Abstract
AMP-activated protein kinase (AMPK) plays a major role in regulating metabolism and has attracted significant attention as a therapeutic target for treating metabolic disorders. AMPK activity is stimulated more than 100-fold by phosphorylation of threonine 172 (Thr172). Binding of AMP to the γ subunit allosterically activates the kinase. Additionally, many small molecules, e.g. 991, have been identified that bind between the kinase domain and the carbohydrate-binding module of the β subunit, stabilising their interaction and leading to activation. It was reported recently that non-phosphorylated Thr172 AMPK is activated by AMP and A769662. We present here the crystal structure of non-phosphorylated Thr172 AMPK in complex with AMP and 991. This structure reveals that the activation loop, as well as the complex overall, is similar to the Thr172 phosphorylated complex. We find that in the presence of AMP and 991 non-phosphorylated Thr172, AMPK is much less active than the Thr172 phosphorylated enzyme. In human cells, the basal level of Thr172 phosphorylation is very low (∼1%), but is increased 10-fold by treatment with 2-deoxyglucose. In cells lacking the major Thr172 kinases, LKB1 and CaMKKβ, Thr172 phosphorylation is almost completely abolished, and AMPK activity is virtually undetectable. Our data show that AMP and 991 binding to non-phosphorylated Thr172 AMPK can induce an ordered, active-like, conformation of the activation loop explaining how AMPK activity can be measured in vitro without Thr172 phosphorylation. However, in a cellular context, phosphorylation of Thr172 is critical for significant activation of AMPK.
Keywords: AMPK, phosphorylation/dephosphorylation, protein–serine–threonine kinases
Introduction
AMP-activated protein kinase (AMPK) maintains the energy status of the cell by promoting ATP-producing pathways and inhibiting ATP-utilising pathways [1–3]. By virtue of the metabolic pathways it controls, including glucose and lipid homeostasis, AMPK has emerged as an important therapeutic target for treating metabolic disorders [4,5]. AMPK is an αβγ heterotrimer that requires phosphorylation of threonine 172 (Thr172) in the activation loop of the kinase domain for maximal activity [6–8]. The negatively charged phosphate group of phospho-Thr172 (pThr172) interacts with residues in the kinase domain, including those from the regulatory αC helix, and a conserved arginine and aspartic acid residue [9,10]. These interactions lead to stabilisation of the activation loop which in turn optimally positions key residues involved in substrate binding and catalysis [11]. Our previous crystal structures of active AMPK show how phosphorylation of Thr172 facilitates the formation of many of these key interactions within the kinase domain that enable effective catalysis [9].
Liver kinase B1 (LKB1) and calcium/calmodulin-dependent protein kinase kinase β (CaMKKβ, also known as CaMKK2) have been identified as the two major upstream kinases capable of phosphorylating Thr172 in mammalian cells [12–17]. AMPK phosphorylated on Thr172 (hereafter, we refer to phosphorylated Thr172 as pThr172 and non-phosphorylated Thr172 as non-pThr172) can be dephosphorylated by protein phosphatases, although it is not known which protein phosphatases carry out this function in vivo. In mammalian cells, AMPK is activated by an increase in the AMP : ATP and ADP : ATP ratio, that occurs in response to a fall in ATP levels [1,18]. AMP and ADP binding to the γ subunit of AMPK promote phosphorylation of Thr172, and AMP and ADP protect pThr172 from dephosphorylation. As well as regulating the phosphorylation status of Thr172, AMP, but not ADP, allosterically activates AMPK. In addition to adenine nucleotides, many small-molecule activators of AMPK have been developed, including A769662 and 991, which both allosterically activate and protect the enzyme from dephosphorylation [9,19–21]. These small-molecule activators bind at an interface formed between the α kinase domain and the carbohydrate-binding module (CBM) of the β subunit [9]. We hypothesised that binding to this pocket promotes the interaction of the kinase domain with the regulatory fragment thus protecting the active enzyme from dephosphorylation and inactivation by protein phosphatases.
A recent study reported that binding of AMP and A769662 could synergistically activate AMPK in the absence of phosphorylation [22]. This finding raises the possibility that AMPK activation can bypass the requirement for Thr172 phosphorylation if AMP is bound at the γ subunit and the kinase domain–CBM interaction is stabilised by binding of a small-molecule activator. As a consequence, this would have significant implications for the development of therapeutic strategies for activating AMPK in the absence of upstream kinase activity. In the present paper, we show a crystal structure of non-pThr172 AMPK in complex with 991. This structure reveals that binding of 991 leads to the ordering of the activation loop of AMPK in the absence of Thr172 phosphorylation. Although recombinant non-pThr172 AMPK expressed in bacteria has catalytic activity in the presence of AMP and A769662, it is much less active than the corresponding pThr172 form. Importantly, in mammalian cells lacking both LKB1 and CaMKKβ, Thr172 phosphorylation is almost completely absent, and phosphorylation of acetyl-CoA carboxylase (ACC), a downstream substrate of AMPK, is virtually undetectable. Our findings suggest that in mammalian cells, Thr172 phosphorylation is essential for significant AMPK activity despite increased AMP levels and the presence of small-molecule activators.
Experimental
Materials and proteins
Recombinant His-tagged AMPK complexes were expressed in Escherichia coli and purified by chromatography on nickel-Sepharose [9]. AMPK was phosphorylated on Thr172 by overnight incubation with MgATP and CaMKKβ, and repurified as previously described [9]. Subsequent incubation with MgATP and CaMKKβ did not increase Thr172 phosphorylation, demonstrating that Thr172 is maximally phosphorylated using these conditions. The following antibodies were from Cell Signaling: rabbit anti-AMPKα1 (#2795), rabbit anti-AMPKα2 (#2757), mouse anti-AMPKα1/2 (#2793), rabbit anti-AMPKβ1/2 (#4150), rabbit anti-ACC (#3676), rabbit anti-pACC (#3661), rabbit anti-AMPKγ1 (#4187), rabbit anti-LKB1 (#3050) and rabbit anti-pThr172 (#2535). Mouse anti-vinculin was from Sigma–Aldrich (V9131). Mouse monoclonal anti-CaMKKβ antibody was a generous gift from Prof. Grahame Hardie (Dundee University).
AMPK assay
AMPK was assayed using the SAMS peptide assay as described previously [23]. Briefly, purified AMPK was assayed in the presence or absence of AMP (10 µM), 991 (1 µM) or both, as indicated in the figure legends.
Western blot analysis
Proteins were resolved by SDS–PAGE on 10% polyacrylamide gels (National Diagnostics) and transferred to Immobilon-FL (Millipore) membrane at 4°C. Membranes were probed with primary antibodies at 1 : 1000 dilution and incubated overnight at 4°C. After extensive washing, membranes were incubated for 30–60 min with LI-COR secondary antibodies at 1 : 10 000 dilution. Blots were imaged on a LI-COR Odyssey CLX. Fluorescence intensity values for individual bands were obtained using Image Studio (LI-COR) to allow quantification of the blots. For estimation of pThr172 levels in cells, the signal obtained for each sample was within the range obtained for the lowest and highest values determined using the recombinant AMPK standards. For capillary western blotting, cell lysates were diluted in HEPES lysis buffer to 0.4 mg/ml. Samples were prepared and analysed according to the manufacturer's instructions (ProteinSimple).
AMPK-binding assays
The binding of 991 to AMPK was determined by monitoring the near-UV CD spectra (340–255 nm) as previously reported [9].
Crystallography
Full-length AMPK, His-α2 (human 1–552), β1 (human 1–270) and γ1 (human, 1–331) were expressed in E. coli and purified using nickel affinity chromatrography and gel filtration as previously described [9]. A stock solution was prepared at 5 mg/ml in 50 mM Tris (pH 8.0), 300 mM NaCl and 1 mM tris(2-carboxyethyl)phosphine, mixed with a 4-fold molar excess of AMP and 1-fold of staurosporine and 991 compound. Crystals were grown by the vapour diffusion technique at 4°C in hanging drops. Drops were prepared by mixing equal volumes of protein complex with reservoir solution containing 12% polyethylene glycol (PEG3350), 300 mM guanidine in 100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) buffer (pH 7.2). Crystals were first transferred into mother liquid with an additional 25% ethylene glycol, before plunging into liquid nitrogen. Diffraction data were collected on a Pilatus 2 M detector (Dectris), Diamond Lightsource, Oxford. Data were integrated using Denzo and scaled with Scalepack. The structure was solved by molecular replacement using Phaser and standard refinement was carried out with Phenix using 4CFE.pdb as search models, with a manual model building with COOT [24]. General crystallographic calculations were carried out using the CCP4 package [25]. Figures were created with Pymol (http://pymol.sourceforge.net/).
Cell culture
HEK293T and A549 cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM, Thermo) supplemented with 10% foetal bovine serum (Sigma–Aldrich). Cells were transferred to serum-free DMEM for 2 h prior to treatment. Cells were incubated with either 0.1% dimethyl sulphoxide (DMSO, as a vehicle control), 991 (5 µM), rotenone (10 µM), phenformin, (2 mM), 2-deoxyglucose (2DG, 12 mM) or 991 (5 µM) plus 2DG (12 mM) for 60 min. Following removal of the cell media, cells were washed rapidly three times with ice-cold phosphate-buffered saline before the addition of lysis buffer [50 mM HEPES (pH 7.4), 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 1 mM ethylenediaminetetraacetic acid, 10% (v/v) glycerol, 1% (v/v) Triton X-100, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 4 µg/ml trypsin inhibitor and 0.1 mM benzamidine].
CRISPR-mediated deletion of CaMKKβ
A549 cells were transfected with plasmids containing Cas9 linked to green fluorescent protein (GFP) via a self-cleaving peptide and guide sequences targeting the first exon of CaMKKβ (GCTAGAGACACATGATGACA, GCAGGGCCTCACAGGGCTTC, GGTGGATGCTCAAGGATGAG, GGGCATGGAGTCCTTCATTG, AGCACAGCCCGGCTCACACT; Horizon Discovery, Cambridge, U.K.). At 24 h post-transfection, cells were sorted based on GFP expression and individual colonies were analysed by western blotting to determine CaMKKβ protein expression.
Nucleotide analysis
Cells were lysed in perchloric acid (5%), insoluble material was removed by centrifugation and the supernatant was extracted twice with an equal volume of 1 : 1 tri-n-octylamine and 1,1,2-trichlorotrifluoroethane. Adenine nucleotides in the aqueous phase were analysed by capillary electrophoresis using a P/ACE MDQ plus (Sciex) in 100 mM sodium acetate buffer (pH 3.5) containing 0.01% hydroxypropylmethylcellulose on a standard fused-silica capillary. The extract was loaded by pressure injection for 5 s at 0.5 psi and separated at 25 kV. Nucleotide peaks were detected by absorbance at 254 nm and integrated peak areas were automatically calculated. Retention times for AMP, ADP and ATP were confirmed by analysing nucleotide standards.
Results
Activation of non-phosphorylated Thr172 AMPK in vitro
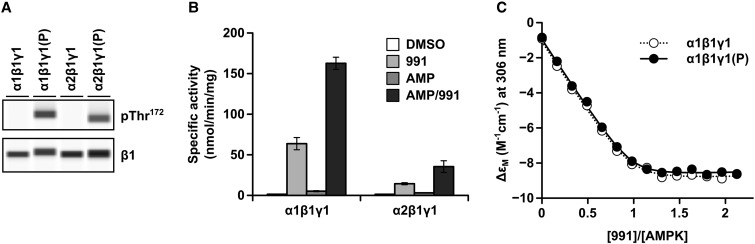
Recombinant AMPK complexes purified from bacteria are intrinsicially non-phosphorylated at Thr172 and can be phosphorylated by incubation with CaMKKβ in vitro (Figure 1A). We determined the activity of both α1 and α2 AMPK complexes and, as previously reported by us and others [6–8,26], found that in the absence of allosteric activators, the activity of the non-pThr172 complexes was less than 1% of the corresponding pThr172 complexes (1.41 ± 0.11 vs. 882.48 ± 21.43 nmol/min/mg for α1β1γ1 non-pThr172 vs. pThr172 and 1.37 ± 0.04 vs. 189.55 ± 15.89 nmol/min/mg for α2β1γ1 non-pThr172 vs. pThr172). The activity of the non-pThr172 complexes was increased markedly in the presence of the allosteric activators 991 and AMP (see Table 1 and Figure 1B), confirming allosteric activation of both α1β1γ1 and α2β1γ1, with the α1β1γ1 complex showing the greater response [22,27]. In our current study, we find that the activity of the non-pThr172 α1β1γ1 complex is ∼10% of the pThr172 complex, when assayed in the presence of both 991 and AMP (162.63 ± 7.52 vs. 1780.07 ± 6.23 nmol/min/mg), a slightly lower value than that reported in the previous studies [22,27]. With the α2β1γ1 complex, the activity of the non-pThr172 enzyme is ∼3% of the pThr172 form (35.44 ± 7.21 vs. 1266.77 ± 37.05). Given that 991 stimulates the activity of non-pThr172 AMPK, we examined its binding to non-pThr172 complexes. As can be seen from the results shown in Figure 1C and Table 2, the affinity of the complexes for 991 is not significantly affected by the phosphorylation status of Thr172 (see below).
Figure 1. Allosteric activation of non-phosphorylated Thr172 AMPK.
(A) AMPK α1β1γ1 and α2β1γ1 complexes were expressed in E. coli, purified and analysed by Western blotting using either an anti-pThr172 antibody or an anti-β antibody. Recombinant complexes that had been phosphorylated with CaMKKβ [α1β1 γ1(P) and α2β1γ1(P)] are shown alongside the non-phosphorylated proteins. (B) AMPK activity of the non-phosphorylated complexes was measured using the SAMS peptide assay in the absence or presence of 991 (1 µM), AMP (10 µM) or both 991 and AMP. Results shown are plotted as nmol/min/mg and are the means ± SEM of three independent experiments. (C) Comparison of circular dichroism signal change measured at 306 nm for non-pThr172 (open symbols) and pThr172 (closed symbols) α1β1γ1 complexes as a function of 991/AMPK.
Table 1. Activity of non-pThr172 AMPK complexes.
AMPK complexes were expressed in E. coli and activity of the purified complexes was measured using the SAMS peptide assay in the absence or presence of the allosteric activators, 991 (1 µM), AMP (10 µM) or both activators together. Fold-activation relative to activity measured in the absence of activator is also shown. In all cases, results shown are the mean (±SEM) determined from at least three independent experiments. In all cases, allosteric activation causes a statistically significant increase in AMPK activity compared with the control value (P < 0.05).
| AMPK complex/activators | Specific activity (nmol/min/mg) | Activation (fold) |
|---|---|---|
| α1β1γ1 | 1.41 (0.11) | 1 |
| α1β1γ1 + 991 | 63.71 (7.52) | 45.7 (6.0) |
| α1β1γ1 + AMP | 5.21 (0.63) | 3.7 (0.4) |
| α1β1γ1 + 991 + AMP | 162.63 (7.52) | 116.6 (17.5) |
| α2β1γ1 | 1.37 (0.04) | 1 |
| α2β1γ1 + 991 | 14.37 (1.28) | 10.5 (1.4) |
| α2β1γ1 + AMP | 3.14 (0.25) | 2.4 (0.12) |
| α2β1γ1 + 991 + AMP | 35.44 (7.21) | 25.6 (4.8) |
Table 2. Equilibrium dissociation constants for 991 binding to AMPK complexes.
AMPK complexes were expressed in E. coli and dissociation constants (Kd) for 991 were determined using circular dichroism (CD). The Kd values are reported as the mean (±SEM) determined from at least three independent experiments. The values for the phosphorylated Thr172 complexes (P) are taken from a previous study (Xiao et al. [9]).
| AMPK complex | CD (306 nm) Kd (µM) |
|---|---|
| α1β1γ1 | 0.13 ± 0.09 |
| α1β1γ1(P) | 0.08 ± 0.02 |
| α2β1γ1 | 0.17 ± 0.03 |
| α2β1γ1(P) | 0.09 ± 0.02 |
Structure of non-phosphorylated Thr172 AMPK
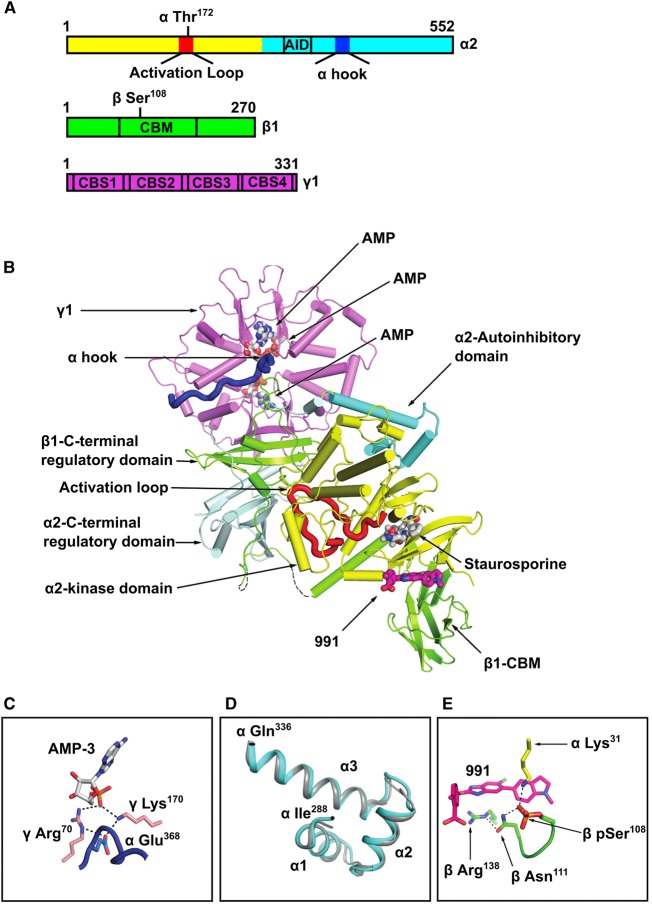
Since 991 both binds to non-pThr172 AMPK and stimulates its activity, we were interested in determining the conformation adopted by the kinase domain in the non-pThr172–991 complex. We obtained the crystal structure of full-length human non-pThr172 α2β1γ1 (Figure 2A), in complex with 991, AMP and staurosporine at a resolution of 2.6 Å (crystallographic statistics are presented in Table 3; co-ordinates deposited in the Protein Databank, PDB ID: 5ISO). The overall arrangement of the heterotrimer is analogous to the structure obtained for the equivalent pThr172 AMPK complex (Figure 2B) [9]. Significantly, key features are preserved, such as the position of the α-hook region (also referred to as the α-RIM2 [28]) interacting with the γ subunit at site 3 with AMP bound and the interaction of the C-lobe of the kinase, via its activation loop, with the regulatory fragment of the enzyme (Figure 2C). The different crystal packing and higher resolution diffraction, relative to our earlier complexes, allows us to assign additional regions of the structure. Of particular note, the structure of the autoinhibitory domain (AID) region is now well resolved (in one of the two molecules in the asymmetric unit) and clearly adopts the three α-helix structure observed for the isolated AID domain (Figure 2D) [29,30].
Figure 2. Structure of non-phosphorylated Thr172 AMPK bound to 991, AMP and stauropsporine.
(A) Bar diagram indicating the three subunits that make up the complex, showing the location of specific regions within the subunits (AID: autoinhibtory domain; CBM: carbohydrate-binding module; CBS: cystathionine-β-synthase domain), as well as the location of Thr172 in the α subunit and Ser108 in the β subunit. (B) Cartoon representation of full-length human α2β1γ1 in complex with 991. The domains of the three subunits are coloured according to (A). The orientation of the figure is similar to our earlier papers, so that the kinase domain is ‘upside-down’ with respect to the classical kinase orientation. 991, which binds at the interface of the kinase domain and CBM, is shown in stick representation, with its carbon atoms coloured magenta. The three AMP molecules bound in the γ1 subunit, and staurosporine bound to the kinase domain, are also shown. The α-hook (also known as αRIM2) region of α2 interacting at site 3 in the γ1 subunit is shown in blue. Thr172 is shown in space-filling representation. (C) A detailed view of the α-hook interaction at site 3 with AMP bound. (D) Comparison of the AID region from the current structure (cyan) with the isolated AID of human α1 (PDB ID:4RED; grey). (E) 991-binding site at the interface of the CBM of β1 and the kinase domain, showing electrostatic interactions with pSer108. The co-ordinates of the non-pThr172 AMPK structure have been deposited in the Protein Databank (PDB ID: 5ISO).
Table 3. Data collection and refinement statistics (molecular replacement).
| Non-pThr172 complex with 991/AMP bound (PDB ID: 5ISO) | |
|---|---|
| Data collection | |
| Space group | P1212 |
| Cell dimensions | |
| a, b, c (Å) | 75.42, 129.30, 139.28 |
| α, β, γ (°) | 90, 92.73, 90 |
| Resolution (Å) | 19.97–2.63 (2.72–2.63)1 |
| Rsym or Rmerge | 0.03 (0.40) |
| Ι/σI | 14.69 (2.20) |
| Completeness (%) | 99.12 (99.85) |
| Redundancy | 3.4 (1.9) |
| Refinement | |
| Resolution (Å) | 19.97–2.63 (2.72–2.63) |
| No. of reflections | 78 451 (7875) |
| Rwork/Rfree | 0.256/0.189 |
| No. of atoms | 15 048 |
| Protein | 14 835 |
| Ligand/ion | 132 |
| Water | 81 |
| B-factors | 82.50 |
| Protein | 82.70 |
| Ligand/ion | 62.80 |
| Water | 73.70 |
| r.m.s deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.22 |
1 Highest resolution shell is shown in parenthesis.
Previous studies have shown that Ser108 within the CBM of the β1 subunit is an autophosphorylation site [31], and that mutation of Ser108 to alanine significantly weakens binding of A769662 and 991 to AMPK [9]. We previously reported that pSer108 in β1 is involved in a network of electrostatic interactions that play a role in stabilising the interaction between the kinase domain and the CBM [9]. Interestingly, Ser108 is phosphorylated in the crystal structure of the non-pThr172 complex (Figure 2E), confirming that AMPK expressed in E. coli, although not phosphorylated on Thr172, is capable of undergoing autophosphorylation on Ser108, consistent with the findings of a previous study [22]. Thus, another important feature conserved in the non-pThr172 AMPK structure is the interaction of the CBM with the N-lobe of the kinase domain facilitated by the salt bridge linking pSer108 on the CBM to the kinase domain. Importantly, these interactions preserve the 991-binding site, and the compound makes identical interactions to those observed in the pThr172 structure. This results in a well-ordered CBM and a kinase domain N-lobe, and provides an explanation for why 991 binding is not affected by the phosphorylation state of Thr172 (Table 2).
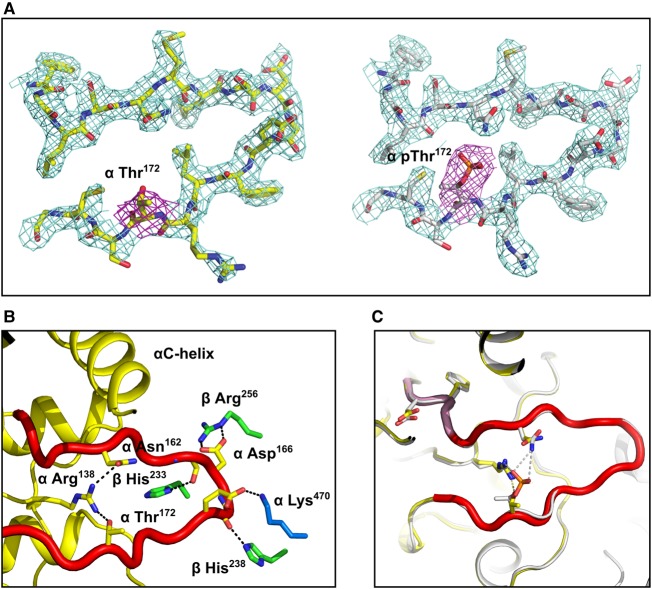
It is clear from the electron density maps that Thr172 is not phosphorylated in the current structure (Figure 3A), but intriguingly, the activation loop is ordered and adopts a similar conformation to that seen in previous pThr172 AMPK structures. Consequently, key residues in the regulatory spine and catalytic spine in the non-pThr172 complex make similar interactions to those observed in the equivalent pThr172 structure (Figure 3A,B). For example, the N-terminus of the activation loop in the present structure makes similar interactions with the regulatory fragment, with the side chain of βHis238 interacting with the main-chain carbonyl of residues αGly167 and βHis233 with the main-chain carbonyl of residue αSer165. Ordering of the activation loop to promote these interactions is important for efficient catalysis [32]. Specifically, Phe158 within the DFG motif adopts an active ‘DFG-in’ conformation (Figure 3C) [32]. Notably, in our previous structures [9], the phosphate of pThr172 made a salt bridge with αArg138 on a loop extending from the αE helix and a charged hydrogen bond with αAsn162 from the N-terminal end of the activation loop. These interactions are thought to stabilise the ‘active conformation’. In the present non-pThr172 structure, the hydroxyl group of Thr172 is able to hydrogen bond with the same αArg138 (in one of the copies in the assymetric unit); however, it is not able to make a further stabilising interaction with αAsn162 (Figure 3C).
Figure 3. Comparison of phosphorylated and non-phosphorylated Thr172 structures.
(A) Omit maps for activation loop residues from non-phosphorylated and phosphorylated Thr172 structures. (B) Overlay of non-pThr172 activation loop (shown in red) and pThr172 (grey). The ‘DFG-in’ conformation is similar in both structures (highlighted in pink for the non-pThr172 structure). (C) Detailed view showing the interaction between the activation loop and residues from the regulatory core of AMPK in the non-pThr172 complex.
The binding of AMP and 991 together with the interactions between the activation loop and the regulatory fragment allows, at least in the crystal, ordering of the activation loop, even in the absence of phosphorylation on Thr172. In solution, this is reflected in the increase in catalytic activity observed for equivalent favourable reaction conditions, i.e. the presence of the allosteric activators 991 and AMP, for the non-pThr172 complex assayed in vitro. These findings raise the intruiging issue of whether this activation, in the absence of Thr172 phosphorylation, could occur in mammalian cells.
Requirement for Thr172 phosphorylation in cells
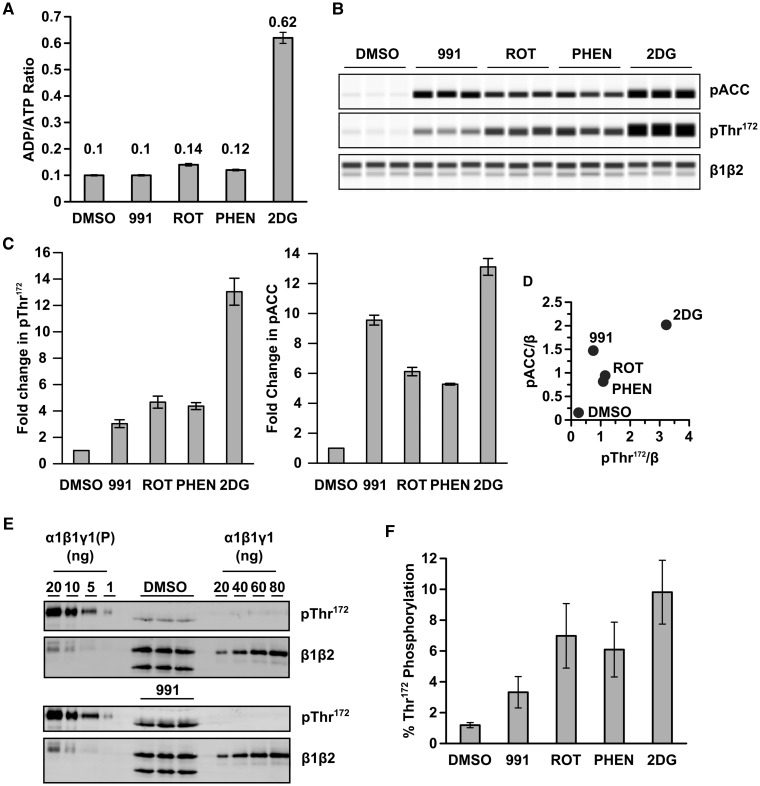
The finding that in the presence of allosteric activators, non-pThr172 AMPK complexes have significant activity relative to the pThr172 complexes raises the important issue of whether non-pThr172 AMPK is active in cells. We monitored phosphorylation of Thr172 on AMPK and of its substrate, ACC, in HEK293T cells following treatment with 991, or compounds that lead to changes in adenine nucleotide levels. For these latter treatments, we used rotenone and phenformin, both of which inhibit complex I of the electron transport chain and 2DG, an inhibitor of glycolysis. The effect of the various treatments on adenine nucleotide levels is shown in Figure 4A. Apart from cells treated with 2DG, the level of AMP was too low to allow accurate determination, and so, we calculated the ADP : ATP ratio and used this as a measure of energy stress [33]. Rotenone and phenformin cause a modest, but not statistically signifcant, increase in the ADP : ATP ratio, whereas incubation with 2DG causes a much larger, statistically significant, increase in this ratio. In addition, treatment with 2DG leads to a clear increase in AMP. As recently reported [34], 991 has no detectable effect on adenine nucleotide levels. All four treatments increase Thr172 and ACC phosphorylation (Figure 4B,C). With the exception of 991 treatment, the levels of Thr172 and ACC phosphorylation are closely correlated in their response to treatment (Figure 4D). A possible explanation for the increased phosphorylation of ACC relative to Thr172 in response to 991 is that activation of AMPK by 991 involves a significant allosteric component [9]. As a consequence, there is an uncoupling between Thr172 and ACC phosphorylation, as has been suggested previously for the effect of A769662 in cell-based assays [35,36]. To estimate the level of pThr172 in cells in response to different treatments, we blotted known amounts of recombinant AMPK with either an anti-pThr172 antibody or an anti-β antibody (raised against a peptide that is completely conserved in both β1 and β2) to generate a standard curve. We titrated different amounts of both non-pThr172 AMPK and AMPK that had been phosphorylated by CaMKKβ on the same blot as the HEK293T lysates (Figure 4E). Our initial experiments indicated that the anti-β antibody that we used was less sensitive than the anti-pThr172 antibody and so we titrated more of the non-phosphorylated complex compared with the phosphorylated form. We noted a slight difference in the migration of pThr172 in the recombinant AMPK α subunit relative to endogenous AMPK in HEK293T cells, which is likely due to the presence of a hexa-histidine tag engineered on the N-terminus of the recombinant protein. The blots were quantified, and the percentage of Thr172 phosphorylation relative to the total β subunit expression (β1 plus β2) in HEK293T cells was determined (Figure 4F). The results we obtained using this method correlate very well with the initial Western blot data shown in Figure 4B. In untreated cells (DMSO control), ∼1% of total AMPK is phosphorylated on Thr172. This level increases to just under 10% in the cells treated with 2DG, which gives the greatest increase in pThr172. These results are consistent with the hypothesis that AMPK exists in a largely inactive, non-phosphorylated Thr172 form under basal conditions, and that only a relatively small change in pThr172 is required to alter phosphorylation of downstream targets [37].
Figure 4. Quantification of Thr172 phosphorylation in HEK293T cells.
HEK293T cells were incubated with 991 (5 µM), rotenone (10 µM), phenformin (2 mM) or 2DG (12 mM), for 60 min. (A) ADP and ATP levels in perchloric acid extracts were determined by capillary electrophoresis, and the ratio of ADP : ATP is shown (mean ± SEM of three independent experiments). (B) A representative capillary western blot of HEK293T cell lysates probed with anti-pACC, anti-pThr172 and pan-β antibody is shown. For each condition, samples were analysed in triplicate. (C) Fold changes in pThr172 : total β-subunit expression and pACC : total β-subunit expression, relative to DMSO control, are shown (mean ± SEM for three independent experiments). (D) The ratio of pACC : total β expression is plotted against the ratio of pThr172 : total β expression. (E) HEK293T cell lysates (samples run in triplicate) were blotted with either anti-pThr172 antibody or pan-β antibody alongside varying amounts of purified, recombinant AMPK, either non-phosphorylated (α1β1γ1) or phosphorylated by CaMKKβ [α1β1γ1(P)] as indicated in the figure. Blots were quantified using the LI-COR-imaging system and the percentage of Thr172 phosphorylation relative to total AMPK plotted in (F). Results shown are the mean ± SEM of three independent experiments.
Lack of upstream kinases abolishes AMPK activity in cells
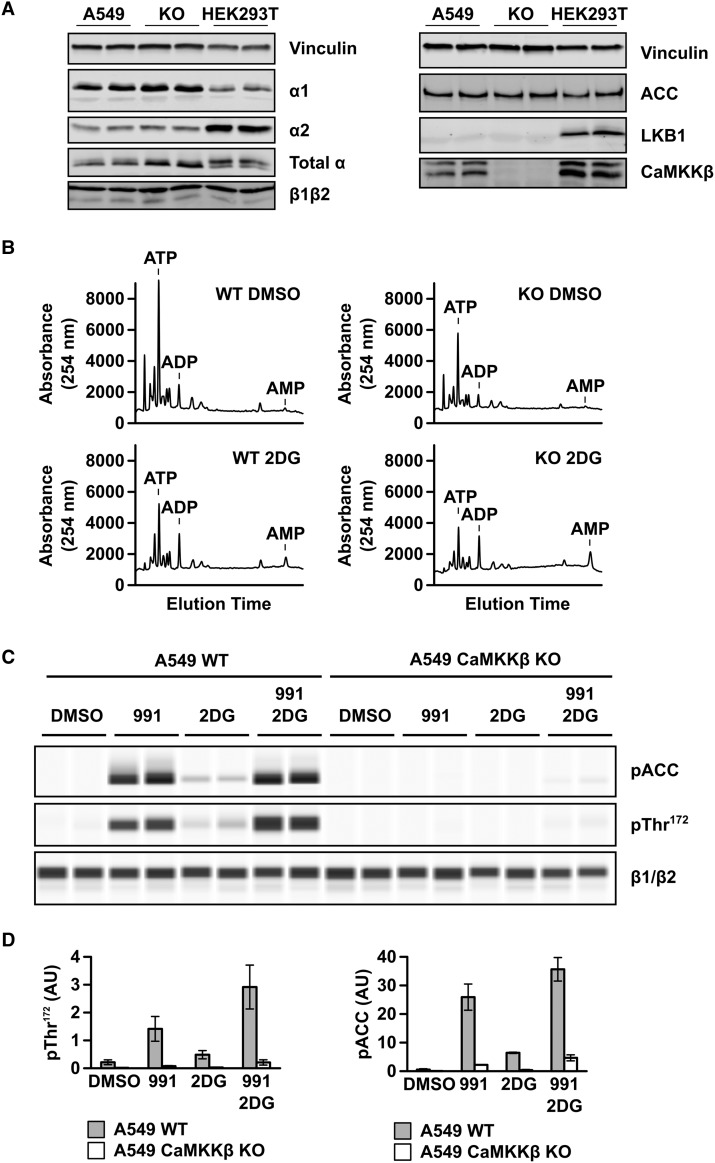
The findings above demonstrate that even low levels of pThr172 are sufficient for significant downstream effects of AMPK (ACC phosphorylation) in cells, but do not address the issue of whether pThr172 is absolutely required for AMPK signalling in cells. To determine this, we deleted CaMKKβ from A549 cells, which lack LKB1 expression (Figure 5A and ref. [38]) using CRISPR-mediated genome editing. As shown in Figure 5A, deletion of CaMKKβ had no detectable effect on the expression levels of ACC and AMPK α or β subunits. AMPKα1 appears to be the major α isoform expressed in A549 cells, whereas in HEK293T cells, the relative expression of α2 is higher (Figure 5A). In both A549 and HEK293T cells, β1 expression is higher than β2 (Figure 5A). We measured adenine nucleotide levels in the CaMKKβ knockout (KO) A549 cells and the parental cells following treatment with 2DG. Incubation with 2DG causes a dramatic increase in the ADP : ATP ratio in both the parental and CaMKKβ KO A549 cells, as well as a marked increase in the peak corresponding to AMP (Figure 5B). In the parental cells, which express CaMKKβ, 2DG treatment results in a modest increase in phosphorylation of both ACC and Thr172, whereas 991 causes more robust increases, which are increased further following dual treatment (Figure 5C,D). In contrast, Thr172 and ACC phosphorylation are almost undetectable in the CaMKKβ KO A549 cells. However, there is a very low level of Thr172 and ACC phosphorylation following dual treatment with 991 and 2DG (Figure 5C,D). These findings demonstrate that Thr172 phosphorylation is by far the dominant factor required for detectable AMPK function, as measured by phosphorylation of ACC, even in the presence of a potent small-molecule activator and increased cellular AMP levels.
Figure 5. Effect of deletion of CaMKKβ on Thr172 and ACC phosphorylation in A549 cells.
The CRISPR–Cas 9 system was used to delete CaMKKβ in A549 cells (which lack endogenous LKB1 expression). (A) Western blot analysis of the parental A549 cells and the CaMKKβ KO cell line with antibodies against ACC, AMPKα1, α2, pan-α, pan-AMPKβ, CaMKKβ and LKB1. For comparison, HEK293T cell lysate is included. Vinculin expression is shown as a total protein loading control. In all cases, duplicate samples from independent cell preparations are analysed. (B) Nucleotide content of prechloric acid extracts of parental or CaMKKβ KO A549 cells, incubated with or without 12 mM 2DG for 60 min, was determined by capillary electrophoresis. In each case, a representative trace showing UV absorbance at 254 nm is shown and the migration of adenine nucleotide standards is indicated on the traces. (C) Parental and CaMKKβ KO A549 cells were treated with 991 (5 µM), 2DG (12 mM) or both 991 and 2DG, for 60 min. Cell lysates were resolved by capillary electrophoresis and the levels of ACC and Thr172 phosphorylation, together with AMPK β-subunit expression, were determined. In each case, a representative blot with two independent samples is shown. (D) Quantification of the blots shown in (C) determined by chemiluminescence. Results are shown as mean ± SEM for three independent experiments and are plotted as arbitrary units (AU).
Discussion
It was reported recently that AMPK, particularly α1β1γ1 complexes, can be allosterically activated in the absence of Thr172 phosphorylation [22,27]. Here, we confirm that these findings show that recombinant non-pThr172 AMPK is allosterically activated by 991 and AMP in cell-free assays. We solved the structure of the non-pThr172 AMPK complex which revealed that the activation loop adopts a similar conformation to that observed in pThr172 AMPK. An important feature of the non-pThr172 enzyme expressed in E. coli is that Ser108 in β1 is phosphorylated, accounting for the finding that 991 binds with a similar affinity to both the non-pThr172 and pThr172 forms of AMPK. Thus, our structural data show how the binding energy of 991 and AMP are capable of inducing an active-like conformation in the non-phosphorylated activation loop, accounting for the low, but quantitative, activity of this species in vitro. Perhaps, more importantly, another finding of our current study is that in the absence of LKB1 and CaMKKβ, Thr172 and ACC phosphorylation are virtually undetectable in human cells, with or without allosteric activators. This finding suggests that Thr172 phosphorylation of AMPK is the critical factor for phosphorylation of its downstream targets in a cellular context.
Activation loop phosphorylation promotes the stabilisation of the active conformation of this loop and the positioning of key residues in the kinase domain. Owing to the less stable conformation of kinases lacking such phosphorylation, it is perhaps not surprising that, in general, there are relatively few examples of such structures available in the Protein Databank. However, there are examples of RD kinases that do not require phosphorylation of the activation loop to achieve partial or full activity [39]. In some cases, this is achieved by the presence of acidic residues in the activation loop that appear to mimic phosphorylation at these sites [39]. In the case of cyclin-dependent kinases (CDKs), bound cyclins interact with the non-phosphorylated activation loop, allowing it to become ordered, giving rise to partial or full activation of the CDK [40]. In our current study, we were able to obtain well-ordered crystals of AMPK in the absence of Thr172 phosphorylation by having 991 bound. The activation loop of AMPK makes many interactions with the regulatory fragment of the enzyme that enables the loop to become ordered in the absence of it being phosphorylated at Thr172. We suggest that binding of the activator (991) provides sufficient energy to stabilise the active-like conformation of the activation loop by driving the binding of the kinase domain, mainly via this loop, onto the regulatory fragment of AMPK.
Around the same time as we solved the crystal structure of the non-pThr172 complex, a study reported that allosteric activation of α1β1γ1 by A769662 led to significant activity without the requirement for Thr172 phosphorylation, and the addition of AMP increased this activation further [22]. We confirmed these findings with 991, although the degree of activation that we observe is not as great as that reported previously. In our hands, the combined activation of non-pThr172 α1β1γ1 by AMP and 991 reached ∼10% of the activity of pThr172 AMPK in a cell-free assay, demonstrating that full activation of AMPK does require phosphorylation of Thr172. Thus, the ordering of the activation loop that we see in the crystal structure with AMP and 991 bound supports the finding of significant, but low, kinase activity in the absence of Thr172 phosphorylation.
As has been suggested previously [22], the ability of allosteric AMPK activators to stimulate non-pThr172 complexes could be highly significant in terms of potential therapeutic strategies for activating AMPK, particularly those diseases where there is a lack of upstream AMPK kinase activity, such as loss of LKB1 in certain cancers [41]. In an attempt to address this important issue, we first estimated the level of Thr172 phosphorylation in response to either 991 or conditions which interfere with cellular ATP production, and which would therefore be anticipated to increase AMP levels. In HEK293T cells, which express both LKB1 and CaMKKβ, the two major Thr172 kinases, we were only able to detect a significant increase in AMP levels following treatment with the glycolysis inhibitor, 2DG. Neither rotenone nor phenformin treatment caused a detectable increase in AMP, although they did lead to measurable increases in ADP. Although we were unable to reliably detect AMP under these conditions, it remains possible that AMP is increased, but falls below the limit of detection using capillary electrophoresis. To resolve this issue, a more sensitive method, such as mass spectrometry, would be required to measure AMP levels. Consistent with the modest changes in adenine nucleotide levels, the increase in Thr172 phosphorylation occurred over a fairly narrow range (between 1 and 10%) following the different treatments. Thus, most of the AMPK within the cell remains in the non-phosphorylated Thr172 form. This is consistent with a model we proposed previously that we based on the nucleotide-binding properties of AMPK in relation to the physiological concentration of adenine nucleotides in mammalian cells [37]. In this model, the majority of AMPK will have Mg·ATP bound and will be in the non-phosphorylated Thr172 form. Small (2–3-fold) changes in AMP and/or ADP levels lead to similar fold increases in pThr172, consistent with our experimental observations in HEK293T cells. It is important to note, however, that although the proportion of AMPK in the pThr172 form is low, following treatment with 2DG the relative increase in Thr172 phosphorylation is substantial (10-fold). There was a good correlation between the increase in phosphorylation of Thr172 and ACC for rotenone, phenformin and 2DG. In contrast, 991 treatment, which caused the smallest increase in Thr172 phosphorylation out of the conditions used, resulted in a disproportionate increase in ACC phosphorylation. A potential explanation for this finding is that 991, in addition to increasing Thr172 phosphorylation, allosterically activates AMPK [9]. As has been noted previously, assessing AMPK activation in cells solely on Thr172 may be misleading, and monitoring downstream substrate phosphorylation provides a more accurate reflection of AMPK activity in cells [35].
A previous study performed a similar analysis for estimating the degree of pThr172 in HEK293 cells [36]. In this earlier study, a much higher level of basal Thr172 phosphorylation (∼27%) was reported. The reason for the apparent difference in the studies is unclear, but may be related to the use of different cell lines (HEK293 vs. HEK293T cells). In HEK293 cells, α1 has been reported to account for ∼95% of total AMPK activity [42], whereas in the HEK293T cells used in our current study, we find that α2 is the predominantly expressed α isoform (Figure 5A). Interestingly, a recent study reported that α2-containing AMPK complexes are significantly more sensitive to dephosphorylation than α1-containing complexes [43], This difference in phosphatase sensitivity could account, at least in part, for the difference in basal Thr172 phosphorylation between HEK293 and HEK293T cells.
The allosteric activation of the non-pThr172 AMPK complex reported in the current study and previously by others [22,27] has potentially significant implications for therapeutic targeting of AMPK. To determine whether AMPK is capable of signalling in the absence of Thr172 phosphorylation in human cells, we generated A549 cells lacking the two major Thr172 kinases, CaMKKβ and LKB1. A549 cells are a human lung adenocarcinoma cell line that lack endogenous expression of LKB1 [38,44]. By deleting CaMKKβ from these cells using the CRISPR–Cas9 system, we were able to establish a human cell line lacking the major AMPK Thr172 kinases. Similar to lung tissue [45], A549 cells express predominantly the α1 and β1 isoforms, making them an appropriate cell line for investigating this mechanism, since it is the non-pThr172 α1β1γ1 complex that is allosterically activated to the greatest extent. Interestingly, a recent study used a pharmacogenetic approach to investigate the role of CaMKK isoforms on downstream signalling in A549 cells, concluding that CaMKKβ is the major CaMKK isoform responsible for Thr172 phosphorylation in these cells [46].
In A549 cells, treatment with 2DG caused a modest increase in phosphorylation of ACC and Thr172 compared with 991 treatment (Figure 5). The modest effect of 2DG treatment is almost certainly due to the lack of LKB1 expression in these cells, since LKB1 has been shown to play the major role in nucleotide-mediated activation of AMPK [12,47]. Dual treatment with 991 and 2DG caused a greater increase in ACC and Thr172 phosphorylation than either treatment alone. In contrast with these results, phosphorylation of ACC and Thr172 was essentially undectable in the CaMKKβ KO A549 cells following any of the treatments. We confirmed that AMP and ADP levels were increased in CaMKKβ KO A549 cells following 2DG treatment. A very low signal for ACC phosphorylation was evident following the dual treatment of the CaMKKβ KO A549 cells, and this correlated with a similarly weak signal for Thr172 phosphorylation. These findings suggest that a low level of Thr172 kinase activity remains in the CaMKKβ KO cells, and that this results in a low level of downstream substrate phosphorylation. Whatever the source of this residual activity, our results clearly show that in the absence of appreciable Thr172 phosphorylation, cellular AMPK has negligible ability to phosphorylate its downstream substrate. Taken together, these findings demonstrate that in the absence of CaMKKβ and LKB1, Thr172 phosphorylation is virtually abolished and ACC phosphorylation prevented. At first sight, this appears to be at odds with the cell-free data, which shows that non-pThr172 AMPK is capable of substrate phosphorylation in vitro. However, the cell-free assays are performed at high substrate concentration, which will be different within the cell. In addition, phosphorylation of downstream targets within a cellular context are dynamic, due to the action of protein phosphatases, which is not the case in a cell-free assay. Furthermore, additional factors are present within the cell, that are not present in the cell-free studies, that influence the activity of the non-pThr172 complex. For example, glycogen has been shown to inhibit AMPK activation in vivo [48]. In addition, AMPK has been shown to interact with other proteins within the cell, and these interactions could affect on AMPK activity, as well as subcellular localisation. For example, recent studies by Lin and colleagues have revealed that under certain conditions, AMPK activation by LKB1 requires the interaction of LKB1 with axin on the surface of the lysosome [49–51]. Cellular AMPK activity is therefore controlled by a combination of factors that are not recapitulated in the cell-free assays. Whatever the reason for the differences, the requirement of developing strategies for activating AMPK that bypass the requirement for Thr172 phosphorylation is unsubstantiated. Moreover, we are not aware of any pathology where both CaMKKβ and LKB1 are inactive and so, the requirement for activation of the non-pThr172 form has not been proved.
Acknowledgements
We are grateful to Dr Philip Walker (Francis Crick Institute) for data collection and to Prof. Grahame Hardie (University of Dundee) for the mouse monoclonal anti-CaMKKβ antibody.
Abbreviations
- 2DG
2-deoxyglucose
- ACC
acetyl-CoA carboxylase
- AID
autoinhibitory domain
- AMPK
AMP-activated protein kinase
- CaMKK
calcium/calmodulin-dependent protein kinase kinase
- CaMKKβ
calcium/calmodulin-dependent protein kinase kinase β
- CBM
carbohydrate-binding module
- CDK
cyclin-dependent kinase
- CRISPR
clustered regularly interspaced short palindromic repeats
- DMEM
Dulbecco's Modified Eagle's Medium
- DMSO
dimethyl sulphoxide
- GFP
green fluorescent protein
- LKB1
liver kinase B1
- non-pThr172
non-phosphorylated Thr172
- pThr172
phospho-Thr172
- SAMS
the synthetic peptide HMRSAMSGLHLVKRR
- Thr172
threonine 172
Author Contribution
R.W., M.J.S., B.X., B.R.P. and S.R.M. carried out the experimental work. All authors helped conceive and design the experiments, and contributed to the writing of the manuscript.
Funding
This work was funded by the Medical Research Council UK, AstraZeneca and the Francis Crick Institute, which receive its core funding from Cancer Research UK [FC001078], the Medical Research Council [FC001078] and the Wellcome Trust [FC001078]. R.W. received a PhD studentship from the Medical Research Council.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Carling D., Mayer F.V., Sanders M.J. and Gamblin S.J. (2011) AMP-activated protein kinase: nature's energy sensor. Nat. Chem. Biol. 7, 512–518 doi: 10.1038/nchembio.610 [DOI] [PubMed] [Google Scholar]
- 2.Carling D., Thornton C., Woods A. and Sanders M.J. (2012) AMP-activated protein kinase: new regulation, new roles? Biochem. J. 445, 11–27 doi: 10.1042/BJ20120546 [DOI] [PubMed] [Google Scholar]
- 3.Steinberg G.R. and Kemp B.E. (2009) AMPK in health and disease. Physiol. Rev. 89, 1025–1078 doi: 10.1152/physrev.00011.2008 [DOI] [PubMed] [Google Scholar]
- 4.Hardie D.G. (2013) AMPK: a target for drugs and natural products with effects on both diabetes and cancer. Diabetes 62, 2164–2172 doi: 10.2337/db13-0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carling D. (2017) AMPK signalling in health and disease. Curr. Opin. Cell Biol. 45, 31–37 doi: 10.1016/j.ceb.2017.01.005 [DOI] [PubMed] [Google Scholar]
- 6.Crute B.E., Seefeld K., Gamble J., Kemp B.E. and Witters L.A. (1998) Functional domains of the α1 catalytic subunit of the AMP-activated protein kinase. J. Biol. Chem. 273, 35347–35354 doi: 10.1074/jbc.273.52.35347 [DOI] [PubMed] [Google Scholar]
- 7.Hawley S.A., Davison M., Woods A., Davies S.P., Beri R.K., Carling D. et al. (1996) Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem. 271, 27879–27887 doi: 10.1074/jbc.271.44.27879 [DOI] [PubMed] [Google Scholar]
- 8.Stein S.C., Woods A., Jones N.A., Davison M.D. and Carling D. (2000) The regulation of AMP-activated protein kinase by phosphorylation. Biochem. J. 345, 437–443 doi: 10.1042/bj3450437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao B., Sanders M.J., Carmena D., Bright N.J., Haire L.F., Underwood E. et al. (2013) Structural basis of AMPK regulation by small molecule activators. Nat. Commun. 4, 3017 doi: 10.1038/ncomms4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao B., Sanders M.J., Underwood E., Heath R., Mayer F.V., Carmena D.J. et al. (2011) Structure of mammalian AMPK and its regulation by ADP. Nature 472, 230–233 doi: 10.1038/nature09932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jura N., Zhang X., Endres N.F., Seeliger M.A., Schindler T. and Kuriyan J. (2011) Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol. Cell 42, 9–22 doi: 10.1016/j.molcel.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawley S.A., Boudeau J., Reid J.L., Mustard K.J., Udd L., Mäkelä T.P. et al. (2003) Complexes between the LKB1 tumor suppressor, STRAD α/β and MO25 α/β are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2, 28 doi: 10.1186/1475-4924-2-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawley S.A., Pan D.A., Mustard K.J., Ross L., Bain J., Edelman A.M. et al. (2005) Calmodulin-dependent protein kinase kinase-β is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2, 9–19 doi: 10.1016/j.cmet.2005.05.009 [DOI] [PubMed] [Google Scholar]
- 14.Hurley R.L., Anderson K.A., Franzone J.M., Kemp B.E., Means A.R. and Witters L.A. (2005) The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J. Biol. Chem. 280, 29060–29066 doi: 10.1074/jbc.M503824200 [DOI] [PubMed] [Google Scholar]
- 15.Shaw R.J., Kosmatka M., Bardeesy N., Hurley R.L., Witters L.A., DePinho R.A. et al. (2004) The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl Acad. Sci. U.S.A. 101, 3329–3335 doi: 10.1073/pnas.0308061100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woods A., Dickerson K., Heath R., Hong S.-P., Momcilovic M., Johnstone S.R. et al. (2005) Ca2+/calmodulin-dependent protein kinase kinase-β acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2, 21–33 doi: 10.1016/j.cmet.2005.06.005 [DOI] [PubMed] [Google Scholar]
- 17.Woods A., Johnstone S.R., Dickerson K., Leiper F.C., Fryer L.G., Neumann D. et al. (2003) LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 13, 2004–2008 doi: 10.1016/j.cub.2003.10.031 [DOI] [PubMed] [Google Scholar]
- 18.Hardie D.G., Carling D. and Gamblin S.J. (2011) AMP-activated protein kinase: also regulated by ADP? Trends Biochem. Sci. 36, 470–477 doi: 10.1016/j.tibs.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 19.Calabrese M.F., Rajamohan F., Harris M.S., Caspers N.L., Magyar R., Withka J.M. et al. (2014) Structural basis for AMPK activation: natural and synthetic ligands regulate kinase activity from opposite poles by different molecular mechanisms. Structure 22, 1161–1172 doi: 10.1016/j.str.2014.06.009 [DOI] [PubMed] [Google Scholar]
- 20.Cool B., Zinker B., Chiou W., Kifle L., Cao N., Perham M. et al. (2006) Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 3, 403–416 doi: 10.1016/j.cmet.2006.05.005 [DOI] [PubMed] [Google Scholar]
- 21.Scott J.W., van Denderen B.J.W., Jorgensen S.B., Honeyman J.E., Steinberg G.R., Oakhill J.S. et al. (2008) Thienopyridone drugs are selective activators of AMP-activated protein kinase β1-containing complexes. Chem. Biol. 15, 1220–1230 doi: 10.1016/j.chembiol.2008.10.005 [DOI] [PubMed] [Google Scholar]
- 22.Scott J.W., Ling N., Issa S.M.A., Dite T.A., O'Brien M.T., Chen Z.-P. et al. (2014) Small molecule drug A-769662 and AMP synergistically activate naive AMPK independent of upstream kinase signaling. Chem. Biol. 21, 619–627 doi: 10.1016/j.chembiol.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 23.Davies S.P., Carling D. and Hardie D.G. (1989) Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studied using a specific and sensitive peptide assay. Eur. J. Biochem. 186, 123–128 doi: 10.1111/j.1432-1033.1989.tb15185.x [DOI] [PubMed] [Google Scholar]
- 24.Emsley P. and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 25.CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 doi: 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 26.Woods A., Vertommen D., Neumann D., Türk R., Bayliss J., Schlattner U. et al. (2003) Identification of phosphorylation sites in AMP-activated protein kinase (AMPK) for upstream AMPK kinases and study of their roles by site-directed mutagenesis. J. Biol. Chem. 278, 28434–28442 doi: 10.1074/jbc.M303946200 [DOI] [PubMed] [Google Scholar]
- 27.Langendorf C.G., Ngoei K.R.W., Scott J.W., Ling N.X.Y., Issa S.M.A., Gorman M.A. et al. (2016) Structural basis of allosteric and synergistic activation of AMPK by furan-2-phosphonic derivative C2 binding. Nat. Commun. 7, 10912 doi: 10.1038/ncomms10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xin F.-J., Wang J., Zhao R.-Q., Wang Z.-X. and Wu J.-W. (2013) Coordinated regulation of AMPK activity by multiple elements in the α-subunit. Cell Res. 23, 1237–1240 doi: 10.1038/cr.2013.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L., Jiao Z.-H., Zheng L.-S., Zhang Y.-Y., Xie S.-T., Wang Z.-X. et al. (2009) Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature 459, 1146–1149 doi: 10.1038/nature08075 [DOI] [PubMed] [Google Scholar]
- 30.Chen L., Xin F.-J., Wang J., Hu J., Zhang Y.-Y., Wan S. et al. (2013) Conserved regulatory elements in AMPK. Nature 498, E8–E10 doi: 10.1038/nature12189 [DOI] [PubMed] [Google Scholar]
- 31.Mitchelhill K.I., Michell B.J., House C.M., Stapleton D., Dyck J., Gamble J. et al. (1997) Posttranslational modifications of the 5′-AMP-activated protein kinase β1 subunit. J. Biol. Chem. 272, 24475–24479 doi: 10.1074/jbc.272.39.24475 [DOI] [PubMed] [Google Scholar]
- 32.Endicott J.A., Noble M.E.M. and Johnson L.N. (2012) The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 81, 587–613 doi: 10.1146/annurev-biochem-052410-090317 [DOI] [PubMed] [Google Scholar]
- 33.Hawley S.A., Ross F.A., Chevtzoff C., Green K.A., Evans A., Fogarty S. et al. (2010) Use of cells expressing γ subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab. 11, 554–565 doi: 10.1016/j.cmet.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johanns M., Lai Y.-C., Hsu M.-F., Jacobs R., Vertommen D., Van Sande J. et al. (2016) AMPK antagonizes hepatic glucagon-stimulated cyclic AMP signalling via phosphorylation-induced activation of cyclic nucleotide phosphodiesterase 4B. Nat. Commun. 7, 10856 doi: 10.1038/ncomms10856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goransson O., McBride A., Hawley S.A., Ross F.A., Shpiro N., Foretz M. et al. (2007) Mechanism of action of A-769662, a valuable tool for activation of AMP-activated protein kinase. J. Biol. Chem. 282, 32549–32560 doi: 10.1074/jbc.M706536200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gowans G.J., Hawley S.A., Ross F.A. and Hardie D.G. (2013) AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab. 18, 556–566 doi: 10.1016/j.cmet.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao B., Heath R., Saiu P., Leone F.C., Leone P., Jing C. et al. (2007) Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature 449, 496–500 doi: 10.1038/nature06161 [DOI] [PubMed] [Google Scholar]
- 38.Carretero J., Medina P.P., Blanco R., Smit L., Tang M., Roncador G. et al. (2007) Dysfunctional AMPK activity, signalling through mTOR and survival in response to energetic stress in LKB1-deficient lung cancer. Oncogene 26, 1616–1625 doi: 10.1038/sj.onc.1209951 [DOI] [PubMed] [Google Scholar]
- 39.Nolen B., Taylor S. and Ghosh G. (2004) Regulation of protein kinases: controlling activity through activation segment conformation. Mol. Cell 15, 661–675 doi: 10.1016/j.molcel.2004.08.024 [DOI] [PubMed] [Google Scholar]
- 40.Jeffrey P.D., Russo A.A., Polyak K., Gibbs E., Hurwitz J., Massagué J. et al. (1995) Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376, 313–320 doi: 10.1038/376313a0 [DOI] [PubMed] [Google Scholar]
- 41.Shackelford D.B. and Shaw R.J. (2009) The LKB1–AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer 9, 563–575 doi: 10.1038/nrc2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross F.A., Jensen T.E. and Hardie D.G. (2016) Differential regulation by AMP and ADP of AMPK complexes containing different γ subunit isoforms. Biochem. J. 473, 189–199 doi: 10.1042/BJ20150910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajamohan F., Reyes A.R., Frisbie R.K., Hoth L.R., Sahasrabudhe P., Magyar R. et al. (2016) Probing the enzyme kinetics, allosteric modulation and activation of α1- and α2-subunit-containing AMP-activated protein kinase (AMPK) heterotrimeric complexes by pharmacological and physiological activators. Biochem. J. 473, 581–592 doi: 10.1042/BJ20151051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carretero J., Medina P.P., Pio R., Montuenga L.M. and Sanchez-Cespedes M. (2004) Novel and natural knockout lung cancer cell lines for the LKB1/STK11 tumor suppressor gene. Oncogene 23, 4037–4040 doi: 10.1038/sj.onc.1207502 [DOI] [PubMed] [Google Scholar]
- 45.Cheung P.C.F., Salt I.P., Davies S.P., Hardie D.G. and Carling D. (2000) Characterization of AMP-activated protein kinase γ-subunit isoforms and their role in AMP binding. Biochem. J. 346, 659–669 doi: 10.1042/bj3460659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujiwara Y., Hiraoka Y., Fujimoto T., Kanayama N., Magari M. and Tokumitsu H. (2015) Analysis of distinct roles of CaMKK isoforms using STO-609-resistant mutants in living cells. Biochemistry 54, 3969–3977 doi: 10.1021/acs.biochem.5b00149 [DOI] [PubMed] [Google Scholar]
- 47.Jeon S.-M., Chandel N.S. and Hay N. (2012) AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature 485, 661–665 doi: 10.1038/nature11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wojtaszewski J.F.P., Jorgensen S.B., Hellsten Y., Hardie D.G. and Richter E.A. (2002) Glycogen-dependent effects of 5-aminoimidazole-4-carboxamide (AICA)-riboside on AMP-activated protein kinase and glycogen synthase activities in rat skeletal muscle. Diabetes 51, 284–292 doi: 10.2337/diabetes.51.2.284 [DOI] [PubMed] [Google Scholar]
- 49.Zhang C.-S., Li M., Ma T., Zong Y., Cui J., Feng J.-W. et al. (2016) Metformin activates AMPK through the lysosomal pathway. Cell Metab. 24, 521–522 doi: 10.1016/j.cmet.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y.-L., Guo H., Zhang C.-S., Lin S.-Y., Yin Z., Peng Y. et al. (2013) AMP as a low-energy charge signal autonomously initiates assembly of AXIN-AMPK-LKB1 complex for AMPK activation. Cell Metab. 18, 546–555 doi: 10.1016/j.cmet.2013.09.005 [DOI] [PubMed] [Google Scholar]
- 51.Zhang C.-S., Jiang B., Li M., Zhu M., Peng Y., Zhang Y.-L. et al. (2014) The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 20, 526–540 doi: 10.1016/j.cmet.2014.06.014 [DOI] [PubMed] [Google Scholar]