
The metabolic defence response can be initiated via a relatively short but strong increase in glucose concentration or by the addition of low concentrations of MG (Fig. 1B and D). Interestingly, these low MG concentrations do not increase intrinsic ROS levels (Fig. 2A). However, MG preconditioning also increases cellular tolerance against H2O2 and vice versa (Supplementary material Fig. 2B). This implicates that in addition to the hormetic response which exists for ROS [20], a similar response also exists for RCS, in particular for the dicarbonyl MG. As RCS are produced before ROS can be released from the respiratory chain, the cell can use RCS as an early indicator for increasing metabolic stress.
Currently, MG is mainly discussed as potentially damaging molecule that is involved in the development of diabetic late complications [57], [58], [59]. Considerable attention has been paid to the main detoxification system of MG, namely the glyoxalase system [23], [26], [58]. Although Glo1 revealed to be involved in the MG-induced hormetic response (Fig. 2A and B), cells were still able to induce MG-tolerance in the absence of Glo1 (Fig. 2D), underlining that MG detoxification via the glyoxalase system is only one part of a complex multifactorial response to deal with critically rising concentrations of RCS.
In yeast, MG has been described to activate the Hog1 pathway leading to induction of Msn2/Msn4 driven transcription, ultimately leading to increased expression of Glo1 [34], [36], [49]. To identify additional transcriptional regulators of the multifactorial defence observed here, our de novo motif analysis identified several highly enriched motifs (Figs. 4B and S4). The top-scoring motif (motif 1) showed the highest motif score for three transcription factors of the YAP family. Activation of Yap1 by MG had been previously described [36], [60]. However, this study suggests that also other members of the Yap family are involved in the MG-induced defence mechanism, for example the paralog of Yap1, Cad1, which arose from whole genome duplication [61], [62], [63], [64]. Surprisingly, the transcription factor Rim101 involved in pH regulation and cell wall assembly [51], was also found to have an effect on MG-tolerance induction. Further studies are required to better understand the role that Rim101 plays in the preconditioning response, but its identification underlines the complexity of the identified defence response.
Recently, a study in C.elegans revealed a regulatory network that mediates induction of dicabonyl detoxification via activation of TRPA-1 channels [65]. Following TRPA-1 activation by dicabonyls, Ca2+-flux is increased leading to the activation of CaM kinase II as well as p38 MAP kinases, ultimately leading to the induction of Nrf2 and increased expression of glyoxalases [65]. This mechanism, however, differs in many aspects from the one described in this study. First, the MG concentrations necessary to activate TRPA-1 channels are much higher than the ones used for MG preconditioning [65], [66]. Second, there is no homologue to TRPA-1 channels or Nrf2 in yeast cells. Nevertheless, both mechanisms involve p38 MAP kinases, as Hog1 is the yeast homologue to p38, indicating that there maybe commonalities between the two mechanisms.
Pathway analysis of ≥ 2-fold up-regulated genes following MG preconditioning revealed that when the defence levels of prevention through metabolic shift (1) and detoxification of reactive metabolites (2) are overwhelmed, level 3 comes into action. Level 3 is comprised of components of the PQS (Fig. 5A). This identifies a direct link between energy flux and the proteotoxic stress response. The current data indicate that different components of the PQS such as molecular chaperones Hsp70 and Btn2 are involved in handling or sorting of MG-modified proteins to specialized cellular protein deposition sites. These deposition sites are cyto-protective, as potentially cytotoxic misfolded proteins can be sequestered into these specific compartments [55], [67]. It has been shown that the aggregase Btn2 is essential for protein sorting into the INQ [55]. Interestingly, Btn2 is induced 8-fold during MG pre-treatment and hence, INQ can be formed more readily in these pre-treated cells. Depletion of Btn2 seems to increase MG sensitivity mildly (Supplementary material Fig. 5C), demonstrating that deposition of aggregates induced by MG stress into specialized protein quality compartments acts cell-protective (Fig. 6D).
Additionally, initial analysis suggests that the observed defence mechanism also exists in mammalian cells, as preconditioning with low MG also led to increased tolerance towards RCS in MCECs and an induction of Hsp70 chaperones (Fig. 7).
5. Conclusion
This study shows a protective role of RCS by initiating a hormetic response upon increasing metabolic stress, indicating that next to mitohormesis there also exists glycohormesis. It also indicates a direct link between metabolic and proteotoxic stress. Targeting different levels or mediators of this response by specific therapeutic interventions, for example manipulation of chaperone systems, might open new fields for drug development and treatment of diseases involving increased RCS and ROS levels, such as diabetes mellitus and neurodegenerative diseases.
Author contribution
Conceptualization, J.Z., P.P.N., S.H. and J.T.; Investigation, J.Z., C.A.F., T.H.F., L.S. and R.A.K.; Formal Analysis, J.Z., C.A.F., T.H.F., A.L. and J.T.; Writing - Original Draft, J.Z.; Writing – Review & Editing, J.Z., T.H.F., A.L., S.H., M.M., B.B., P.P.N. and J.T.; Funding Acquisition, P.P.N. and J.T.; Supervision, P.P.N. and J.T.
Conflict of interest
The authors declare that there is no conflict of interest.
Acknowledgements
We thank Deutsche Forschungsgemeinschaft (CRC1118) and Dietmar Hopp foundation for supporting this study. Mass spectrometry/Proteomics was performed at the ZMBH Core facility for mass spectrometry and proteomics. Fluorescence microscopy was done at the ZMBH Imaging Facility. RNASeq was carried out at the EMBL Genomics Core Facility. We also thank members of the Nawroth/Herzig/Tyedmers laboratory as well as of the CRC1118 for helpful comments and discussions.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.08.007.
Contributor Information
Johanna Zemva, Email: johanna.zemva@med.uni-heidelberg.de.
Jens Tyedmers, Email: j.tyedmers@zmbh.uni-heidelberg.de.
Appendix A. Supplementary material
Fig. S1.
MG and MG-H1 concentrations after MG treatment. (A) Wild-type yeast cells were treated with increasing MG concentrations (2.5 mM, 5 mM, 10 mM, 15 mM, 20 mM and 25 mM), which were added to standard YPD medium while cells were growing in log phase. Samples were withdrawn after 0, 30, 60 and 90 min and intra- as well as extracellular MG concentrations were measured. Values are shown as means ± SD (n = 3). (B) Intracellular MG-H1 levels were measured directly after MG preconditioning with 2.5 mM MG for 45 min as well as after additional incubation for 60 min with 0, 10 or 20 mM MG using LC-MS/MS. Cells without MG preconditioning served as respective controls. Values are shown as means ± SD (n = 3).
Fig. S2.
Role of oxidative stress and ROS formation during MG preconditioning and MG tolerance induction. (A) DHE (Dihydroethidium) positive wild-type cells were counted using Flow Cytometry after MG preconditioning with 2.5 mM and high MG treatment with 20 mM versus respective controls. (B) Effects of preconditioning with low levels of H2O2 (0.3 mM) for 45 min on ROS and MG tolerance induction were analysed and compared to effects of MG preconditioning.
Fig. S3.
Top motifs resulting from de novo motif analysis of promoters in ≥ 2-fold down-regulated genes.
Fig. S4.
(A) KEGG pathway analysis of ≥ 2-fold up-regulated genes after MG preconditioning. Shown are logP values of significantly enriched terms of genetic information processing. (B) KEGG pathway analysis of ≥ 2-fold down-regulated genes after MG preconditioning. Shown are logP values of significantly enriched terms of metabolism and genetic information processing.
Fig. S5.
(A) Cell survival and cell growth of wt cells as compared to Δhsp12, Δhsp26, ΔΔhsp31, Δhsp78, Δhsp82 cells after MG preconditioning with 2.5 mM MG and MG treatment with 15 mM. (B) Fluorescence microscopy of wt cells expressing Hsp104-GFP after MG preconditioning and high MG treatment. In order to visualize foci localisation, cells were additionally stained with DAPI. Microscopy images represent the maximum projection of z-stacks with a step width of 0,2 µm. Scale bar = 2 µm. (C) Cell survival of wt as compared to Δbtn2 cells after MG preconditioning with 2.5 mM MG and additional incubation with 0 or 20 mM MG.
Supplementary material Table_S1_Diff_expressed_MG_ctr_DESeq2
Supplementary material Table_S2_KEGG_pathway_categories_MG_UP
Supplementary material Table_S3_KEGG_pathway_categories_MG_DOWN
References
- 1.Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 2.Harman D. The biologic clock: the mitochondria? J. Am. Geriatr. Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 3.Gredilla R., Sanz A., Lopez-Torres M., Barja G. Caloric restriction decreases mitochondrial free radical generation at complex I and lowers oxidative damage to mitochondrial DNA in the rat heart. FASEB J. 2001;15:1589–1591. doi: 10.1096/fj.00-0764fje. [DOI] [PubMed] [Google Scholar]
- 4.Sanz A., Fernandez-Ayala D.J., Stefanatos R.K., Jacobs H.T., Mitochondrial ROS production correlates with, but does not directly regulate lifespan in Drosophila. Aging (Albany NY) 2010;2:200–223. doi: 10.18632/aging.100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melov S., Ravenscroft J., Malik S., Gill M.S., Walker D.W., Clayton P.E., Wallace D.C., Malfroy B., Doctrow S.R., Lithgow G.J. Extension of life-span with superoxide dismutase/catalase mimetics. Science. 2000;289:1567–1569. doi: 10.1126/science.289.5484.1567. [DOI] [PubMed] [Google Scholar]
- 6.Moskovitz J., Bar-Noy S., Williams W.M., Requena J., Berlett B.S., Stadtman E.R. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl. Acad. Sci. USA. 2001;98:12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruan H., Tang X.D., Chen M.L., Joiner M.L., Sun G., Brot N., Weissbach H., Heinemann S.H., Iverson L., Wu C.F., Hoshi T. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc. Natl. Acad. Sci. USA. 2002;99:2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishii N., Senoo-Matsuda N., Miyake K., Yasuda K., Ishii T., Hartman P.S., Furukawa S. Coenzyme Q10 can prolong C. elegans lifespan by lowering oxidative stress. Mech. Ageing Dev. 2004;125:41–46. doi: 10.1016/j.mad.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Huang T.T., Naeemuddin M., Elchuri S., Yamaguchi M., Kozy H.M., Carlson E.J., Epstein C.J. Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxide dismutase. Hum. Mol. Genet. 2006;15:1187–1194. doi: 10.1093/hmg/ddl034. [DOI] [PubMed] [Google Scholar]
- 10.Zou S., Sinclair J., Wilson M.A., Carey J.R., Liedo P., Oropeza A., Kalra A., de Cabo R., Ingram D.K., Longo D.L., Wolkow C.A. Comparative approaches to facilitate the discovery of prolongevity interventions: effects of tocopherols on lifespan of three invertebrate species. Mech. Ageing Dev. 2007;128:222–226. doi: 10.1016/j.mad.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quick K.L., Ali S.S., Arch R., Xiong C., Wozniak D., Dugan L.L. A carboxyfullerene SOD mimetic improves cognition and extends the lifespan of mice. Neurobiol. Aging. 2008;29:117–128. doi: 10.1016/j.neurobiolaging.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Shibamura A., Ikeda T., Nishikawa Y. A method for oral administration of hydrophilic substances to Caenorhabditis elegans: effects of oral supplementation with antioxidants on the nematode lifespan. Mech. Ageing Dev. 2009;130:652–655. doi: 10.1016/j.mad.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Doonan R., McElwee J.J., Matthijssens F., Walker G.A., Houthoofd K., Back P., Matscheski A., Vanfleteren J.R., Gems D. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008;22:3236–3241. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mesquita A., Weinberger M., Silva A., Sampaio-Marques B., Almeida B., Leao C., Costa V., Rodrigues F., Burhans W.C., Ludovico P. Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proc. Natl. Acad. Sci. USA. 2010;107:15123–15128. doi: 10.1073/pnas.1004432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Raamsdonk J.M., Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz T.J., Zarse K., Voigt A., Urban N., Birringer M., Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Schmeisser S., Priebe S., Groth M., Monajembashi S., Hemmerich P., Guthke R., Platzer M., Ristow M. Neuronal ROS signaling rather than AMPK/sirtuin-mediated energy sensing links dietary restriction to lifespan extension. Mol. Metab. 2013;2:92–102. doi: 10.1016/j.molmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kujoth G.C., Hiona A., Pugh T.D., Someya S., Panzer K., Wohlgemuth S.E., Hofer T., Seo A.Y., Sullivan R., Jobling W.A., Morrow J.D., Van Remmen H., Sedivy J.M., Yamasoba T., Tanokura M., Weindruch R., Leeuwenburgh C., Prolla T.A., Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 19.Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly Y.M., Gidlof S., Oldfors A., Wibom R., Tornell J., Jacobs H.T., Larsson N.G. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 20.Ristow M., Schmeisser K. Mitohormesis: promoting health and lifespan by increased levels of reactive oxygen species (ROS) Dose Response. 2014;12:288–341. doi: 10.2203/dose-response.13-035.Ristow. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Brien P.J., Siraki A.G., Shangari N. Aldehyde sources, metabolism, molecular toxicity mechanisms, and possible effects on human health. Crit. Rev. Toxicol. 2005;35:609–662. doi: 10.1080/10408440591002183. [DOI] [PubMed] [Google Scholar]
- 22.Rabbani N., Ashour A., Thornalley P.J. Mass spectrometric determination of early and advanced glycation in biology. Glycoconj. J. 2016;33:553–568. doi: 10.1007/s10719-016-9709-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabbani N., Thornalley P.J. Methylglyoxal, glyoxalase 1 and the dicarbonyl proteome. Amino Acids. 2012;42:1133–1142. doi: 10.1007/s00726-010-0783-0. [DOI] [PubMed] [Google Scholar]
- 24.Calabrese E.J. Preconditioning is hormesis part I: documentation, dose-response features and mechanistic foundations. Pharmacol. Res. 2016;110:242–264. doi: 10.1016/j.phrs.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 25.Calabrese E.J. Preconditioning is hormesis part II: how the conditioning dose mediates protection: dose optimization within temporal and mechanistic frameworks. Pharmacol. Res. 2016;110:265–275. doi: 10.1016/j.phrs.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 26.Rabbani N., Xue M., Thornalley P.J. Activity, regulation, copy number and function in the glyoxalase system. Biochem. Soc. Trans. 2014;42:419–424. doi: 10.1042/BST20140008. [DOI] [PubMed] [Google Scholar]
- 27.Aguilera J., Prieto J.A. The Saccharomyces cerevisiae aldose reductase is implied in the metabolism of methylglyoxal in response to stress conditions. Curr. Genet. 2001;39:273–283. doi: 10.1007/s002940100213. [DOI] [PubMed] [Google Scholar]
- 28.Morgenstern J., Fleming T., Schumacher D., Eckstein V., Freichel M., Herzig S., Nawroth P. Loss of glyoxalase 1 induces compensatory mechanism to achieve dicarbonyl detoxification in mammalian schwann cells. J. Biol. Chem. 2017;292:3224–3238. doi: 10.1074/jbc.M116.760132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rae C., Berners-Price S.J., Bulliman B.T., Kuchel P.W. Kinetic analysis of the human erythrocyte glyoxalase system using 1H NMR and a computer model. Eur. J. Biochem. 1990;193:83–90. doi: 10.1111/j.1432-1033.1990.tb19307.x. [DOI] [PubMed] [Google Scholar]
- 30.Abordo E.A., Minhas H.S., Thornalley P.J. Accumulation of alpha-oxoaldehydes during oxidative stress: a role in cytotoxicity. Biochem. Pharmacol. 1999;58:641–648. doi: 10.1016/s0006-2952(99)00132-x. [DOI] [PubMed] [Google Scholar]
- 31.Gomes R.A., Sousa Silva M., Vicente Miranda H., Ferreira A.E., Cordeiro C.A., Freire A.P. Protein glycation in Saccharomyces cerevisiae. Argpyrimidine formation and methylglyoxal catabolism. FEBS J. 2005;272:4521–4531. doi: 10.1111/j.1742-4658.2005.04872.x. [DOI] [PubMed] [Google Scholar]
- 32.Gomes R.A., Vicente Miranda H., Silva M.S., Graca G., Coelho A.V., Ferreira A.E., Cordeiro C., Freire A.P. Yeast protein glycation in vivo by methylglyoxal. Molecular modification of glycolytic enzymes and heat shock proteins. FEBS J. 2006;273:5273–5287. doi: 10.1111/j.1742-4658.2006.05520.x. [DOI] [PubMed] [Google Scholar]
- 33.Maeta K., Izawa S., Inoue Y. Methylglyoxal, a metabolite derived from glycolysis, functions as a signal initiator of the high osmolarity glycerol-mitogen-activated protein kinase cascade and calcineurin/Crz1-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:253–260. doi: 10.1074/jbc.M408061200. [DOI] [PubMed] [Google Scholar]
- 34.Inoue Y., Tsujimoto Y., Kimura A. Expression of the glyoxalase I gene of Saccharomyces cerevisiae is regulated by high osmolarity glycerol mitogen-activated protein kinase pathway in osmotic stress response. J. Biol. Chem. 1998;273:2977–2983. doi: 10.1074/jbc.273.5.2977. [DOI] [PubMed] [Google Scholar]
- 35.Aguilera J., Rodriguez-Vargas S., Prieto J.A. The HOG MAP kinase pathway is required for the induction of methylglyoxal-responsive genes and determines methylglyoxal resistance in Saccharomyces cerevisiae. Mol. Microbiol. 2005;56:228–239. doi: 10.1111/j.1365-2958.2005.04533.x. [DOI] [PubMed] [Google Scholar]
- 36.Maeta K., Izawa S., Okazaki S., Kuge S., Inoue Y. Activity of the Yap1 transcription factor in Saccharomyces cerevisiae is modulated by methylglyoxal, a metabolite derived from glycolysis. Mol. Cell. Biol. 2004;24:8753–8764. doi: 10.1128/MCB.24.19.8753-8764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoon S., Gebbia M., Costanzo M., Davis R.W., Giaever G., Nislow C. A global perspective of the genetic basis for carbonyl stress resistance. G3(Bethesda). 2011;1:219–231. doi: 10.1534/g3.111.000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 39.Wishart J.A., Hayes A., Wardleworth L., Zhang N., Oliver S.G. Doxycycline, the drug used to control the tet-regulatable promoter system, has no effect on global gene expression in Saccharomyces cerevisiae. Yeast. 2005;22:565–569. doi: 10.1002/yea.1225. [DOI] [PubMed] [Google Scholar]
- 40.Cooper A.A., Gitler A.D., Cashikar A., Haynes C.M., Hill K.J., Bhullar B., Liu K., Xu K., Strathearn K.E., Liu F., Cao S., Caldwell K.A., Caldwell G.A., Marsischky G., Kolodner R.D., Labaer J., Rochet J.C., Bonini N.M., Lindquist S. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 42.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Boersema P.J., Raijmakers R., Lemeer S., Mohammed S., Heck A.J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 47.McLellan A.C., Phillips S.A., Thornalley P.J. The assay of methylglyoxal in biological systems by derivatization with 1,2-diamino-4,5-dimethoxybenzene. Anal. Biochem. 1992;206:17–23. doi: 10.1016/s0003-2697(05)80005-3. [DOI] [PubMed] [Google Scholar]
- 48.Thornalley P.J., Battah S., Ahmed N., Karachalias N., Agalou S., Babaei-Jadidi R., Dawnay A. Quantitative screening of advanced glycation endproducts in cellular and extracellular proteins by tandem mass spectrometry. Biochem. J. 2003;375:581–592. doi: 10.1042/BJ20030763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aguilera J., Prieto J.A. Yeast cells display a regulatory mechanism in response to methylglyoxal. FEMS Yeast Res. 2004;4:633–641. doi: 10.1016/j.femsyr.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 50.Thornalley P.J. Use of aminoguanidine (Pimagedine) to prevent the formation of advanced glycation endproducts. Arch. Biochem. Biophys. 2003;419:31–40. doi: 10.1016/j.abb.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 51.Castrejon F., Gomez A., Sanz M., Duran A., Roncero C. The RIM101 pathway contributes to yeast cell wall assembly and its function becomes essential in the absence of mitogen-activated protein kinase Slt2p. Eukaryot. Cell. 2006;5:507–517. doi: 10.1128/EC.5.3.507-517.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakurai H., Ota A. Regulation of chaperone gene expression by heat shock transcription factor in Saccharomyces cerevisiae: importance in normal cell growth, stress resistance, and longevity. FEBS Lett. 2011;585:2744–2748. doi: 10.1016/j.febslet.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 53.Erjavec N., Larsson L., Grantham J., Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21:2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller S.B., Mogk A., Bukau B. Spatially organized aggregation of misfolded proteins as cellular stress defense strategy. J. Mol. Biol. 2015;427:1564–1574. doi: 10.1016/j.jmb.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Miller S.B., Ho C.T., Winkler J., Khokhrina M., Neuner A., Mohamed M.Y., Guilbride D.L., Richter K., Lisby M., Schiebel E., Mogk A., Bukau B. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 2015;34:778–797. doi: 10.15252/embj.201489524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alberti S. Molecular mechanisms of spatial protein quality control. Prion. 2012;6:437–442. doi: 10.4161/pri.22470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jensen T.M., Vistisen D., Fleming T., Nawroth P.P., Rossing P., Jorgensen M.E., Lauritzen T., Sandbaek A., Witte D.R. Methylglyoxal is associated with changes in kidney function among individuals with screen-detected Type 2 diabetes mellitus. Diabet. Med. 2016;33:1625–1631. doi: 10.1111/dme.13201. [DOI] [PubMed] [Google Scholar]
- 58.Rabbani N., Xue M., Thornalley P.J. Dicarbonyls and glyoxalase in disease mechanisms and clinical therapeutics. Glycoconj. J. 2016;33:513–525. doi: 10.1007/s10719-016-9705-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ito K., Sakata N., Nagai R., Shirakawa J.I., Watanabe M., Mimata A., Abe Y., Yasuno T., Sasatomi Y., Miyake K., Ueki N., Hamauchi A., Nakashima H. High serum level of methylglyoxal-derived AGE, Ndelta-(5-hydro-5-methyl-4-imidazolone-2-yl)-ornithine, independently relates to renal dysfunction. Clin. Exp. Nephrol. 2016 doi: 10.1007/s10157-016-1301-9. [DOI] [PubMed] [Google Scholar]
- 60.Delaunay A., Pflieger D., Barrault M.B., Vinh J., Toledano M.B. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 61.Mortimer R.K., Schild D., Contopoulou C.R., Kans J.A. Genetic map of Saccharomyces cerevisiae, edition 10. Yeast. 1989;5:321–403. doi: 10.1002/yea.320050503. [DOI] [PubMed] [Google Scholar]
- 62.Fernandes L., Rodrigues-Pousada C., Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Byrne K.P., Wolfe K.H. The yeast gene order browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cohen B.A., Pilpel Y., Mitra R.D., Church G.M. Discrimination between paralogs using microarray analysis: application to the Yap1p and Yap2p transcriptional networks. Mol. Biol. Cell. 2002;13:1608–1614. doi: 10.1091/mbc.01-10-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaudhuri J., Bose N., Gong J., Hall D., Rifkind A., Bhaumik D., Peiris T.H., Chamoli M., Le C.H., Liu J., Lithgow G.J., Ramanathan A., Xu X.Z., Kapahi P. A Caenorhabditis elegans model elucidates a conserved role for TRPA1-Nrf signaling in reactive alpha-dicarbonyl detoxification. Curr. Biol. 2016;26:3014–3025. doi: 10.1016/j.cub.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eberhardt M.J., Filipovic M.R., Leffler A., de la Roche J., Kistner K., Fischer M.J., Fleming T., Zimmermann K., Ivanovic-Burmazovic I., Nawroth P.P., Bierhaus A., Reeh P.W., Sauer S.K. Methylglyoxal activates nociceptors through transient receptor potential channel A1 (TRPA1): a possible mechanism of metabolic neuropathies. J. Biol. Chem. 2012;287:28291–28306. doi: 10.1074/jbc.M111.328674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaganovich D., Kopito R., Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Table_S1_Diff_expressed_MG_ctr_DESeq2
Supplementary material Table_S2_KEGG_pathway_categories_MG_UP
Supplementary material Table_S3_KEGG_pathway_categories_MG_DOWN