ABSTRACT
The ability to detect and measure danger from an environmental signal is paramount for bacteria to respond accordingly, deploying strategies that halt or counteract potential cellular injury and maximize survival chances. Type VI secretion systems (T6SSs) are complex bacterial contractile nanomachines able to target toxic effectors into neighboring bacteria competing for the same colonization niche. Previous studies support the concept that either T6SSs are constitutively active or they fire effectors in response to various stimuli, such as high bacterial density, cell-cell contact, nutrient depletion, or components from dead sibling cells. For Serratia marcescens, it has been proposed that its T6SS is stochastically expressed, with no distinction between harmless or aggressive competitors. In contrast, we demonstrate that the Rcs regulatory system is responsible for finely tuning Serratia T6SS expression levels, behaving as a transcriptional rheostat. When confronted with harmless bacteria, basal T6SS expression levels suffice for Serratia to eliminate the competitor. A moderate T6SS upregulation is triggered when, according to the aggressor-prey ratio, an unbalanced interplay between homologous and heterologous effectors and immunity proteins takes place. Higher T6SS expression levels are achieved when Serratia is challenged by a contender like Acinetobacter, which indiscriminately fires heterologous effectors able to exert lethal cellular harm, threatening the survival of the Serratia population. We also demonstrate that Serratia’s RcsB-dependent T6SS regulatory mechanism responds not to general stress signals but to the action of specific effectors from competitors, displaying an exquisite strategy to weigh risks and keep the balance between energy expenditure and fitness costs.
KEYWORDS: Serratia, type VI secretion system, bacterial competition
IMPORTANCE
Serratia marcescens is among the health-threatening pathogens categorized by the WHO as research priorities to develop alternative antimicrobial strategies, and it was also recently identified as one major component of the gut microbiome in familial Crohn disease dysbiosis. Type VI secretion systems (T6SSs) stand among the array of survival strategies that Serratia displays. They are contractile multiprotein complexes able to deliver toxic effectors directed to kill bacterial species sharing the same niche and, thus, competing for vital resources. Here, we show that Serratia is able to detect and measure the extent of damage generated through T6SS-delivered toxins from neighboring bacteria and responds by transcriptionally adjusting the expression level of its own T6SS machinery to counterattack the rival. This strategy allows Serratia to finely tune the production of costly T6SS devices to maximize the chances of successfully fighting against enemies and minimize energy investment. The knowledge of this novel mechanism provides insight to better understand bacterial interactions and design alternative treatments for polymicrobial infections.
INTRODUCTION
Serratia is a Gram-negative genus that belongs to the family Enterobacteriaceae and encompasses species that can colonize a wide variety of environmental niches, ranging from water and soil to air. In addition to environmental ubiquity, among Serratia species, S. marcescens constitutes an emergent health-threatening nosocomial pathogen (1), with increasing reports of multidrug-resistant-strain outbreaks and high incidences in intensive and neonatal care units (2–4). S. marcescens has also recently been identified as one of the most abundant microbial species that colonizes the dysbiotic gut of Crohn patients, in detriment of beneficial bacteria (5). Furthermore, S. marcescens can interact either symbiotically or pathogenically with plants and insects. These features denote S. marcescens’ plasticity in adapting to changing ambient conditions and the deployment of strategies that allow the pathogen to colonize and thrive in complex polymicrobial niches.
Type VI secretion systems (T6SSs) have been found to be encoded in the genomes of a wide variety of Gram-negative bacteria, including both environmental and pathogenic microorganisms. These systems are assembled by a needlelike appendage that is able to export bacterial effectors and translocate them into an adjacent eukaryotic or prokaryotic cell. A T6SS assembles as a contractile device that displays homology to the tail spike complex of the T4 bacteriophage (6). Across different bacterial species, T6SSs differ in their capacity to liberate a variety of effectors, which are commonly associated with two conserved proteins encompassed within the estimated 13 core T6SS components: Hcp (hemolysin coregulated protein) a protein that forms the ejectable inner sheath of the nanotubular device, and VgrG (valine-glycine repeat G protein). VgrG trimmers form the spike of the needle, frequently associated with PAAR (proline, alanine, alanine, arginine) repeat motif-containing proteins at the tip of the spike (7). In order to prevent self or sister cell intoxication, T6SS+ organisms encode not only effectors but also immunity proteins to neutralize their cognate antibacterial effectors. Therefore, T6SSs are important components of the attack/defense force that bacteria deploy to antagonize other bacterial partners (intra- or intergenera) in order to survive and/or colonize a niche. In addition, some species rely on T6SSs to defeat the defenses of the invaded host (8–10).
While there is an increasing number of reports that shed light on detailed structural aspects of the apparatus assembly of the T6SS components and the mode of action of exported effectors across diverse bacterial species (11–13), the regulatory mechanisms that govern the expression of such complex and energetically costly devices are far from being understood.
The work of Iguchi et al. (14) provided evidences that the T6SS is part of the evolutionarily conserved weaponry of S. marcescens, dedicated to surmounting the challenge of surviving in a variety of environments. Nonetheless, in Serratia, the T6SS has been shown to be used in attacking bacterial competitors, and activity against eukaryotic targets has not been yet detected (15, 16). S. marcescens strain Db10 possesses a single T6SS with potent antibacterial activity, delivering at least six antibacterial effector proteins, including the peptidoglycan hydrolases Ssp1 and Ssp2 (15–18).
Previous work indicated that the expression of the T6SS in S. marcescens Db10 depends on one large transcriptional unit that encompasses the whole cluster and is constitutively active, as it was found to be independent of the growth phase, the growth medium composition, or contact with other bacterial cells assayed (16). PppA (phosphatase) and PpkA (kinase) proteins compose a reversible phosphorylation system that posttranslationally modifies Fha (forkhead associated), a component required for the assembly of the system (9). In some bacteria, such as Pseudomonas aeruginosa, this protein pair acts as part of a transduction cascade that functions as a posttranslational molecular switch to control T6SS assembly and activity upon detection of environmental and killed-sister-cell-derived signals (19, 20). However, although the PppA-PpkA switch controls the phosphorylation status of Fha in Db10, its activity was found to be at most modestly influenced by cell-cell contact, and it was postulated to finely adjust the bacterial killing capacity of the bacteria (21). More recently, a study by Gerc et al. (22) using single-cell techniques supported a model of a randomly triggered assembly of S. marcescens T6SS machinery, ready for attack on circumstantial prey.
Signal transduction systems and specialized secretory devices are key for bacteria to detect and adequately counteract the effects of noxious environmental conditions and defeat competitors. In our previous work, we have shown that in S. marcescens clinical isolate RM66262 (23), the Rcs system is key in controlling several traits of the bacterium. The Rcs signal transduction system is essentially composed of three proteins that belong to the two-component family: two inner membrane sensor proteins, RcsC (a bifunctional kinase/phosphatase sensor) and RcsD, and the cognate cytoplasmic transcriptional regulator RcsB. The phosphorylation status of RcsB modulates the binding of this regulator to promoter regions of the target genes, activating or repressing their transcription. Three additional components can be part of the Rcs-dependent signaling cascade: RcsF, a membrane-anchored lipoprotein that can channel stimuli to the RcsC sensor; IgaA, which is able to repress RcsC activity; and RcsA, which coregulates subsets of genes together with RcsB (24, 25).
Rcs system activity finely tunes the motile phenotypes (swimming and swarming) of Serratia by regulating the expression of FlhDC, the master regulator of the transcriptional flagellar cascade (26, 27). As a consequence, the Rcs system also modulates the capacity of Serratia to produce the PhlA phospholipase, which is transcriptionally regulated by FliA (the sigma factor that determines the secondary wave of the flagellar cascade expression) and exported through the flagellar secretome (26, 27). In addition, we have demonstrated that RcsB also controls the expression of S. marcescens ShlA, which belongs to the pore-forming toxin family of proteins (28). ShlA elicits an early autophagy response prior to Serratia’s internalization in nonphagocytic cells (28). Once Serratia cells have replicated inside a specialized vacuole, this effector triggers nonlytic egress of the bacteria from the infected cell by promoting an exocytic process (29). We have also shown that RcsB is involved in the regulation of Serratia outer membrane vesicle production (30). To summarize, we have demonstrated that the Rcs system is implicated in governing the expression of multiple phenotypes that are key to Serratia’s virulence potential.
In this work, we show for the first time that the T6SS expression levels of S. marcescens RM66262 are transcriptionally controlled by direct interaction of the RcsB response regulator with the promoter region of the T6SS gene cluster, a feature that appears to be highly conserved in S. marcescens strains, irrespective of the host or environmental source of the strain. We show that the RcsB-dependent modulation of the killing capacity of RM66262 against competitor bacteria is activated upon encountering rival bacteria. We reveal that RcsB-dependent induction of Serratia T6SS expression occurs in response to the detection of damage exerted by specific T6SS effectors translocated by competitors rather than to broad-spectrum envelope injury. Collectively, our results demonstrate that in Serratia, RcsB-controlled upregulation of the T6SS over basal expression levels constitutes a survival strategy triggered by specific lethal threats posed by interbacterial competition.
RESULTS
RcsB can alter S. marcescens’ interbacterial competition capacity.
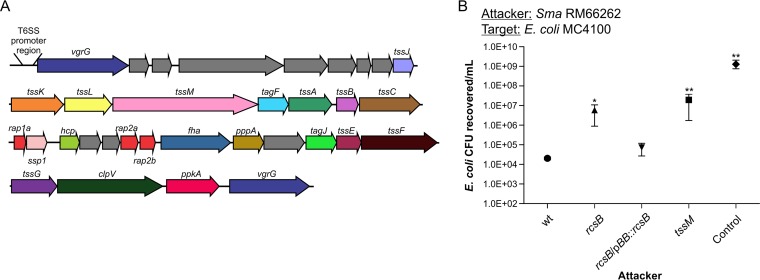
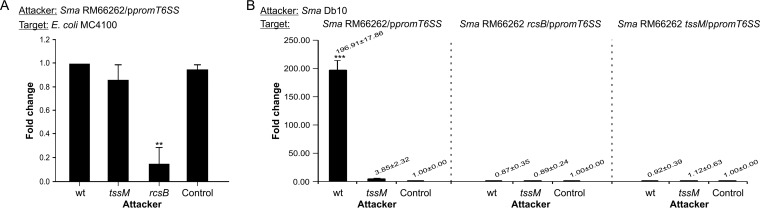
Our bioinformatics analysis of the annotated genome of the S. marcescens clinical strain RM66262 (23) showed that it harbors a single canonical T6SS-encoding cluster, similar to those of previously analyzed Serratia strains (Fig. 1A) (14, 16). Because the Rcs signal transduction system regulates the expression of key virulence factors of S. marcescens (27, 28, 30), we examined whether this system affects the ability of Serratia to antagonize other bacteria. We carried out competition assays using RM66262 and rcsB and tssM mutants (unable to assemble the T6SS) derived from it as attacker strains and Escherichia coli MC4100, a strain naturally devoid of T6SS, as the prey. As shown by the results in Fig. 1B, tssM and rcsB strains were equally unable to kill the E. coli prey. This killing deficiency was restored to wild-type levels when the rcsB mutant was complemented in trans by the expression of RcsB from the pBB::rcsB plasmid. These results suggested that S. marcescens’ T6SS expression is under the control of RcsB.
FIG 1 .
S. marcescens (Sma) kills E. coli MC4100 in a T6SS- and RcsB-dependent manner. (A) Schematic representation of the T6SS gene cluster of S. marcescens RM66262. (B) Recovery of viable E. coli MC4100 cells after coculture with the indicated S. marcescens RM66262 strains for 4 h at 37°C, with an initial 5:1 (attacker/target) ratio. E. coli DH5α mixed with E. coli MC4100 at a 5:1 ratio was used as the control. S. marcescens RM66262 rcsB and tssM mutant strains were 300-fold and 1,000-fold less proficient, respectively, than the wt strain in the ability to kill E. coli. The average values ± standard errors of the means (SEM) from four independent experiments are shown (*, P < 0.05; **, P < 0.01). wt, wild type.
Serratia marcescens’ T6SS expression is under RcsB regulation.
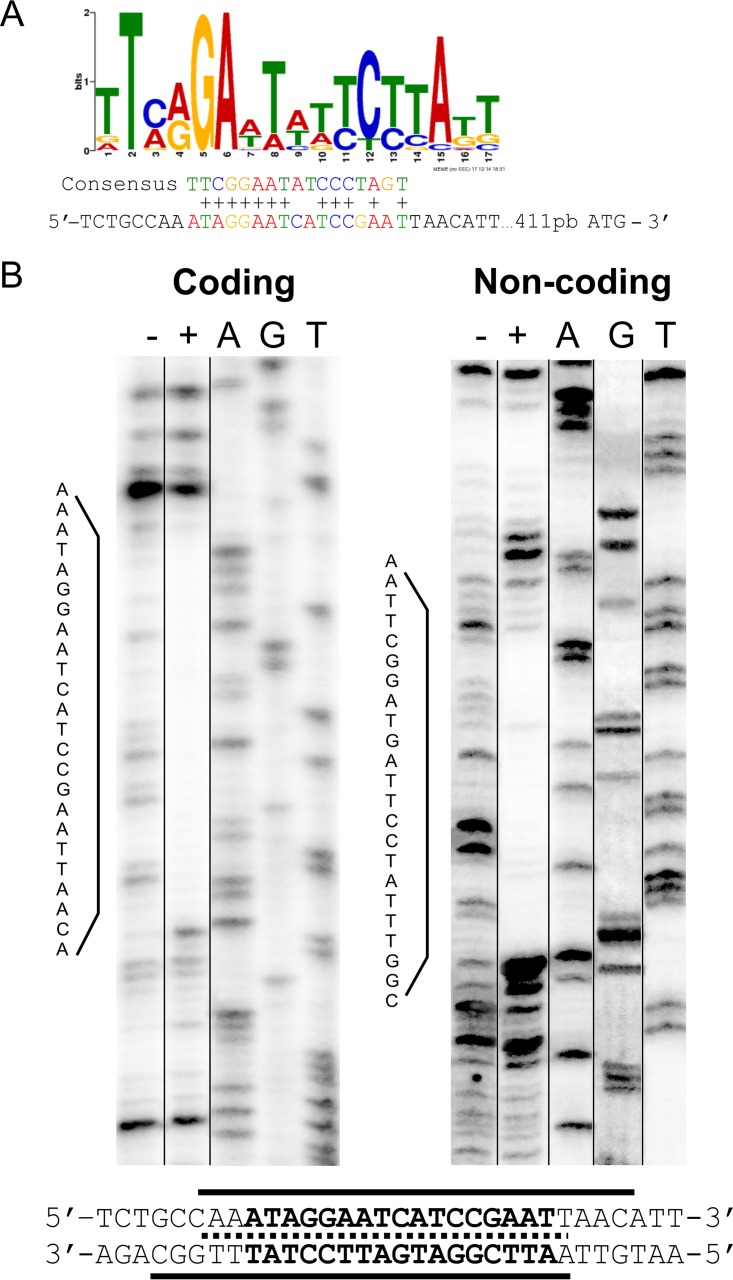
We next performed an in silico search for a potential RcsB-binding motif upstream from vgrG, the first gene of the S. marcescens RM66262 T6SS cluster. By the use of the MEME/MAST motif detection programs (41, 42), we found the presence of a putative RcsB-binding motif upstream from vgrG’s ATG (Fig. 2A). This RcsB-binding sequence showed 12 of 17 conserved bases in the logo constructed by the search engine, using previously identified promoter regions that contain bona fide RcsB recognition sequences (Fig. 2A).
FIG 2 .
RcsB directly interacts with an RcsB-binding motif. (A) The logo shows the consensus motif for the RcsB-binding site, and the predicted RcsB-binding site sequence in the promoter region of S. marcescens RM66262 T6SS is indicated below the logo. The conserved residues are marked with “+” in the alignment with the consensus motif. (B) DNA footprinting analysis was performed on coding and noncoding strands of the T6SS promoter region. The DNA fragments were incubated with increasing amounts of purified RcsB. Results for 0 (−) and 25 (+) (the minimal protein concentration that showed protection) pmol of purified RcsB are shown. Ladders and the corresponding nucleotides are shown. The gels were sliced (noncontiguous lanes from a single gel are indicated by black lines) to exclude nonoptimal protein concentrations assayed or ladder lanes that resulted in smeared patterns. The nucleotide sequences of the RcsB-protected regions are indicated, and the protected DNA regions are underlined. The overlap of the protected sequences is marked with a dotted line.
To assess whether there is direct interaction of RcsB with the vgrG promoter region and to better define the RcsB recognition sequence, DNase I protection assays were performed on both coding and noncoding strands. A nucleotide sequence encompassing 500 bp upstream from the vgrG ATG site and purified 6×His-tagged RcsB protein (RcsB-6×His) phosphorylated in the presence of acetyl phosphate were used, as described previously (28). We have previously corroborated that the 6×His tag does not alter either RcsB’s regulatory ability or its in vitro DNA-binding capacity (28). RcsB-6×His protected an overlapping region from nucleotide 418 to 438 relative to the vgrG translational ATG start site (Fig. 2B). This protected region encompassed the in silico-predicted RcsB-binding consensus motif.
An in silico analysis of upstream region sequences of the vgrG genes in available Serratia genomes deposited in the NCBI database was performed. As shown by the results in Fig. S1, a conserved motif with 100% sequence identity to that of RM66262 was found in the Serratia genomes that display synteny with the cluster borne by RM66262, in which vgrG was found to be the first gene of the T6SS cluster. This result is strongly indicative of conserved RcsB-dependent control of T6SS expression along the Serratia genus.
The RcsB-binding motif is conserved in the Serratia T6SS promoter region. Download FIG S1, PDF file, 2.3 MB (2.4MB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
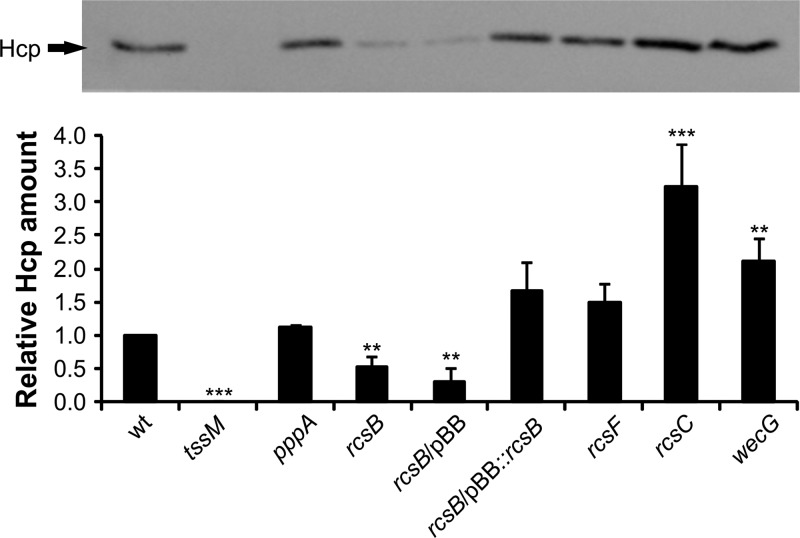
Because Hcp composes the puncture device, which is expelled to the medium as it disassembles, the presence of Hcp in the culture supernatant is indicative of the dynamic assembly of the T6SS (33). To examine whether RcsB-dependent regulation is reflected at the level of protein expression, we determined the Hcp levels in strains with genetic backgrounds that alter Rcs function. Total culture supernatants from planktonic RM66262 wild-type (wt) or mutant strains grown in LB were trichloroacetic acid (TCA) precipitated and subjected to SDS-PAGE analysis and Coomassie blue staining or transferred to nitrocellulose and immunodetected with anti-Hcp polyclonal antibodies. Hcp expression levels were determined by densitometry (Fig. 3; Fig. S2). As predicted, Hcp levels were downregulated in the rcsB background and restored to wild-type levels when the rcsB strain was complemented in trans by the expression of RcsB from the pBB::rcsB plasmid (Fig. 3). Hcp was undetectable in the tssM mutant strain, while it was not affected by the pppA mutant background compared to its levels in the wild-type strain. In the wecG mutant strain, in which the Rcs system is activated (27), Hcp secretion was increased. rcsF inactivation did not alter Hcp secretion levels, while an rcsC background resulted in an increase in Hcp secretion levels (Fig. 3). These results were also in agreement with the results of reverse transcription-quantitative PCR (RT-qPCR) assays in which the transcriptional levels of vgrG (which codes for VgrG) and hcp (which codes for Hcp), distant from each other in the Serratia T6SS gene cluster, were analyzed in wild-type or rcsB planktonic bacteria grown in LB medium (Fig. S3A and B). To summarize, our results demonstrate that T6SS expression is modulated by RcsB at the transcriptional level in S. marcescens RM66262 and suggest that this regulatory mechanism might have been subjected to strong selective pressure along evolution in Serratia.
FIG 3 .
RcsB controls secretion of Hcp. Filtered supernatants from saturated cultures of the indicated S. marcescens RM66262 strains were precipitated and loaded into 15% SDS-PAGE gels. Hcp levels were determined by immunodetection using Hcp antisera followed by densitometry. (Top) Representative image of the assay. (Bottom) Average values ± standard deviations (SD) from three independent experiments are shown (**, P < 0.01; ***, P < 0.001). See Fig. S2 in the supplemental material for the densitometry normalization procedure employed.
Hcp densitometry. Download FIG S2, PDF file, 0.8 MB (840.8KB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RcsB transcriptionally modulates T6SS expression. Download FIG S3, PDF file, 0.6 MB (657.2KB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RcsB controls S. marcescens T6SS-mediated intraspecies competition.
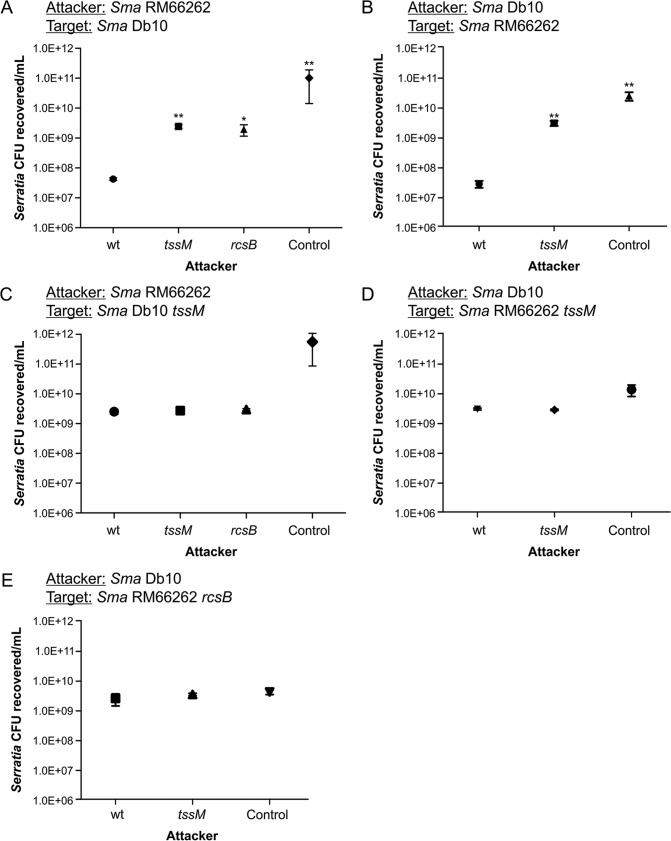
S. marcescens T6SS has been described as functioning essentially in targeting competitor bacteria (21, 22, 34). Using the S. marcescens Db10 strain, previous reports have shown that the intraspecies killing of competing Serratia bacteria occurs irrespective of the functionality of the T6SS of the prey, discarding a counterattack, such as a “tit-for-tat” type of reciprocal response, for Serratia (22, 33). To evaluate the influence of RcsB-dependent regulation on the functionality of Serratia T6SS in intraspecies antagonism, we performed killing assays between S. marcescens strains Db10 and RM66262 and their tssM derivatives, as described in Materials and Methods. At a 5:1 attacker-to-prey ratio, both strains were able to kill their wild-type counterparts in a T6SS-dependent fashion, as demonstrated by the differences in the CFU counts of the prey recovered after attack by either the wild type or the tssM mutant (Fig. 4A and B). In these assays, the RM66262 rcsB strain showed a decrease in killing ability similar to that seen for the tssM strain compared to that of the wild-type strain (Fig. 4A).
FIG 4 .
RcsB controls Serratia intraspecies T6SS-mediated competition. (A to E) Recovery of viable S. marcescens cells after 6 h of coculture with the indicated attacker strain at 37°C, with an initial ratio of 5:1 (attacker/target). E. coli DH5α mixed with S. marcescens at a 5:1 ratio was used for controls. S. marcescens RM66262 tssM or S. marcescens Db10 tssM mutants were 57-fold or 107-fold less proficient than their respective parental strains (wt) in outcompeting wild-type counterpart preys. S. marcescens RM66262 rcsB showed a 46-fold reduction in killing ability compared to that of the wild-type strain. Average values ± SEM from four independent experiments are shown (*, P < 0.05; **, P < 0.01).
However, neither the RM66262 nor the Db10 wild-type strain could kill its tssM counterpart strain in statistically significant numbers, and the same recovered CFU levels were obtained using either RM66262 tssM or Db10 tssM as the aggressor strain (Fig. 4C and D). Moreover, in the encounter with a tssM target strain, the results for an RM66262 rcsB attacker were also indistinguishable from the results for the wild-type or tssM attacker strain (Fig. 4C).
These results suggested that, in an intraspecific encounter, one S. marcescens strain will not attack another S. marcescens strain if the latter does not express an active T6SS. This is further supported by the observation that an S. marcescens RM66262 rcsB prey was not targeted by either a wild-type S. marcescens Db10 or a tssM attacker (Fig. 4E). This result also reinforces the involvement of RcsB in the control of Serratia T6SS expression.
S. marcescens T6SS promoter activity is induced by kin T6SS+ competitors in an RcsB-dependent manner.
To analyze T6SS RcsB-dependent regulation upon exposure to T6SS-expressing (T6SS+) or T6SS-deficient (T6SS−) competitor strains, we cloned a 500-bp sequence upstream from vgrG (promoter) harboring the RcsB-binding motif in the pPROBE(NT') vector (31), resulting in the ppromT6SS construct. This construct allows the quantitative detection of transcriptional activation by measuring the fluorescence of green fluorescent protein (GFP) expression driven by the upstream cloned sequence. As shown by the results in Fig. S3C, an expected rcsB-dependent response of the reporter was verified by introducing ppromT6SS into the S. marcescens genetic backgrounds used in the Hcp secretion assays described above. In addition, we tested the ppromT6SS response in an S. marcescens-versus-E. coli competition assay. Both wild-type and tssM S. marcescens strains showed promT6SS expression levels that were equivalent to the value showed by the wild-type strain alone used as control. These basal expression levels depended on rcsB integrity (Fig. 5A).
FIG 5 .
Intraspecies competition triggers Serratia T6SS activity in an RcsB-dependent manner. (A) S. marcescens RM66262/ppromT6SS and the indicated mutant strains were incubated with E. coli MC4100 in a 5:1 (attacker/target) ratio for 4 h. S. marcescens RM66262/ppromT6SS alone was used as the control. Serial dilutions were plated onto ampicillin-LB plates for S. marcescens RM66262 selection. GFP fluorescence/CFU was calculated. Fold change in transcriptional induction was calculated relative to wild-type strain values. Average values ± SD for four independent experiments are shown (**, P < 0.01). (B) S. marcescens Db10 wt or tssM mutant strains were incubated with S. marcescens RM66262 wt or mutant strains carrying the ppromT6SS in a 5:1 (attacker/target) ratio for 6 h at 37°C. GFP fluorescence/CFU was determined. E. coli DH5α mixed with target bacteria at a 5:1 ratio was used for controls. Fold change in transcriptional induction was calculated relative to the control value. Average values ± SD from three independent experiments are shown (***, P < 0.001).
Next, RM66262 strains harboring ppromT6SS (prey) were challenged with either a Db10 wild-type or tssM strain (attackers). The fluorescence levels obtained from ppromT66S contained in the RM66262 wild-type, rcsB, or tssM strain when coincubated with the control E. coli DH5α strain were measured. As shown by the results in Fig. 5B (left), the activity driven by the T6SS promoter increased ∼200-fold when the wild-type RM66262/ppromT6SS strain was challenged by the wild-type Db10 strain. In contrast, the Db10 tssM strain was not able to induce T6SS promoter expression in RM66262/ppromT6SS. Lack of RcsB expression resulted in the inability of the T6SS promoter to be induced when RM66262 rcsB/ppromT6SS was challenged by either the wild-type or tssM Db10 strain (Fig. 5B, middle). In agreement with the incapacity of a wild-type strain to target either an rcsB or a tssM strain, the reporter was not induced from either the rcsB or the tssM background upon this encounter (Fig. 5B, right).
The latter results demonstrate that, in a Serratia intraspecific competition, the induction of a prey’s T6SS expression is RcsB dependent and occurs only when both attacker and prey carry a functional T6SS.
To further understand the S. marcescens competition strategy, we tested whether the stimulus detected by a T6SS+ S. marcescens strain in the encounter with a kin T6SS+ strain would be enough to enable the former strain to kill an otherwise undetected T6SS− strain. As shown by the results in Fig. S4, both T6SS+ and T6SS− preys are killed equally well when mixed with a T6SS+ attacker. Together, these results indicate that, as far as the expression of T6SS is triggered by the encounter with a threatening T6SS+ opponent, there is neither preferential nor directional aiming of a bystander prey, reinforcing the concept of a transcriptionally based response.
Serratia-induced T6SS can target preys in a nondirectional mode. Download FIG S4, PDF file, 1.8 MB (1.8MB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Interspecies competition induces Serratia T6SS expression in an RcsB-dependent manner.
To further understand the signals that activate T6SS in Serratia, we employed competition assays between Serratia and Acinetobacter baumannii strain ATCC 17978. T6SS expression in A. baumannii ATCC 17978 is regulated by a plasmid that is spontaneously lost in part of the population. A strain without this plasmid possesses a constantly active T6SS (32). In all our experiments, we used either A. baumannii ATCC 17978 wild type or derivative strains lacking the repressing plasmid.
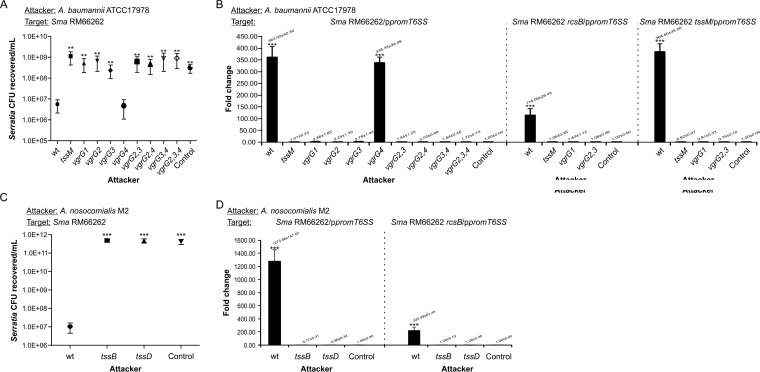
We challenged S. marcescens RM66262 with either A. baumannii ATCC 17978 wild type or a tssM (T6SS−) strain in a 5:1 attacker-to-prey ratio. As shown by the results in Fig. 6A, the A. baumannii wild-type strain was able to kill S. marcescens RM66262 in a T6SS-dependent manner. In counterpart killing assays, also using a 5:1 attacker-to-prey ratio, the Serratia strain was unsuccessful at eliminating either T6SS+ or T6SS− A. baumannii (Fig. S5A and B).
FIG 6 .
Acinetobacter killing capacity and RcsB-dependent induction of Serratia T6SS expression. (A, C) Recovery of viable S. marcescens RM66262 wt or mutant strains cocultured with wt or mutant strains of A. baumannii ATCC 17978 (A) or A. nosocomialis M2 (C) for 4 h at 37°C, with an initial 5:1 (attacker/target) ratio. E. coli DH5α mixed with target bacteria at a 5:1 ratio was used for controls. A. baumannii wt or vgrG4 strains were 200-fold more proficient in outcompeting Serratia than the other vgrG single, double, or triple mutant strains analyzed. A. nosocomialis wt was ∼5 orders of magnitude more proficient in outcompeting Serratia than the mutant strains analyzed. Average values ± SEM from four independent experiments are shown (**, P < 0.01; ***, P < 0.001). (B, D) Wild-type or mutant strains of A. baumannii ATCC 17978 (B) or A. nosocomialis M2 (D) were coincubated with wt or mutant strains S. marcescens RM66262 carrying ppromT6SS at an initial 5:1 (attacker/target) ratio for 4 h at 37°C. GFP fluorescence/CFU was calculated. E. coli DH5α mixed with target bacteria at a 5:1 ratio was used for controls. Fold change in transcriptional induction was calculated relative to the control value. Average values ± SD from four independent experiments are shown (***, P < 0.001).
Serratia-Acinetobacter interspecific competition assays. Download FIG S5, PDF file, 1.5 MB (1.5MB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Challenge with wild-type A. baumannii caused an ∼400-fold induction of the expression of the Serratia T6SS promoter. This induction was dependent on the presence of an active T6SS system, as the A. baumannii tssM strain was not able to induce T6SS expression in Serratia (Fig. 6B, left, compare results from A. baumannii wild-type and tssM strains). To assess whether this induction depends on RcsB, the S. marcescens RM66262 rcsB/ppromT6SS strain was also tested as prey. The abrogation of RcsB expression led to a 75% reduction of promT6SS fold induction (Fig. 6B, middle), showing the dependence on RcsB for the activation of Serratia’s T6SS transcriptional activity upon encountering Acinetobacter. This activation was independent of the T6SS status of the prey, as both wild-type and tssM S. marcescens strains upregulated their T6SS as a consequence of the Acinetobacter attack. Our results show that Acinetobacter kills and also induces ppromT6SS expression when coincubated with either a wild-type or tssM S. marcescens strain (Fig. 6A and B). Because the Acinetobacter strain used expresses its T6SS constitutively (32), the latter results indicate that, in this encounter, only Serratia follows a counterattack mode of action.
To verify that the phenotypes were not restricted to the interaction of Serratia with a unique Acinetobacter strain, we also determined that Acinetobacter nosocomialis strain M2 was able to kill S. marcescens RM66262, reducing the CFU count by 5 orders of magnitude (Fig. 6C). Only A. nosocomialis strain M2 carrying an active T6SS, and not isogenic tssB or tssD T6SS− strains, was able to induce the transcriptional activity of RM66262 T6SS, by ∼1,300-fold in an RcsB-dependent fashion (Fig. 6D). It is important to highlight that a correlation between killing capacity and ppromT6SS induction levels can be observed when the results from A. baumannii ATCC 17978 and A. nosocomialis M2 versus S. marcescens RM66262 are compared (Fig. 6A, C, B, and D, compare CFU values in killing assays and promT6SS induction levels). Remarkably, if we also include S. marcescens intraspecies competition results in this comparative analysis (Fig. 4 and 5B), the correspondence between killing capacity and levels of promT6SS upregulation can also be observed.
Which signals are able to induce Serratia T6SS expression?
The components of A. baumannii’s T6SS have been comprehensively analyzed in a previous work by Weber et al. (13), showing that the T6SS gene cluster encodes four VgrG proteins with their associated putative effectors. VgrG1 has an essential role in Hcp export and, therefore, is a functional T6SS apparatus component. VgrG2 and VgrG3 would be required for the proper secretion of Tde and Tse effectors, respectively, which play a role in killing E. coli strains. Vgr4 and its cognate Tae effector were found to be dispensable for E. coli killing (13). As shown by the results in Fig. 6A, while Vgr4 was dispensable, VgrG2 and VgrG3 were simultaneously required for A. baumannii to kill S. marcescens RM66262. In correlation with this result, only A. baumannii wild-type and vgrG4 strains were able to induce promT6SS activity (Fig. 6B). Interestingly, the vgrG2 vgrG3 vgrG4 triple mutant strain, despite proficiency in displaying a fully assembled T6SS (13), was inefficient in either killing Serratia or inducing promT6SS activity (Fig. 6A and B). These results show that the simultaneous action of VgrG2 and VgrG3 cognate effectors is required both for killing and for RcsB-dependent T6SS induction. In agreement with the latter result, the sole inactivation of the Tse3 gene sufficed to make the wild type unable to kill Serratia or upregulate S. marcescens T6SS expression (Fig. S5C and D). Therefore, we ruled out the possibility that activation of the T6SS could simply be caused by a membrane perturbation provoked when S. marcescens encounters an Acinetobacter cell that carries a functional T6SS but is unable to deliver toxic effectors into Serratia. We then hypothesized that a component released by the killed Serratia subpopulation could act as a trigger of RcsB-dependent activation in the survivors. This hypothesis was supported by recent work in Pseudomonas indicating that kin cell lysate components are danger signals that can be detected by the prey (20).
Different approaches were designed to examine this possibility, as follows: (i) total lysate of wild-type S. marcescens RM66262 was incubated with RM66262/ppromT6SS, and the transcriptional induction of the reporter was measured (Fig. S6A), and (ii), to more accurately obtain S. marcescens by-products released after a killing assay of A. baumannii against Serratia, the supernatant devoid of intact bacteria was collected and subsequently incubated with the reporter strain (Fig. S6B). In parallel, competition assays using A. nosocomialis M2 or E. coli DH5α (attackers) and wild-type S. marcescens RM66262 (prey) were carried out and a filter was placed on top where S. marcescens RM66262/ppromT6SS strain was seeded, incubated for 4 h, and then collected to measure fluorescence (see Fig. S6C for a scheme of the approach). In addition, we performed a triple competition assay employing an attacker, a prey, and a reporter strain to determine whether self or nonself components released by dead cells in situ during the killing assay could induce T6SS expression. S. marcescens Db10 wild type or tssM (attacker) strains and S. marcescens RM66262 or E. coli MC4100 (prey) were coincubated with S. marcescens RM66262 tssM/ppromT6SS (reporter) at a 5:1:1 ratio (Fig. S6D). None of these interactions induced the expression of the reporter, providing evidence that endogenous Serratia molecules released as the result of bacterial lysis or T6SS-provoked death are not able to trigger T6SS upregulation.
Bacterial lysis or derived products of T6SS-provoked killing are unable to activate S. marcescens T6SS expression. Download FIG S6, PDF file, 1 MB (1MB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, because the Rcs system is responsive to envelope stress, we also analyzed whether the actions of bacterial-envelope-damaging agents other than specific T6SS-dependent effectors were able to promote T6SS expression. To this end, and to exclude by-products of lethal action, the RM66262/ppromT6SS reporter strain was incubated with sublethal concentrations of polymyxin B, deoxycholate, or SDS. To verify that each agent caused the expected perturbations of the bacterial envelope, their actions against the Serratia outer membrane were measured by β-lactamase (periplasmic) leakage and inner membrane disturbances by β-galactosidase (cytoplasmic) escape, using nitrocefin or o-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate, respectively (35). None of these challenges induced ppromT6SS activity (Fig. S7A), although as expected, sublethal concentrations of each assayed agent were able to perturb bacterial membrane integrity (Fig. S7B to D).
Nonspecific envelope damage does not activate T6SS expression. Download FIG S7, PDF file, 1 MB (1MB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The latter results indicate that nonspecific envelope damage does not suffice to induce T6SS expression, strongly indicating that damage provoked by the specific action of T6SS effectors on Serratia cells generates the signal detected by the Rcs system that in turn upregulates T6SS expression.
DISCUSSION
Interbacterial competition for a shared niche has evolutionarily selected refined mechanisms of defense and attack. T6SS is a potent weapon that endows bacteria with the ability to dynamically antagonize their competitors, turning kin and nonkin interactions into attacker-prey survival battles. To design new ways to fight polymicrobial infections, it is crucial to understand the dynamics of mixed bacterial populations that define the balance between species or determine the successful establishment and propagation of one dominant pathogen.
S. marcescens, as a typical opportunist and adaptable pathogen, is frequently isolated from polymicrobial infections in patients suffering from diseases such as pneumonia, meningitis, or urinary tract infections. Mixed bacterial populations include recalcitrant multiresistant Acinetobacter species, among other pathogens that can be Serratia cohabitants or contenders in the infected host (36–38).
In this work, we focused on understanding the mechanism that governs the firing of S. marcescens’ T6SS expression in response to intra- or interspecies encounters with rival bacteria. Our results show for the first time that in S. marcescens, the T6SS expression levels can be modulated at the transcriptional level. This regulation was found to be dependent on RcsB, the transcriptional regulator of the RcsCDB signal transduction system. We determined that RcsB is able to directly interact with a conserved RcsB-binding motif present in the promoter region of the T6SS gene cluster, governing the expression levels of both vgrG and hcp, as determined by quantitative RT-PCR assays. Taking into account previous work indicating that the S. marcescens T6SS gene cluster codes for one transcriptional unit (16), these results showed that RcsB controls S. marcescens’ T6SS expression. Detection of secreted Hcp in Serratia strains whose genetic backgrounds lead to the inhibition or activation of RcsB corroborated the idea that RcsB-dependent regulation affects the expression and assembly of Serratia T6SS. Interestingly, by in silico analysis, we found that the RcsB box motif has been highly conserved in the T6SS promoter region of a wide variety of Serratia strains, isolated either from hosts or ambient sources, indicating that a strong selective pressure might have evolutionarily preserved RcsB-dependent regulatory control over Serratia T6SS expression.
When S. marcescens strain RM66262 and its close relative S. marcescens strain Db10 were assayed in intraspecific competition experiments, an rcsB mutant strain displayed a killing deficiency similar to the one determined for a tssM mutant strain, which is unable to assemble the T6SS. This demonstrates that functional RcsB is necessary for Serratia to exhibit its killing ability against a kin competitor.
The results obtained while monitoring transcriptional expression from the RM66262 T6SS promoter during competition against Db10 clearly showed that T6SS expression is upregulated upon bacterial encounter. Furthermore, we demonstrated that this induction is RcsB dependent and occurs provided the two rival strains simultaneously hold a functional T6SS. These results reveal that, in Serratia-versus-Serratia antagonism, a strain activates the RcsB-dependent transcriptional expression and subsequent assembly of its T6SS weaponry when it detects signals of an aggressive T6SS+ opponent. It is tempting to associate this observed intraspecies behavior with the so-called “tit-for-tat” type of counterattack model originally described for Pseudomonas by Basler et al. (8). In that work, it was demonstrated that Pseudomonas T6SS assembly and counterpart killing were activated by signals elicited by the opponent’s T6SS apparatus, deploying the TagQRST/PppA-PpkA posttranslational signal/response cascade (it is pertinent to mention that Serratia has no TagQRST protein homologues encoded in its genome, and we found no T6SS transcription or Hcp secretion alterations in an S. marcescens pppA strain). However, we show herein that S. marcescens exhibits a transcriptional mode of competition strategy in which the signal elicited by the encounter with a T6SS+ contender enables the induced bacteria to target contiguous, otherwise harmless T6SS− bacteria that share the same niche. In contrast to the tit-for-tat action, the S. marcescens strategy allows the menaced bacteria to display nonspatially oriented targeting of bystander preys.
Therefore, the competition-dependent S. marcescens T6SS firing behavior differs from that in previous reports postulating that, in S. marcescens Db10, the assembly of the T6SS is stochastic and ready for a circumstantial attack, irrespective of whether the encounter is with either aggressive T6SS+ or innocuous T6SS− bacteria (22). These apparently contradictory results can be attributed to dissimilarities in the sets of genes encoding T6SS and, therefore, in the interplay among the particular array of effectors and immunity proteins expressed by each S. marcescens strain used for competition assays. Consequently, different Serratia strains would, in turn, generate distinct, specific signals for each encounter. We favor the hypothesis that Serratia expresses basal (or constitutive) T6SS levels, which depend on RcsB, and that these basal levels are sufficient to eliminate competitor bacteria devoid of T6SS effectors and immunity defenses, such as E. coli strain K-12-derived strains. This is based on the results shown in Fig. S3 in the supplemental material, demonstrating that T6SS expression in nonchallenged, planktonic Serratia cells is dependent on RcsB, and in Fig. 5A, showing that when Serratia cells encounter E. coli MC4100 cells, either a wild-type or a tssM S. marcescens strain displays RcsB-dependent T6SS expression levels that are equivalent to those obtained with an unchallenged Serratia strain.
We also determined that wild-type Serratia (either S. marcescens RM66262 or Db10) is not able to kill a tssM strain, a result that puts forth Serratia’s inability to detect and fire the T6SS machinery when an apparently harmless T6SS− kin strain is encountered. In contrast to the results for S. marcescens RM66262 upon encountering E. coli, wild-type RM66262 or a tssM or rcsB mutant displayed an equivalent inability to kill a S. marcescens Db10 tssM mutant competitor, demonstrating that in an intraspecies encounter, only a T6SS+ prey is able to be detected by the aggressor and trigger T6SS upregulation in a RcsB-dependent manner. Serratia strains defective in T6SS assembly would, however, be protected against constitutive levels of T6SS effectors of its kin rival by the exposure of immunity proteins that specifically neutralize the lethal action of cognate effectors. In agreement with this result, both a tssM and an rcsB Serratia rival were equivalently unable to upregulate a wild-type S. marcescens strain’s expression of T6SS. Up to this moment, the comparison between S. marcescens RM66262 and Db10 genomes shows 12 proteins with amino acid sequence homology that are identified as T6SS effectors or immunity proteins (Fig. S8A). However, this is far from being an exhaustive analysis. Apart from those that do not have apparent counterparts, unassigned gene sequences that might code for T6SS effectors and/or immunity protein pairs or orphan ones in both strains could be found in the future. Taking into account that killing assays were carried out in a 5:1 attacker-to-prey relationship, we can conclude that upon encountering danger signals from S. marcescens T6SS+ opponents, even when partially sharing a similar set of effectors and immunity proteins, the expression of the T6SS system of a Serratia prey strain is transcriptionally upregulated over basal expression levels by RcsB in an attempt to counteract an enemy whose killing capacity outweighs the neutralizing ability of the prey.
(A) Schematic representation of the in silico comparison between S. marcescens Db10 and S. marcescens RM66262 gene clusters coding for T6SS effectors and immunity proteins. (B) Neither the PhoP/PhoQ nor the CpxRA system exerts a regulatory effect on T6SS expression. (C) Representative confocal z-slices of a competition assay between A. nosocomialis M2 and S. marcescens RM66262 wt/ppromT6SS. Download FIG S8, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
On the other hand, when we examined the interspecific interaction of S. marcescens with Acinetobacter strains, we found that either A. baumannii ATCC 17978 or A. nosocomialis M2 was able to efficiently kill S. marcescens strains. Strikingly, Serratia was unable to kill Acinetobacter, even when a 5:1 ratio was used, indicating a clear dominancy of Acinetobacter over Serratia under the conditions tested.
Previous results showed that in A. baumannii, a vgrG1 mutant strain is impaired for Hcp secretion, while individual mutant strains with mutations in the other three vgrG genes (vgrG2, vgrG3, and vgrG4) do not display significant effects on Hcp secretion levels (13). As expected, an A. baumannii vgrG1 strain was as attenuated as the tssM mutant strain in its capacity to kill Serratia. Moreover, while wild-type A. baumannii induced S. marcescens’ T6SS promoter transcription 400-fold, neither a tssM nor a vgrG1 strain was able to upregulate its expression. A similar result was obtained when A. baumannii vgrG2 and vgrG3 strains were assayed. An estimated 100-fold induction was detected when a Serratia rcsB mutant was assayed as prey, suggesting that in this harsh encounter, an RcsB-independent, unknown regulatory mechanism is also put into play, partially contributing to the induction of Serratia’s T6SS expression.
The combinatorial effect of vgrG2, vgrG3, and vgrG4 mutations indicated that Tde (a putative nuclease) and Tse (with unknown activity), the two cognate effectors whose secretion is facilitated by VgrG2 and VgrG3, respectively, were simultaneously required for killing and also for stimulating S. marcescens’ T6SS expression. Indeed, a mutant strain with a single Tse3 gene mutation was unable to either kill S. marcescens or upregulate its T6SS. Importantly, the fact that either double vgrG2 vgrG3 or triple vgrG2 vgrG3 vgrG4 mutant strains that fully assemble A. baumannii T6SS (13) were not able to upregulate the T6SS expression of S. marcescens allows us to rule out the idea that mere contact with an Acinetobacter T6SS apparatus that is assembled but incapable of translocating toxic effectors can constitute a signal for activating S. marcescens T6SS expression. It is worth emphasizing that A. nosocomialis strain M2 was found to be more aggressive than A. baumannii against Serratia and that a higher killing capacity correlated with an enhanced ability to activate Serratia’s T6SS expression, by up to 1,300-fold. This observation indicated that the extent of Serratia’s T6SS transcriptional induction is linked to the opponent’s capability of damaging the prey.
The RcsCDB phosphorelay is exclusively present in Enterobacteriaceae (24) and is known to act as a global regulatory network, controlling multiple cellular pathways, including capsule synthesis, cell division, motility, biofilm formation, and virulence mechanisms (25). The identity of specific signal(s) that activate the RcsCDB phosphorelay has remained elusive. However, a variety of environmental stimuli (high osmolarity and chlorpromazine treatment) and genetic mutations (igaA, dsbA, rfa, wec, mdoH, and opg) and the overexpression of certain proteins (DjlA, LolA, and OmpG) promote changes in the properties of the cell envelope, inducing the activity of the system in diverse bacterial species (25, 27). Nevertheless, we show that diverse agents, used in sublethal concentrations but causing measurable damage to either the outer (polymyxin B and sodium deoxycholate) or inner (SDS) membrane of S. marcescens, are not able to induce T6SS transcription in S. marcescens. This clearly indicates that cellular alterations generated by specific effectors are the source of the signal or signals able to trigger RcsB-mediated induction of Serratia T6SS promoter activity. In other enterobacteria, the Rcs and Cpx systems both respond to envelope stress, and their regulons partially overlap (39). As well, in Edwardsiella tarda, the PhoP/PhoQ system was shown to regulate T6SS expression (40). Our assays also indicate that neither the Cpx nor the PhoP/PhoQ system affects T6SS expression levels (Fig. S8B).
In Pseudomonas aeruginosa, LeRoux et al. (20) have shown that, upon killing by rival bacteria, the surviving subpopulation of Pseudomonas is able to detect its siblings’ lysis products by the Gac/Rsm signaling pathway and, in a posttranscriptional response, mounts the T6SS-dependent attack. Our assays showed that exposure of the T6SS reporter strain to Serratia cellular lysates, after-killing diffusible products, or cellular self components liberated in situ during the competition assay was not capable of triggering the T6SS promoter activity.
Our results converge to demonstrate that, in S. marcescens, the specific action of either homologous or heterologous T6SS-derived effectors is recognized as a trigger signal and transduced to activate RcsB. In turn, activated RcsB interacts with a conserved recognition motif within the promoter region of T6SS and upregulates the expression of the killing machinery. As schematized in Fig. 7, we propose a model of action for the S. marcescens T6SS in which the extent of damage commands the response mechanism that is put into play. Challenge with E. coli MC4100, a natural T6SS− strain that is efficiently killed by Serratia, was unable to induce the reporter’s transcriptional activity, indicating that a basal, uninduced level of T6SS expression is efficacious enough to get rid of defenseless rivals. When T6SS+ S. marcescens undergoes competition from kin T6SS+ S. marcescens, the extent of injury can be balanced by both shared offensive effectors and protective immunity proteins. This represents a moderate survival threat for the Serratia populations involved that would lead to one strain dominating, depending on the initial attacker-prey ratio. In this response, the cellular damage is weak and so is the RcsB-mediated induction of T6SS expression. A predator like Acinetobacter that does not discriminate T6SS+ or T6SS− rivals exerts overwhelming cellular harm to S. marcescens in competition at a 5:1 ratio. The cellular damage inflicted by Acinetobacter under this condition cannot be countered by Serratia immunity proteins or halted by the killing action of Serratia effectors. Indeed, in a T6SS-deficient (tssM) S. marcescens strain, the RcsB-dependent induction is also triggered by an encounter with Acinetobacter. As a consequence, the RcsB-dependent response is amplified and T6SS expression is enhanced to high levels in an attempt to fight an antagonist that is a large-scale threat to the Serratia population’s survival. We predict that this response would be advantageous for Serratia in complex polymicrobial natural environments in which aggressor-prey ratios could be quite distinct from the conditions used in the experiments described herein. One nonlethal activating signal from a competitor would boost a Serratia survivor subpopulation with an improved capacity to fight other surrounding rivals.
FIG 7 .
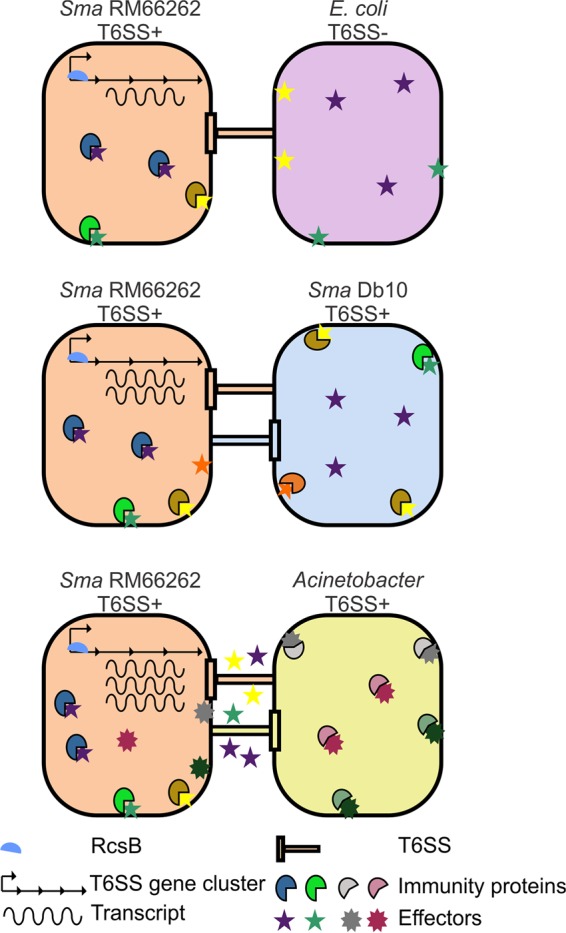
Schematic representation of the proposed model for the regulatory mechanism of the S. marcescens RM66262 T6SS (see Discussion for a full description).
Taken together, our results allow us to postulate that the detection-response system deployed by Serratia would enable this bacterium to weigh population damage versus energy expenditure at the time of expressing a complex machinery like the T6SS. In addition, the understanding of how this pathogen finely tunes the expression of its antibacterial armory will open new avenues for strategies that can be deployed to restrain Serratia infections.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The strains, plasmids, and primers used in this study are listed in Table S1 in the supplemental material.
Bacterial strains, plasmids, and primers used in this study. Download TABLE S1, DOCX file, 0.04 MB (47.7KB, docx) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antibacterial competition assays.
Competition assays were performed as described previously (16), with modifications as follows. Bacterial cells grown overnight were normalized to an optical density at 600 nm (OD600) of 0.5 and mixed at a 5:1 or 10:1 (attacker/target) ratio as indicated in each figure legend. Twenty-five microliters of this mixture was spotted onto a prewarmed agar plate and incubated at 37°C for 4 h or 6 h, as indicated. Cells were recovered from the spot and resuspended in 1 ml LB broth. Serial dilutions were plated out on antibiotic selection medium, using streptomycin for E. coli strain MC4100, tetracycline for S. marcescens strain Db10, ampicillin for S. marcescens strain RM66262, and chloramphenicol for A. baumannii strain ATCC 17978. Controls consisted of E. coli strain DH5α mixed with target bacteria at a 5:1 ratio. The recovery of viable cells is reported as the total number recovered per coculture spot. The results for each experiment are the average values of an assay performed in triplicate and independently repeated at least four times.
Antibacterial competition assay with GFP transcriptional reporter.
The competition assays with a GFP transcriptional reporter were performed as described above, with modifications as follows. The target or attacker strain carried the ppromT6SS construct as indicated. Controls consisted of E. coli DH5α mixed with target bacteria at a 5:1 ratio. Serial dilutions were plated out on kanamycin or ampicillin for selection of the reporter strain. Cells were washed three times with phosphate-buffered saline (PBS), and GFP fluorescence (excitation [λexc] at 485 nm and emission [λem] at 528 nm) was determined using a 96-microwell plate reader (Synergy2). Transcriptional induction was calculated as fluorescence divided by CFU. The fold change in transcriptional induction was calculated relative to the value obtained for the control or for the wild-type strain as indicated. We verified by simultaneous staining with propidium iodide that in competition assays, GFP fluorescence immediately extinguishes in dead bacteria and, therefore, GFP fluorescence detection corresponds only to live cells (see Fig. S8C and Text S1). The results for each experiment are the average values of an assay performed in triplicate and independently repeated at least three times.
Supplemental methods are provided. Download TEXT S1, DOCX file, 0.1 MB (68.9KB, docx) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistical analysis.
One-way analysis of variance (ANOVA) and the Tukey-Kramer multiple-comparison test with an overall significance level of 0.05 were used. In the figures, asterisks denote the values among the treatment groups in which a statistically significant difference was determined.
ACKNOWLEDGMENTS
We are grateful to Rodrigo Vena and Marina Avecilla for excellent technical assistance. We thank Alejandro J. Vila for the kind gift of nitrocefin. We are grateful to Brent Weber, Juvenal López, and Gisela Di Venanzio for technical support in the construction of Acinetobacter strains. We thank Fernando C. Soncini, Brent Weber, Seth Hennon, and Pek Man Ly for useful advice during the preparation of the manuscript. We also thank the anonymous reviewers that contributed with their suggestions to improve the quality of the manuscript.
E.G.V. is a Career Investigator of Consejo de Investigaciones Científicas y Tecnológicas (CONICET), Argentina. M.L. has a fellowship from CONICET and from Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), Argentina. This work was supported by a grant from ANPCyT, grant number PICT 2012-1403, to E.G.V. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Lazzaro M, Feldman MF, García Véscovi E. 2017. A transcriptional regulatory mechanism finely tunes the firing of type VI secretion system in response to bacterial enemies. mBio 8:e00559-17. https://doi.org/10.1128/mBio.00559-17.
REFERENCES
- 1.WHO 27 February 2017. WHO publishes list of bacteria for which new antibiotics are urgently needed. News release. http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/.
- 2.Gastmeier P. 2014. Serratia marcescens: an outbreak experience. Front Microbiol 5:81. doi: 10.3389/fmicb.2014.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimont PA, Grimont F. 1978. The genus Serratia. Annu Rev Microbiol 32:221–248. [DOI] [PubMed] [Google Scholar]
- 4.Mahlen SD. 2011. Serratia infections: from military experiments to current practice. Clin Microbiol Rev 24:755–791. doi: 10.1128/CMR.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, Poulain D, Sendid B, Ghannoum MA. 2016. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio 7:e01250-16. doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A 106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. 2013. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang YW, Rettberg LA, Ortega DR, Jensen GJ. 2017. In vivo structures of an intact type VI secretion system revealed by electron cryotomography. EMBO Rep 18:1090–1099. doi: 10.15252/embr.201744072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vettiger A, Basler M. 2016. Type VI secretion system substrates are transferred and reused among sister cells. Cell 167:99-110.e12. doi: 10.1016/j.cell.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 13.Weber BS, Hennon SW, Wright MS, Scott NE, de Berardinis V, Foster LJ, Ayala JA, Adams MD, Feldman MF. 2016. Genetic dissection of the type VI secretion system in Acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. mBio 7:e01253-16. doi: 10.1128/mBio.01253-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iguchi A, Nagaya Y, Pradel E, Ooka T, Ogura Y, Katsura K, Kurokawa K, Oshima K, Hattori M, Parkhill J, Sebaihia M, Coulthurst SJ, Gotoh N, Thomson NR, Ewbank JJ, Hayashi T. 2014. Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol Evol 6:2096–2110. doi: 10.1093/gbe/evu160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.English G, Trunk K, Rao VA, Srikannathasan V, Hunter WN, Coulthurst SJ. 2012. New secreted toxins and immunity proteins encoded within the type VI secretion system gene cluster of Serratia marcescens. Mol Microbiol 86:921–936. doi: 10.1111/mmi.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. 2011. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol 193:6057–6069. doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.English G, Byron O, Cianfanelli FR, Prescott AR, Coulthurst SJ. 2014. Biochemical analysis of TssK, a core component of the bacterial type VI secretion system, reveals distinct oligomeric states of TssK and identifies a TssK-TssFG subcomplex. Biochem J 461:291–304. doi: 10.1042/BJ20131426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srikannathasan V, English G, Bui NK, Trunk K, O’Rourke PE, Rao VA, Vollmer W, Coulthurst SJ, Hunter WN. 2013. Structural basis for type VI secreted peptidoglycan DL-endopeptidase function, specificity and neutralization in Serratia marcescens. Acta Crystallogr D Biol Crystallogr 69:2468–2482. doi: 10.1107/S0907444913022725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu F, Schwarz S, Mougous JD. 2009. TagR promotes PpkA-catalysed type VI secretion activation in Pseudomonas aeruginosa. Mol Microbiol 72:1111–1125. doi: 10.1111/j.1365-2958.2009.06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeRoux M, Kirkpatrick RL, Montauti EI, Tran BQ, Peterson SB, Harding BN, Whitney JC, Russell AB, Traxler B, Goo YA, Goodlett DR, Wiggins PA, Mougous JD. 2015. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. Elife 4:e05701. doi: 10.7554/eLife.05701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fritsch MJ, Trunk K, Diniz JA, Guo M, Trost M, Coulthurst SJ. 2013. Proteomic identification of novel secreted antibacterial toxins of the Serratia marcescens type VI secretion system. Mol Cell Proteomics 12:2735–2749. doi: 10.1074/mcp.M113.030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerc AJ, Diepold A, Trunk K, Porter M, Rickman C, Armitage JP, Stanley-Wall NR, Coulthurst SJ. 2015. Visualization of the Serratia type VI secretion system reveals unprovoked attacks and dynamic assembly. Cell Rep 12:2131–2142. doi: 10.1016/j.celrep.2015.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruna RE, Revale S, García Véscovi E, Mariscotti JF. 2015. Draft whole-genome sequence of Serratia marcescens strain RM66262, isolated from a patient with a urinary tract infection. Genome Announc 3:e01423-15. doi: 10.1128/genomeA.01423-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang YH, Ferrières L, Clarke DJ. 2006. The role of the Rcs phosphorelay in Enterobacteriaceae. Res Microbiol 157:206–212. doi: 10.1016/j.resmic.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 26.Castelli ME, Fedrigo GV, Clementín AL, Ielmini MV, Feldman MF, García Véscovi E. 2008. Enterobacterial common antigen integrity is a checkpoint for flagellar biogenesis in Serratia marcescens. J Bacteriol 190:213–220. doi: 10.1128/JB.01348-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castelli ME, Véscovi EG. 2011. The Rcs signal transduction pathway is triggered by enterobacterial common antigen structure alterations in Serratia marcescens. J Bacteriol 193:63–74. doi: 10.1128/JB.00839-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Venanzio G, Stepanenko TM, García Véscovi E. 2014. Serratia marcescens ShlA pore-forming toxin is responsible for early induction of autophagy in host cells and is transcriptionally regulated by RcsB. Infect Immun 82:3542–3554. doi: 10.1128/IAI.01682-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Venanzio G, Lazzaro M, Morales ES, Krapf D, García Véscovi E. 2017. A pore-forming toxin enables Serratia a nonlytic egress from host cells. Cell Microbiol 19:e12656. doi: 10.1111/cmi.12656. [DOI] [PubMed] [Google Scholar]
- 30.McMahon KJ, Castelli ME, Garcia VE, Feldman MF. 2012. Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can be induced by activation of the Rcs phosphorelay system 7. J Bacteriol 194:3241–3249. doi: 10.1128/JB.00016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller WG, Leveau JH, Lindow SE. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol Plant Microbe Interact 13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 32.Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. 2015. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci U S A 112:9442–9447. doi: 10.1073/pnas.1502966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcoforado Diniz J, Coulthurst SJ. 2015. Intraspecies competition in Serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J Bacteriol 197:2350–2360. doi: 10.1128/JB.00199-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arias M, Vogel HJ. 2017. Fluorescence and absorbance spectroscopy methods to study membrane perturbations by antimicrobial host defense peptides. Methods Mol Biol 1548:141–157. doi: 10.1007/978-1-4939-6737-7_10. [DOI] [PubMed] [Google Scholar]
- 36.Su LH, Ou JT, Leu HS, Chiang PC, Chiu YP, Chia JH, Kuo AJ, Chiu CH, Chu C, Wu TL, Sun CF, Riley TV, Chang BJ, Infection Control Group . 2003. Extended epidemic of nosocomial urinary tract infections caused by Serratia marcescens. J Clin Microbiol 41:4726–4732. doi: 10.1128/JCM.41.10.4726-4732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu YM, Hsu PC, Yang CC, Chang HJ, Ye JJ, Huang CT, Lee MH. 2013. Serratia marcescens meningitis: epidemiology, prognostic factors and treatment outcomes. J Microbiol Immunol Infect 46:259–265. doi: 10.1016/j.jmii.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Ye JJ, Lin HS, Yeh CF, Wu YM, Huang PY, Yang CC, Huang CT, Lee MH. 2016. Tigecycline-based versus sulbactam-based treatment for pneumonia involving multidrug-resistant Acinetobacter calcoaceticus-Acinetobacter baumannii complex. BMC Infect Dis 16:374. doi: 10.1186/s12879-016-1717-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rowley G, Spector M, Kormanec J, Roberts M. 2006. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol 4:383–394. doi: 10.1038/nrmicro1394. [DOI] [PubMed] [Google Scholar]
- 40.Chakraborty S, Li M, Chatterjee C, Sivaraman J, Leung KY, Mok YK. 2010. Temperature and Mg2+ sensing by a novel PhoP-PhoQ two-component system for regulation of virulence in Edwardsiella tarda. J Biol Chem 285:38876–38888. doi: 10.1074/jbc.M110.179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey TL, Elkan C. 1995. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3:21–29. [PubMed] [Google Scholar]
- 42.Bailey TL, Gribskov M. 1998. Methods and statistics for combining motif match scores. J Comput Biol 5:211–221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The RcsB-binding motif is conserved in the Serratia T6SS promoter region. Download FIG S1, PDF file, 2.3 MB (2.4MB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hcp densitometry. Download FIG S2, PDF file, 0.8 MB (840.8KB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RcsB transcriptionally modulates T6SS expression. Download FIG S3, PDF file, 0.6 MB (657.2KB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Serratia-induced T6SS can target preys in a nondirectional mode. Download FIG S4, PDF file, 1.8 MB (1.8MB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Serratia-Acinetobacter interspecific competition assays. Download FIG S5, PDF file, 1.5 MB (1.5MB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial lysis or derived products of T6SS-provoked killing are unable to activate S. marcescens T6SS expression. Download FIG S6, PDF file, 1 MB (1MB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nonspecific envelope damage does not activate T6SS expression. Download FIG S7, PDF file, 1 MB (1MB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Schematic representation of the in silico comparison between S. marcescens Db10 and S. marcescens RM66262 gene clusters coding for T6SS effectors and immunity proteins. (B) Neither the PhoP/PhoQ nor the CpxRA system exerts a regulatory effect on T6SS expression. (C) Representative confocal z-slices of a competition assay between A. nosocomialis M2 and S. marcescens RM66262 wt/ppromT6SS. Download FIG S8, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains, plasmids, and primers used in this study. Download TABLE S1, DOCX file, 0.04 MB (47.7KB, docx) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental methods are provided. Download TEXT S1, DOCX file, 0.1 MB (68.9KB, docx) .
Copyright © 2017 Lazzaro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.