ABSTRACT
Salmonella enterica serovar Typhimurium genome encodes 13 fimbrial operons. Most of the fimbriae encoded by these operons are not produced under laboratory conditions but are likely to be synthesized in vivo. We used an in vivo expression technology (IVET) strategy to identify four fimbrial operons, agf, saf, sti, and stc that are expressed in the spleen. When any three of these operons were deleted, the strain retained wild-type virulence. However, when all four operons were deleted, the resulting strain was completely attenuated, indicating that these four fimbriae play functionally redundant roles critical for virulence. In mice, oral doses of as low as 1 × 105 CFU of the strain with four fimbrial operons deleted provided 100% protection against challenge with 1 × 109 CFU of wild-type S. Typhimurium. We also examined the possible effect of these fimbriae on the ability of a Salmonella vaccine strain to deliver a guest antigen. We modified one of our established attenuated vaccine strains, χ9088, to delete three fimbrial operons while the fourth operon was constitutively expressed. Each derivative was modified to express the Streptococcus pneumoniae antigen PspA. Strains that constitutively expressed saf or stc elicited a strong Th1 response with significantly greater levels of anti-PspA serum IgG and greater protective efficacy than strains carrying saf or stc deletions. The isogenic strain in which all four operons were deleted generated the lowest anti-PspA levels and did not protect against challenge with virulent S. pneumoniae. Our results indicate that these fimbriae play important roles, as yet not understood, in Salmonella virulence and immunogenicity.
KEYWORDS: Agf, Saf, Stc, Sti, fimbriae, in vivo expression, recombinant attenuated Salmonella vaccine
IMPORTANCE
Salmonella enterica is the leading cause of bacterial food-borne infection in the United States. S. Typhimurium is capable of producing up to 13 distinct surface structures called fimbriae that presumably mediate its adherence to surfaces. The roles of most of these fimbriae in disease are unknown. Identifying fimbriae produced during infection will provide important insights into how these bacterial structures contribute to disease and potentially induce protective immunity to Salmonella infection. We identified four fimbriae that are produced during infection. Deletion of all four of these fimbriae results in a significant reduction in virulence. We explored ways in which the expression of these fimbriae may be exploited for use in recombinant Salmonella vaccine strains and found that production of Saf and Stc fimbriae are important for generating a strong immune response against a vectored antigen. This work provides new insight into the role of fimbriae in disease and their potential for improving the efficacy of Salmonella-based vaccines.
INTRODUCTION
Bacterial pathogens produce adhesins, often associated with fimbrial structures on the cell surface, to facilitate their initial interactions with host tissues (1). The chromosome of Salmonella enterica serovar Typhimurium contains 13 fimbrial operons, agf (csg), bcf, fim, lpf, pef, saf, stb, stc, std, stf, sth, sti, and stj (2–4). While the functions of a few of these fimbriae, including type 1 fimbriae (Fim), have been characterized (1, 5), the functions of most fimbriae are unknown. This is due, in part, to the fact that only type 1 and Agf fimbriae are produced under laboratory growth conditions (6). Type 1 fimbriae are produced when cells are grown at 37°C, and Agf fimbriae are produced when cells are grown at 26°C (7). While it is possible that some of these other fimbriae may be required for life outside a host (8), it is likely that many play an as yet undiscovered role in host interactions.
The agf operon encodes thin aggregative fimbriae (9) in Salmonella, and these fimbriae were later found to be similar to the fibronectin-binding surface structure known as curli (10) originally described in Escherichia coli (11). Thin aggregative fimbriae (hereafter Agf fimbriae) and curli are not produced in vitro at 37°C (11). Production of Agf fimbriae is typically induced in laboratory settings by growing cells at 26°C. Pef fimbriae mediate adherence to the murine small intestine and are required for fluid accumulation in infant mice. Expression of pef genes is regulated by DNA methylation (12). Stf fimbriae share homology with MR/P fimbriae of Proteus mirabilis and E. coli Pap fimbriae (13), and expression of stfA is induced during infection of bovine ileal loops (14). Long polar fimbriae (Lpf) are important for colonization of Peyer’s patches in mice by mediating adherence to M cells (5). Lpf also plays a role in the early stages of biofilm formation on host epithelial cells (15) and is involved in intestinal persistence (16). Lpf synthesis is regulated by an on-off switch mechanism (phase variation) to avoid host immune responses (17).
Some S. enterica fimbriae have been shown to serve functions beyond those required for interactions at the intestinal mucosal surface. For example, the Agf fimbriae are required for biofilm formation in the gallbladder (18, 19). In addition, the Stg fimbriae of S. enterica serovar Typhi, required for adherence to epithelial cells, also serves to inhibit phagocytosis (20). In S. Typhimurium, most fimbriae are produced in vivo, since mice immunized with S. Typhimurium produce antibodies against fimbrial subunits AgfA, BcfA, FimA, LpfA, PefA, StbA, StcA, StdA, StfA, SthA, and StiA (6). Thus, it is likely that some of these uncharacterized fimbriae may be synthesized in extraintestinal tissues.
To investigate potential roles for S. Typhimurium fimbriae in the host, we utilized an in vivo expression technology (IVET) strategy (21). We identified four fimbrial operons that are actively expressed in the spleen, only one of which, agf, is synthesized during in vitro growth (at 26°C). We characterized the impact of deletion and constitutive expression of all four fimbriae on virulence and immunogenicity.
RESULTS
Identification of fimbrial operons expressed in the spleen by IVET.
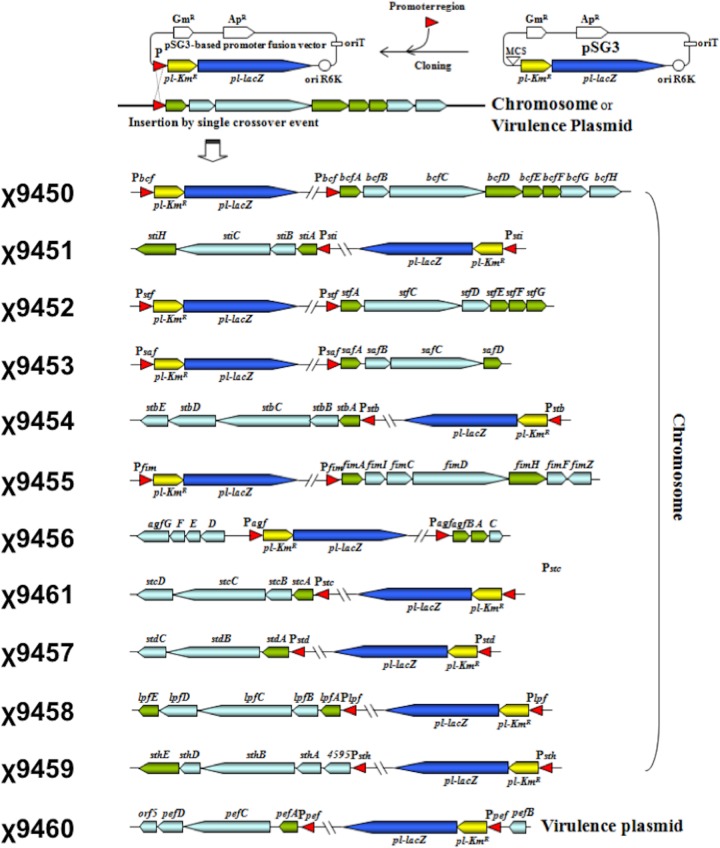
We constructed 12 S. Typhimurium strains, each harboring chromosomal transcriptional fusions of fimbrial promoter regions with aph lacZ reporter genes (Fig. 1). The stj operon is incomplete due to the apparent absence of any identifiable fimbrial subunit genes, so it was not included in our study (2). However, it is likely that this operon encodes a nonfimbrial or fibrillar structure (4). A mixture of all 12 fusion strains were orally administered to BALB/c mice. After infection, mice were treated orally and intraperitoneally with three doses of kanamycin to select for S. Typhimurium clones expressing the aph reporter gene in vivo. The experiment was performed twice, and 96 clones were obtained from pooled spleen samples in each experiment. Clones were identified by PCR using specific primers (see Table S2 in the supplemental material). In both experiments, we recovered the same four S. Typhimurium strains, strains χ9451, χ9453, χ9456, and χ9461, which contain aph lacZ reporter genes fused to the promoter regions of stiABCH, safABCD, agfBAC, and stcABCD operons, respectively (Table 1). Each of these strains was sensitive to kanamycin when grown at 37°C on LB agar plates, indicating that these four fimbrial operons are expressed in the mouse host.
FIG 1 .
Construction of pSG3-based aph lacZ fusions in S. Typhimurium fimbrial operons. Promoters were isolated as PCR products ranging from 271 to 391 bp and cloned into plasmid pSG3 to construct chromosomal fusions of each fimbrial operon promoter to aph. The resulting promoter fusions are illustrated. Strains are resistant to kanamycin only when the corresponding fimbrial promoter is active.
TABLE 1 .
Identification of S. Typhimurium fimbrial operons expressed in spleen using in vivo expression technology
| Strain | Relevant reporter fusion for IVET | No. of PCR-positive clones (%) detected in spleen after kanamycin treatment (n = 96) |
|
|---|---|---|---|
| Expt 1 | Expt 2 | ||
| χ9450 | Pbcf::pl-aph pl-lacZ | 0 | 0 |
| χ9451 | Psti::pl-aph pl-lacZ | 42 (44) | 10 (10) |
| χ9452 | Pstf::pl-aph pl-lacZ | 0 | 0 |
| χ9453 | Psaf::pl-aph pl-lacZ | 19 (20) | 59 (61) |
| χ9454 | Pstb::pl-aph pl-lacZ | 0 | 0 |
| χ9455 | Pfim::pl-aph pl-lacZ | 0 | 0 |
| χ9456 | Pagf::pl-aph pl-lacZ | 26 (27) | 7 (7) |
| χ9457 | Pstd::pl-aph pl-lacZ | 0 | 0 |
| χ9458 | Plpf::pl-aph pl-lacZ | 0 | 0 |
| χ9459 | Psth::pl-aph pl-lacZ | 0 | 0 |
| χ9460 | Ppef::pl-aph pl-lacZ | 0 | 0 |
| χ9461 | Pstc::pl-aph pl-lacZ | 5 (5) | 17 (18) |
| Unidentified | 4 | 3 | |
Virulence and immunogenicity of S. Typhimurium fimbrial mutants in BALB/c mice.
Because these fimbrial promoters are active in the spleen, we hypothesized that the fimbriae may be important for virulence. Therefore, we constructed strains harboring single and multiple deletions of the four fimbrial operons, ΔstiABCH1225, ΔsafABCD31, Δ(agfC-agfG)-999, and ΔstcABCD36. BALB/c mice were orally administered graded doses of bacteria and monitored for 4 weeks. All single deletion mutants retained wild-type virulence (data not shown). Strains with any three of the four fimbrial operons deleted were also virulent (Table 2). In contrast, two independently constructed quadruple deletion mutants, χ11484 and χ11599, were fully attenuated, with no deaths or disease symptoms occurring at the highest dose tested (50% lethal dose [LD50] > ~1 × 109 to 2 × 109 CFU).
TABLE 2 .
Virulence of S. Typhimurium fimbrial mutants in BALB/c micea
| Strain | Relevant genotype | Oral LD50 (CFU) |
|
|---|---|---|---|
| Expt 1 | Expt 2 | ||
| χ3761 | Wild-type | 3.5 × 102 | NTb |
| χ11467 | Δ(agfC-agfG)-999 ΔsafABCD31 ΔstcABCD36 | 4.2 × 103 | 5.1 × 102 |
| χ11483 | ΔsafABCD31 ΔstiABCH1225 Δ(agfC-agfG)-999 | 1.0 × 103 | 4.1 × 102 |
| χ11505 | ΔsafABCD31 ΔstiABCH1225 ΔstcABCD36 | 5.6 × 102 | 4.0 × 102 |
| χ11507 | Δ(agfC-agfG)-999 ΔstiABCH1225 ΔstcABCD36 | 1.8 × 102 | 1.2 × 103 |
| χ11484 | ΔsafABCD31 ΔstiABCH1225 Δ(agfC-agfG)-999 ΔstcABCD36 | >1.2 × 109 | >2.0 × 109 |
| χ11599 | ΔsafABCD31 ΔstiABCH1225 ΔstcABCD36 Δ(agfC-agfG)-999 | >1.6 × 109 | >1.3 × 109 |
BALB/c mice were orally administered graded doses of the indicated strains and monitored for 4 weeks.
NT, not tested.
We evaluated the immunogenicity of one of the strains, χ11484, by determining its ability to confer protection against challenge with the virulent S. Typhimurium UK-1 strain χ3761. The mice used in the virulence assay (above), which received graded doses of strain χ11484 were challenged 4 weeks after immunization with S. Typhimurium χ3761 (Table 3). A control group was given sterile buffer. Protection was achieved at all doses, and all mice that were immunized with at least 1.4 × 105 CFU of χ11484 survived challenge with the virulent S. Typhimurium strain (Table 3). Even mice inoculated with a single dose of only 8.4 × 102 CFU were partially protected, indicating that this avirulent S. Typhimurium fimbrial quadruple mutant is highly immunogenic.
TABLE 3 .
Immunogenicity of S. Typhimurium fimbrial quadruple mutant in BALB/c micea
| Strain | Dose of χ11484 (CFU) |
No. of mice alive after inoculation with χ11484/total no. |
No. of mice alive after challenge with χ3761/ total no. (% survival) |
|---|---|---|---|
| χ11484 | 1.4 × 109 | 6/6 | 6/6 (100) |
| 1.4 × 107 | 6/6 | 6/6 (100) | |
| 1.4 × 105 | 6/6 | 6/6 (100) | |
| 8.4 × 102 | 6/6 | 4/6 (67) | |
| None (control) | 0/3 (0) | ||
BALB/c mice were immunized orally with the indicated dose of strain χ11484 (all mice survived) and challenged 4 weeks after immunization with ~1 × 109 CFU of S. Typhimurium wild-type strain (χ3761).
Colonization by S. Typhimurium fimbrial quadruple mutants.
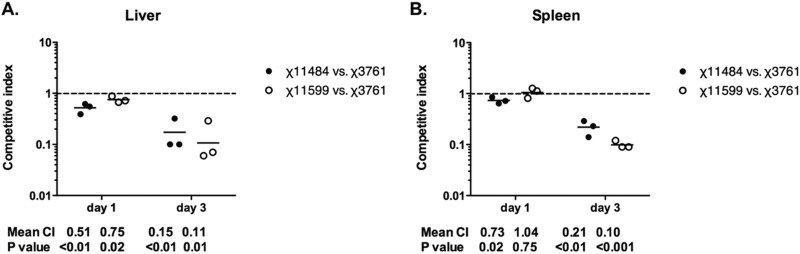
To evaluate the impact of the quadruple deletion on colonization, mice were orally inoculated with either strain χ11484 or strain χ11599. Peyer’s patches, spleens, and livers were harvested 5 days later, and the bacteria in each tissue were enumerated. Both quadruple mutants colonized all tested organs as well as wild-type χ3761 strain did (data not shown). To look more closely at spleen and liver colonization, we performed a competition assay. We chose to inoculate by the intraperitoneal route to eliminate any differences between strains that might be due to passage through the gastrointestinal tract. Thus, mice were inoculated parenterally with a mixture of S. Typhimurium wild-type χ3761 and either χ11484 or χ11599. Each strain was marked with a stable low-copy-number chloramphenicol-resistant plasmid (pHSG576) or kanamycin-resistant plasmid (pWSK129). Groups of mice were euthanized on days 1 and 3 postinfection. Samples of the spleens and livers were plated for enumeration of Salmonella. The total numbers of Salmonella recovered from each organ were consistent from mouse to mouse, between 104 and 106 CFU per g of tissue (data not shown). The ratio of the two strains in each organ was determined and compared to the input ratio to determine the competitive index (CI). On day 1 postinfection, there were no differences in spleen colonization between wild-type and mutant strains (Fig. 2B), while strain χ11484, but not χ11599, was outcompeted by the wild-type strain in the liver (Fig. 2A) (P < 0.01). By day 3, the wild type had outcompeted both quadruple mutants in both the spleen and liver (P < 0.05), indicating an important role for saf, sti, stc, and agf in colonization of the spleen and liver in mice. In preliminary competition experiments comparing single deletion mutants and the wild type, no significant differences were observed between strains (data not shown), indicating that no single fimbria is responsible for this phenotype.
FIG 2 .
Effect of saf sti stc agf quadruple deletion on the colonization of mouse liver (A) and spleen (B) by S. Typhimurium. The competitive indexes were determined from mixed intraperitoneal infection with S. Typhimurium wild-type strain (χ3761) and one of two fimbrial quadruple mutants (χ11484 and χ11599). Each symbol represents the value for an organ from an individual mouse at the indicated day following the infection. The geometric means of the competitive indexes (mean CI) and the P values from a Student’s t test are given below the graphs.
Recombinant attenuated S. Typhimurium vaccine (RASV) strains producing fimbriae (Saf+, Sti+, Stc+, and Agf+) in a constitutive manner.
Our results showing that the saf, sti, stc and agf fimbrial operons are expressed in vivo led us to speculate as to whether these fimbriae could be exploited to enhance the immunogenicity and protective efficacy of Salmonella vaccine strains. For this work, we constructed derivatives of attenuated S. Typhimurium strain χ9088 [ΔPfur33::TT araC PBAD fur Δpmi-2426 Δ(gmd-fcl)-26 ΔasdA33] (22) in which three fimbrial operons were deleted and the fourth was expressed from the constitutive PmurA promoter (23). Consequently, the resulting strains, strains χ11595, χ11850, and χ11851, have a genetic background that includes attenuating mutations, deletions in three fimbrial operons, and one deletion-insertion mutation (Table 4). The agf genes are expressed from two divergent operons, agfDEFG and agfBAC, necessitating a different strategy. In this case, we introduced the previously described agfD812 mutation (24) to drive constitutive expression of the agf operon. Strain χ12038 constitutively produced Agf fimbriae as indicated by the red, dry, and rough (rdar) colony morphology when grown on Congo red plates (7) at 37°C (data not shown). For a control, we also constructed strain χ11606, which harbors deletions of all four fimbrial operons (agf, saf, sti, and stc).
TABLE 4 .
Key bacterial strains and plasmids used in this study
| Bacterial strain or plasmid |
Relevant characteristic(s) | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1; used for general cloning |
51 |
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3); used for protein overproduction | Novagen |
| χ6212 | F− λ− φ80 Δ(lacZYA-argF) endA1 recA1 hsdR17 deoR thi-1 glnV44 gyrA96 relA1 ΔasdA4 | 52 |
| χ7213 | thi-1 thr-1 leuB6 glnV44 fhuA21 lacY1 recA1 RP4-2-Tc::Mu(λpir) ΔasdA4 Δ(zhf-2::Tn10) | 46 |
| S. Typhimurium strains | ||
| χ3761 | Wild-type UK-1 | 45 |
| χ9088 | Δpmi-2426 Δ(gmd-fcl)-26 ΔPfur33::TT araC PBAD fur ΔasdA33 | 22 |
| χ11467 | Δ(agfC-agfG)-999 ΔsafABCD31 ΔstcABCD36 | χ11466 |
| χ11483 | ΔsafABCD31 ΔstiABCH1225 Δ(agfC-agfG)-999 | χ11468 |
| χ11484 | ΔsafABCD31 ΔstiABCH1225 Δ(agfC-agfG)-999 ΔstcABCD36 | χ11483 |
| χ11505 | ΔsafABCD31 ΔstiABCH1225 ΔstcABCD36 | χ11468 |
| χ11507 | Δ(agfC-agfG)-999 ΔstiABCH1225 ΔstcABCD36 | χ11506 |
| χ11595 | Δpmi-2426 Δ(gmd-fcl)-26 ΔPfur33::TT araC PBAD
fur ΔasdA33 ΔsafABCD31 Δ(agfC-agfG)-999 ΔstcABCD36 ΔPstiA52::PmurA stiA52 |
χ11594 |
| χ11599 | ΔsafABCD31 ΔstiABCH1225 ΔstcABCD36 Δ(agfC-agfG)-999 | χ11505 |
| χ11606 | Δpmi-2426 Δ(gmd-fcl)-26 ΔPfur33::TT araC PBAD
fur ΔasdA33 ΔsafABCD31 ΔstiABCH1225 Δ(agfC-agfG)-999 ΔstcABCD36 |
χ11597 |
| χ11850 | Δpmi-2426 Δ(gmd-fcl)-26 ΔPfur33::TT araC PBAD
fur ΔasdA33 ΔstiABCH1225 Δ(agfC-agfG)-999 ΔstcABCD36 ΔPsafA55::PmurA safA55 |
χ11594 |
| χ11851 | Δpmi-2426 Δ(gmd-fcl)-26 ΔPfur33::TT araC PBAD
fur ΔasdA33 ΔsafABCD31 ΔstiABCH1225 Δ(agfC-agfG)-999 ΔPstcA53::PmurA stcA53 |
χ11597 |
| χ12038 | Δpmi-2426 Δ(gmd-fcl)-26 ΔPfur33::TT araC PBAD
fur ΔasdA33 ΔsafABCD31 ΔstiABCH1225 ΔstcABCD36 agfD812 |
χ11562 |
| S. pneumoniae strain WU2 | Wild-type; virulent; encapsulated type 3 | 41 |
| Plasmids | ||
| pSG3 | IVET vector; promoterless aph lacZ mobRP4; R6K ori; Apr Gmr | 43 |
| Plasmids used for production of recombinant proteins |
||
| pET28b | Expression vector; T7 promoter 6xHis lacI; f1 pBR ori; Kmr | Novagen |
| pYA4085 | pET30a derivative for overproduction of rPspA | 49 |
| pYA4088 | pYA3493 derivative for production of rPspA (amino acids 3 to 285) fused to β-lactamase signal sequence |
25 |
To study the ability of these strains to elicit protective immune responses against heterologous antigens in mice, we introduced plasmid pYA4088 (25), carrying the gene encoding the Streptococcus pneumoniae protein PspA, into each strain. This pneumococcal protein has been extensively studied by our group (26) and others (27, 28) and shown to elicit protective immunity against virulent S. pneumoniae challenge. For clarity, we will refer to these strains as χ11595(pYA4088) (Sti+), χ11850(pYA4088) (Saf+), χ11851(pYA4088) (Stc+), χ12038(pYA4088) (Agf+), and χ11606(pYA4088) (Δ4). All strains were grown to mid-log phase in LB with appropriate supplements. Western blot analysis with specific anti-recombinant PspA (anti-rPspA) antibodies showed that all strains produced similar amounts of PspA (Fig. S1).
Production of PspA antigen in S. Typhimurium RASV strains. Western blot showing whole-cell lysates obtained from mid-log-phase cultures, electrophoresed on a 12% SDS-polyacrylamide gel, transferred onto nitrocellulose, and probed with anti-rPspA serum. Download FIG S1, TIF file, 0.2 MB (258.4KB, tif) .
Copyright © 2017 Łaniewski et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antibody responses in mice immunized with RASV strains constitutively producing individual fimbriae (Saf+, Sti+, Stc+, and Agf+).
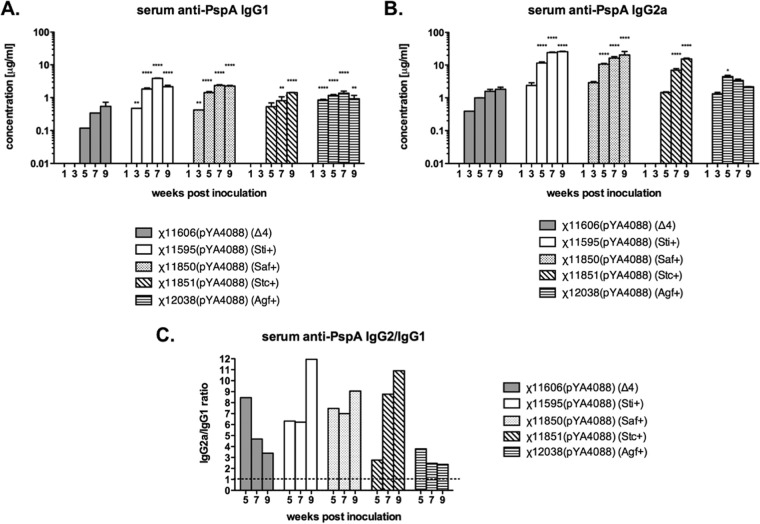
BALB/c mice were orally primed and boosted 6 weeks later with identical doses of ~1 × 108 CFU of each strain. A control group was given sterile buffer instead of vaccine. All mice immunized with RASVs expressing pspA produced anti-rPspA serum IgG1 (Fig. 3A) and IgG2a (Fig. 3B). No anti-rPspA IgG1 or IgG2a was detected in sera from control mice treated with phosphate-buffered saline (PBS). The anti-PspA serum IgG1 titers in all immunized mice were significantly higher than the titers in mice immunized with strain χ11606(pYA4088) (Δ4) by week 3 (Fig. 3A). The IgG2a subclass concentrations were also greater than in the χ11606(pYA4088) group in all cases except the group immunized with strain χ12038(pYA4088) (Agf+) (Fig. 3B). By week 9, IgG2a concentrations were 8- to 14-fold higher in mice immunized with strains χ11595(pYA4088) (Sti+), χ11850(pYA4088) (Saf+), and χ11851(pYA4088) (Stc+) than in mice immunized with strain χ11606(pYA4088) (Δ4) (P < 0.0001) (Fig. 3B). The IgG1 subclass concentrations for these three strains at week 9 were also elevated (2.6- to 4.2-fold) compared to those of χ11606(pYA4088) (Δ4) (P < 0.0001) (Fig. 3A). Comparing the anti-PspA IgG2a/IgG isotype ratios showed that immunization with each strain induced a mixed Th1/Th2 response, with a strong Th1 bias (Fig. 3C). At week 9, mice immunized with χ11595(pYA4088) (Sti+), χ11850(pYA4088) (Saf+), and χ11851(pYA4088) (Stc+) showed the highest IgG2a-to-IgG1 ratios, ranging from 9 to 12 (Fig. 3C). In contrast, mice immunized with χ11606(pYA4088) (Δ4) or χ12038(pYA4088) (Agf+) showed only two- to threefold differences in IgG2a-to-IgG1 titers. However, these differences were not statistically significant (P > 0.05).
FIG 3 .
Serum IgG1 and IgG2a responses to PspA in mice immunized with RASV strains expressing fimbriae in a constitutive manner (Saf+, Sti+, Stc+, and Agf+) and producing PspA antigen. The kinetics of serum IgG1 and IgG2a responses to PspA in mice are shown. The data represent the concentrations of anti-PspA IgG1 (A) and IgG2a (B) in pooled serum samples from eight mice measured in duplicate. Error bars show the differences between the duplicates (standard deviations). All samples from immunized mice were significantly different from those from the control group given PBS (P < 0.05). Values that are significantly different from the values for the S. Typhimurium χ11606(pYA4088) group are indicated by asterisks as follows: *, P < 0.05; **, P < 0.01, ****, P < 0.0001. (C) Calculated IgG2a/IgG1 ratios based on the data shown in panels A and B.
Protection of mice immunized with RASV strains constitutively producing individual fimbriae (Saf+, Sti+, Stc+, and Agf+) against S. pneumoniae challenge.
Four weeks after the boost, mice were injected intraperitoneally with ~40 times the LD50 of virulent S. pneumoniae strain WU2. Immunization with strain χ11850(pYA4088) (Saf+) and strain χ11851(pYA4088) (Stc+) provided the highest level of protection (52.6%) compared to nonimmunized control mice (P < 0.001) (Table 5). Strain χ11595(pYA4088) (Sti+) also provided significant protection against pneumococcal challenge (31.6%; P < 0.05). Vaccination with strain χ12038(pYA4088) (Agf+) resulted in 26.3% survival, but this result was not significantly different from that with the nonimmunized control group. In addition, mice vaccinated with strain χ11606(pYA4088) (Δ4) were not protected (10.5% survival). Two independent protection experiments were performed. All deaths occurred 4 to 6 days postinfection.
TABLE 5 .
Protective efficacy of RASV strains expressing fimbriae in a constitutive manner (Saf+, Sti+, Stc+, and Agf+) and producing PspA antigena
| Strain | Constitutively expressed fimbrial gene | No. of mice alive/total no. (% survival) |
||
|---|---|---|---|---|
| Expt 1 | Expt 2 | Combinedb | ||
| χ11595(pYA4088) | sti | 4/8 (50)c | 2/11 (18.2) | 6/19 (31.6)c |
| χ11850(pYA4088) | saf | 5/8 (62.5)d | 5/11 (45.5) | 10/19 (52.6)e |
| χ11851(pYA4088) | stc | 5/8 (62.5)d | 5/11 (45.5) | 10/19 (52.6)d |
| χ12038(pYA4088) | agf | 3/8 (37.5) | 2/11 (18.2) | 5/19 (26.3) |
| χ11606(pYA4088) (Δ4) | 1/8 (12.5) | 1/11 (9.1) | 2/19 (10.5) | |
| χ9088(pYA4088) | NTf | 3/11 (27.3) | 3/11 (27.3) | |
| None (PBS) (control) | 0/8 (0) | 0/11 (0) | 0/18 (0) | |
Seven-week-old BALB/c mice were immunized orally with ~1 × 108 CFU of the indicated of S. Typhimurium vaccine strains and boosted with the same dose 6 weeks later. All mice were challenged by intraperitoneal inoculation 4 weeks after the booster dose with ~1 × 104 CFU of virulent S. pneumoniae strain WU2. Deaths were recorded until 3 weeks postinfection.
Combined percent survival from two independent experiments.
Significantly different (P < 0.05) from value obtained for the control (PBS) group.
Significantly different (P < 0.01) from value obtained for the control (PBS) group.
Significantly different (P < 0.001) rom value obtained for the control (PBS) group.
NT, not tested.
DISCUSSION
Fimbrial genes are widely distributed among bacteria, but only a few fimbriae are produced under standard laboratory conditions. Most bacterial fimbriae serve to present adhesins that assist in the adherence of bacteria to biotic and abiotic surfaces (1) and are produced in response to the appropriate environmental cues. Of the 13 known fimbriae in S. Typhimurium, only two, type 1 fimbriae and curli (Agf) are readily produced when grown in the laboratory. However, in one study, cells were coaxed to produce Pef, Bcf, Stb, Stc, Std, and Sth fimbriae after static growth in CFA broth at 32°C and Agf, Pef, Lpf, Stc, Stf, and Sth fimbriae in LB at pH 5.1 at 37°C, although the levels were low, as fimbriae were detected on <7% of the cells by a highly sensitive flow cytometry method (14). In the same study, fimbrial expression was further enhanced to around 10% of cells after growth for 8 h in bovine ileal loops. Type 1 fimbriae were detected in >20% of the cells under all three growth conditions. Thus, it is likely that a majority of the known S. Typhimurium fimbrial operons are expressed inside a mammalian host.
In the current study, we demonstrated that sti, saf, stc, and agf fimbrial genes are actively expressed in the mouse spleen (Table 1). In vivo expression of these genes is consistent with a previous study in which CBA mice inoculated with S. Typhimurium developed antibodies against recombinant His-tagged StiA, StcA, and AgfA (6). The mice were not evaluated for antibody responses against Saf fimbrial components. In another study, mice were protected from challenge with S. Typhimurium after injection with a mixture of purified recombinant His-tagged SafB, a putative chaperone, and recombinant SafD, the Saf adhesin, both produced in E. coli (29, 30). In addition, transcription of saf fimbrial genes has been detected in blood samples from patients infected with S. Typhi (31) and S. Paratyphi A (32), supporting a role for these fimbriae in the human host.
SafA monomeric fimbriae were assembled in vitro in the presence of the chaperone protein SafB and crystallized (33). Subsequent crystallographic analysis showed that Saf fimbriae are composed of highly flexible fibers formed by globular subunits organized in a “beads on a string” arrangement (33). Characterization of the safABCD operon protein sequences suggest that SafA is the major structural protein, SafB is the periplasmic chaperone, and SafC is an outer membrane usher (29). The SafD protein is homologous to several other fimbrial adhesins and so is likely to be the Saf adhesin, believed to be present only at the tip of the fiber (29, 33). In addition, the major fimbrial protein, SafA, exhibits similarity to the λ phage-encoded Bor protein that has been implicated in serum resistance of λ-infected hosts (34). Thus, it is possible that the saf fimbriae play a role in serum resistance.
Our results with agf seem to run counter to a previous report. Using a bioluminescence imaging technique, White et al. showed that agfB was not expressed during infection (35). The authors concluded that Agf fimbriae are not produced in vivo. However, their observations were based on results obtained from a single time point, while in our study, the bacteria were under constant selective pressure for 3 days. Thus, it is possible that there is a temporal component to agfB expression. Our data suggesting the in vivo production of Agf is also supported by the study we cite above in which anti-AgfA antibodies were detected in S. Typhimurium-infected mice (6).
In a previous study, strains with either agfAB or stcABCD deleted exhibited wild-type levels of spleen colonization in genetically resistant CBA mice (16). Consistent with those results, we observed that strains in which any single fimbrial operon (agf, saf, stc, or sti) or combination of three operons was deleted had no effect on virulence, while deletion of all four fimbrial operons resulted in a complete loss of virulence when mutant strains were administered by the oral route to genetically sensitive BALB/c mice (Table 2). Our results suggest that these four fimbriae serve functionally redundant roles in mouse virulence. Interestingly, while a ΔstcABCD strain exhibits wild-type spleen colonization, it exhibits reduced fecal shedding, indicating a role for this fimbriae in long-term intestinal carriage (16).
Strain χ11484 with sti, saf, stc, and agf deleted was immunogenic, protecting mice from a high-dose challenge with wild-type S. Typhimurium after a single immunizing dose as low as 1.4 × 105 CFU (Table 3). We expanded our analysis of the roles of these genes in immunogenicity by examining the effect of constitutive production of each fimbriae individually in a previously characterized vaccine strain background (χ9088) in which we had also deleted the other three fimbrial operons. These vaccine strains were used to deliver the heterologous antigen, PspA. Our results indicate that constitutive production of Sti, Saf, or Stc, but not Agf, significantly enhanced protective immunity (Table 5), although they each had different impacts on the immune system.
Th1-type dominant immune responses are frequently observed after immunization with attenuated Salmonella (36), and most of the fimbrial deletion strains elicited a Th1-biased response. However, mice immunized with strain χ12038(pYA4088) (Agf+) produced more of a mixed Th1/Th2 humoral response, indicating that overproduction of Agf fimbriae resulted in a reduced ability to stimulate Th1 helper cells to direct IgG class switching to IgG2a (37). IgG2a is the isotype with the greatest capacity to mediate complement deposition onto the surfaces of bacteria, and an increase in anti-PspA IgG2a has been correlated with increased C3 deposition on the S. pneumoniae cell surface (38).
The immune responses to PspA were examined by measuring the levels of IgG isotype subclasses. The anti-PspA IgG2a titers were higher than the IgG1 titers in all groups, indicating that all of the Salmonella vaccines induced a Th1-biased response against PspA (Fig. 3). Strain χ11850(pYA4088) (Saf+) elicited high levels of anti-PspA IgG with a strong Th1 bias (Fig. 3). Thus, the strong Th1 responses observed in mice vaccinated with strain χ11850(pYA4088) (Saf+) can explain why this strain was highly protective (Fig. 3 and Table 3). Strain χ11595(pYA4088) (Sti+) produced a strong Th1 response by week 9 (Fig. 3A). In contrast, the strains that provided the weakest protection, χ11606(pYA4088) (Δ4) and χ12038(pYA4088) (Agf+), were deficient in either strong Th1-biased antibody responses.
The strong protection observed for mice immunized with strain χ11851(pYA4088) (Stc+) does not fit as neatly into this interpretation, as this strain did not elicit a strong Th1 response at early time points (Fig. 3). However, by week 9, this strain elicited the greatest IgG2a/IgG1 ratio (Fig. 3C), which may have provided a humoral response that was adequate to control the S. pneumoniae challenge. This result, along with the results for strain χ12038(pYA4088) (Agf+), which stimulated a low IgG2a/IgG1 ratio, indicates that production of IgG2a is the most important parameter for protection against pneumococcal challenge in this model.
Deletion of all four fimbrial operons in strain χ11484 resulted in complete attenuation (Table 2), while preserving its ability to elicit a protective response against challenge with wild-type S. Typhimurium at immunizing doses as low as 8.4 × 102 CFU (Table 3). In contrast, deletion of these same four fimbrial operons in the χ9088 background vectoring PspA compromised the ability of the strain to elicit protection against streptococcal challenge (Table 5). Since the Δ4 deletion is attenuating, combining these mutations with additional attenuating mutations could have resulted in overattenuation of the Salmonella vector strain, possibly due to a reduction in the ability of strain χ11606(pYA4088) to colonize the spleen or other lymphoid organs. While the basis of this overattenuation is not clear, it does indicate that one must carefully consider the background genotype before combining Δ4 with other attenuating mutations.
This study demonstrates that in vivo-induced fimbriae play a role in spleen colonization and may be used to augment the immunogenicity of orally administered, live attenuated Salmonella vaccines. This represents a novel strategy for modulating host immune responses to strengthen Th1-biased immune responses and enhance protective immunity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 4 and Table S1 in the supplemental material. Escherichia coli and Salmonella enterica serovar Typhimurium strains were routinely cultured at 37°C in LB broth (39) or on LB agar. Cultures of S. Typhimurium strain χ9088 (22) and its derivatives were supplemented with 0.05% mannose (for Δpmi-2426) and 0.2% arabinose (for ΔPfur33::TT araC PBAD fur). Diaminopimelic acid (DAP) (50 µg/ml) was added to LB medium for growing Δasd mutant strains. The following antibiotics were used as needed at the indicated concentrations: ampicillin, 100 μg/ml; chloramphenicol, 15 μg/ml; gentamicin, 20 μg/ml; kanamycin, 50 μg/ml; tetracycline, 10 μg/ml. Carbohydrate-free nutrient broth (NB) was used for growth when determining lipopolysaccharide (LPS) profiles. LB agar without sodium chloride and with 7.5% sucrose was employed for sacB-based counterselection. MacConkey agar plates with 1% mannose were used to indicate sugar fermentation.
For animal experiments, S. Typhimurium strains were grown in LB broth with appropriate supplements. Overnight cultures were diluted 1:100 and grown with shaking (200 rpm) to an optical density at 600 nm of ~0.8. Then, bacteria were centrifuged at 5,000 × g for 15 min at room temperature and resuspended in phosphate-buffered saline (PBS) or buffered saline with 0.01% gelatin (BSG) (40). LB or Salmonella Shigella (SS) agar plates were used to enumerate S. Typhimurium recovered from tissues. Selenite cystine broth was employed to enrich samples for S. Typhimurium. Streptococcus pneumoniae WU2 was cultured on brain heart infusion agar containing 5% sheep blood or in Todd-Hewitt broth with 0.5% yeast extract (41). All media, antibiotics, and chemicals were purchased from BD Difco (Franklin Lakes, NJ) or Sigma-Aldrich (St. Louis, MO).
General DNA procedures.
DNA manipulations, including plasmid and genomic DNA isolation, restriction enzyme digestions, ligations, and other DNA-modifying reactions, were conducted as described previously (42) or were performed according to the manufacturers’ instructions (New England Biolabs, Ipswich, MA; Qiagen, Valencia, CA; Promega, Madison, WI). Synthesis of primers (Table S2) and DNA sequencing were performed by Integrated DNA Technologies (Coralville, IA) and the DNA Laboratory at Arizona State University (Tempe, AZ), respectively. PCRs were conducted with Klentaq LA polymerase (DNA Polymerase Technology, St. Louis, MO), possessing proofreading activity. Recombinant plasmids were introduced into E. coli and S. Typhimurium cells by transformation and electroporation, respectively.
Construction of transcriptional aph-lacZ fusions.
DNA fragments containing the promoter regions of 12 fimbrial operons were amplified from the S. Typhimurium χ3761 genome by PCR using the appropriate primers (Table S2). The PCR products were digested with ApaI and BamHI and cloned into the unique ApaI/BamHI sites of aph-lacZ fusion suicide vector pSG3 (43). The resulting plasmids were introduced by conjugation into S. Typhimurium strain χ3761 to obtain fusions of selected promoter regions with aph-lacZ genes by a single-crossover event as previously described (43).
In vivo expression technology (IVET).
Each S. Typhimurium aph-lacZ fusion strain was grown statically in LB broth at 37°C for 20 h. Bacterial cells were harvested by centrifugation at 5,000 × g for 15 min at room temperature. The pellets were resuspended in BSG buffer. BALB/c mice were inoculated orally with ~1 × 109 CFU of the mixture of the 12 aph-lacZ fusion strains. Mice were treated with kanamycin at 3, 24, and 48 h postinoculation by oral administration (2 mg in 20 µl) and intraperitoneal (10 mg in 100 µl) injection. Three days after inoculation, the spleens were collected from the treated mice and homogenized. Dilutions of the homogenate were made in BSG and plated onto LB agar plates supplemented with gentamicin and incubated overnight at 37°C. Finally, selected clones were identified by PCR using specific primers (Table S2).
Construction of suicide plasmids for introduction of deletions or ΔPfimbrial operon::PmurA deletion/insertions of fimbrial operons.
To construct the ΔstiABCH1225, ΔsafABCD31, and ΔstcABCD36 deletions, two-step PCR mutagenesis was used. First, two DNA fragments flanking fimbrial operons were amplified from the S. Typhimurium χ3761 genome using appropriate primer sets: PstiF/PstiR (P stands for primer, F stands for forward, and R stands for reverse) and d-stiAH-F/d-stiAH-R (d stands for deletion) (for ΔstiABCH1225), PsafF/PsafR and d-safAD-F/d-safAD-R (for ΔsafABCD31), and PstcF/PstcR and d-stcAD-F/d-stiAD-R (for ΔstiABCH1225) (Table S2). Thereafter, the mixes of two PCR products flanking each fimbrial operon were used as the templates in the next amplification reactions with PstiF/d-stiAH-R, PsafF/d-safAD-R, and PstcF/d-stcAD-R primers, respectively. The DNA fragments obtained were digested with ApaI/SacI restriction enzymes and cloned into suicide plasmid vector pCHSUI-1. The resulting plasmids, pYA4584, pYA4586, and pYA5007, carried deletions of the entire stiABCH, safABCD, and stcABCD operons, respectively. Plasmids pYA3490 and pYA4941 for introduction of the agfD812 and Δ(agfC-agfG)-999 mutations were described previously (24, 44).
To construct the ΔPstiA52::PmurA stiA52, ΔPstcA53::PmurA stcA53, and ΔPsafA55::PmurA safA55 deletion/insertion mutations, two-step PCR mutagenesis was also used. DNA fragments containing the upstream regions of the stiA, stcA, and safA promoters were amplified from the S. Typhimurium χ3761 genome using PmurA-stiA-F/PmurA-stiA-R, PmurA-stcA-F/PmurA-stcA-R, and PmurA-safA-F/PmurA-safA-R primer pairs (Table S2), respectively. The PCR products were digested with BglII. A 65-bp murA promoter region was amplified from E. coli K-12 using primers Ec_PmurA-F and Ec_PmurA-R (Ec stands for E. coli). This PCR product was digested with BglII and NcoI. DNA fragments containing downstream regions of the stiA, stcA, and safA promoters were amplified from the S. Typhimurium χ3761 genome using PmurA-stiA-F1/PmurA-stiA-R1, PmurA-stcA-F1/PmurA-stcA-R1, and PmurA-safA-F1/PmurA-safA-R1 primer pairs, respectively. The PCR products were digested with NcoI. Two digested PCR products containing flanking regions of each fimbrial operon and a PCR product containing the murA promoter were combined by ligation and used as the templates for PCR to amplify the combined DNA fragments using PmurA-stiA-F/PmurA-stiA-R, PmurA-stcA-F/PmurA-stcA-R1, and PmurA-safA-F/PmurA-safA-R1 primer pairs, respectively. These final PCR products were digested with KpnI and SacI and cloned into the unique KpnI/SacI sites of suicide vector pRE112 to generate pYA5052, pYA5053, and pYA5054.
Construction of S. Typhimurium mutants.
All S. Typhimurium mutants were derived from the highly virulent parent strain χ3761 (45). The genealogy of constructed strains is shown in Table S1. All gene replacements were introduced by conjugational transfer of suicide plasmids using donor E. coli strain χ7213 (46). All mutations were verified by PCR. We confirmed arabinose-regulated Fur production by Western blotting. The Δpmi mutation was confirmed by white colony phenotype on mannose-MacConkey agar. Lipopolysaccharide (LPS) profiles were examined by silver staining of 12% polyacrylamide gels as described previously (47).
Additional strains and plasmids used in this study. Download TABLE S1, DOCX file, 0.03 MB (30.8KB, docx) .
Copyright © 2017 Łaniewski et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S2, DOCX file, 0.02 MB (20.1KB, docx) .
Copyright © 2017 Łaniewski et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SDS-PAGE and Western blotting.
SDS-PAGE and Western blotting were performed by standard techniques. The blots were developed with nitroblue tetrazolium chloride/5-bromo-4-chloro-3′-indolyl phosphate (Amresco, Solon, OH) or Pierce ECL Western blotting substrate (Thermo Scientific), using rabbit polyclonal anti-rPspA serum as primary antibodies and mouse anti-rabbit IgG alkaline phosphatase conjugate (Sigma-Aldrich) as secondary antibodies.
Animal supply and housing.
Female BALB/c mice (6 to 8 weeks old) were obtained from Charles River Laboratories (Wilmington, MA). Animals were allowed to acclimate for 1 week after arrival before starting the experiments. All animal procedures were carried out in compliance with the Institutional Animal Care and Use Committee (IACUC) at Arizona State University and the Animal Welfare Act.
Colonization of the mouse spleen and determination of the competitive index.
BALB/c mice were inoculated intraperitoneally with a mixture containing ~1 × 104 of S. Typhimurium wild-type strain (χ3761) and either strain χ11484 or strain χ11599 suspended in 100 μl of PBS. Wild-type and mutant strains were marked with low-copy-number chloramphenicol- or kanamycin-resistant plasmids: pHSG576 and pWSK129, respectively. On days 1 and 3 postinoculation, three mice in each group were euthanized, and the spleens and livers were collected to determine the colonization levels. The competitive index (CI) for each strain compared to the wild type was calculated by dividing the ratio of two strains from an organ divided by the same ratio in the suspension used for the infection.
Determination of the 50% lethal dose.
Freshly grown bacterial cultures were pelleted by centrifugation at 5,000 × g for 15 min at room temperature. Bacterial pellets were resuspended in BSG and adjusted to achieve a dose of ~102 to ~109 CFU in a volume of 20 μl for orally inoculating BALB/c mice. Animals were observed for typhoid symptoms for 3 weeks postinoculation. Deaths were recorded daily. The 50% lethal dose (LD50) was calculated using the Reed and Muench method (48).
Immunization and pneumococcal challenge.
BALB/c mice were inoculated orally with 20 μl of PBS containing ~1 × 108 CFU of the appropriate S. Typhimurium strain and boosted with the same strain and dose 6 weeks later. No food or water was provided for ~4 h prior to immunizations. Groups of mice inoculated with PBS served as a control. At week 10 (i.e., 4 weeks after the booster), all mice were challenged by intraperitoneal injection with ~1 × 104 CFU of S. pneumoniae WU2 in 100 µl of BSG (equivalent to 40 times the LD50). Mice were monitored daily for 3 weeks.
Antigen preparation and ELISA.
Recombinant PspA (rPspA) protein was purified from E. coli BL21(DE3)(pYA4085) as described previously (49). Antibody titers in serum and vaginal washes were determined by enzyme-linked immunosorbent assay (ELISA) as described previously (50).
Statistical analyses.
All statistical analyses were performed using GraphPad Prism 6 (GraphPad Software, San Diego, CA). The significance of the different values obtained was appraised using two-way analysis of variance (ANOVA) followed by Dunnett’s tests (for ELISA). For challenge experiments, log rank (Mantel-Cox) test was used to determine the significant differences between the survival curves. For CI assays, the geometric means of the CIs were determined, and a Student’s t test was used to determine whether the logarithmically transformed ratios differed significantly from zero. P values of <0.05 were considered significant.
ACKNOWLEDGMENTS
We thank Soo-Young Wanda for constructing some of the strains and plasmids used in this study. We also thank Crystal Willingham and Jacquelyn Kilbourne for their expert technical assistance and the Arizona State University Department of Animal Care and Technology for taking outstanding care of the animals used in this study.
The funding agency had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
This work was funded by grants R01 AI60557 and R01 AI56289 from the National Institutes of Health.
Footnotes
Citation Łaniewski P, Baek C-H, Roland KL, Curtiss R, III. 2017. Analysis of spleen-induced fimbria production in recombinant attenuated Salmonella enterica serovar Typhimurium vaccine strains. mBio 8:e01189-17. https://doi.org/10.1128/mBio.01189-17.
REFERENCES
- 1.Hultgren S, Jones C, Normark S. 1996. Bacterial adhesins and their assembly, p 2730–2756. In Neidhardt F, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. [Google Scholar]
- 2.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 3.Yue M, Rankin SC, Blanchet RT, Nulton JD, Edwards RA, Schifferli DM. 2012. Diversification of the Salmonella fimbriae: a model of macro- and microevolution. PLoS One 7:e38596. doi: 10.1371/journal.pone.0038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nuccio SP, Bäumler AJ. 2007. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev 71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäumler AJ, Tsolis RM, Heffron F. 1996. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer’s patches. Proc Natl Acad Sci U S A 93:279–283. doi: 10.1073/pnas.93.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humphries A, Deridder S, Bäumler AJ. 2005. Salmonella enterica serotype Typhimurium fimbrial proteins serve as antigens during infection of mice. Infect Immun 73:5329–5338. doi: 10.1128/IAI.73.9.5329-5338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Curtiss R III. 2001. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol Microbiol 41:349–363. doi: 10.1046/j.1365-2958.2001.02529.x. [DOI] [PubMed] [Google Scholar]
- 8.Barak JD, Jahn CE, Gibson DL, Charkowski AO. 2007. The role of cellulose and O-antigen capsule in the colonization of plants by Salmonella enterica. Mol Plant Microbe Interact 20:1083–1091. doi: 10.1094/MPMI-20-9-1083. [DOI] [PubMed] [Google Scholar]
- 9.Collinson SK, Emody L, Muller KH, Trust J, Kay WW. 1991. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol 173:4773–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sukupolvi S, Lorenz RG, Gordon JI, Bian Z, Pfeifer JD, Normark SJ, Rhen M. 1997. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect Immun 65:5320–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsén A, Jonsson A, Normark S. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 12.Nicholson B, Low D. 2000. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol Microbiol 35:728–742. doi: 10.1046/j.1365-2958.2000.01743.x. [DOI] [PubMed] [Google Scholar]
- 13.Morrow BJ, Graham JE, Curtiss R III. 1999. Genomic subtractive hybridization and selective capture of transcribed sequences identify a novel Salmonella typhimurium fimbrial operon and putative transcriptional regulator that are absent from the Salmonella typhi genome. Infect Immun 67:5106–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphries AD, Raffatellu M, Winter S, Weening EH, Kingsley RA, Droleskey R, Zhang S, Figueiredo J, Khare S, Nunes J, Adams LG, Tsolis RM, Bäumler AJ. 2003. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol Microbiol 48:1357–1376. doi: 10.1046/j.1365-2958.2003.03507.x. [DOI] [PubMed] [Google Scholar]
- 15.Ledeboer NA, Frye JG, McClelland M, Jones BD. 2006. Salmonella enterica serovar Typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect Immun 74:3156–3169. doi: 10.1128/IAI.01428-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weening EH, Barker JD, Laarakker MC, Humphries AD, Tsolis RM, Bäumler AJ. 2005. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect Immun 73:3358–3366. doi: 10.1128/IAI.73.6.3358-3366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norris TL, Kingsley RA, Bäumler AJ. 1998. Expression and transcriptional control of the Salmonella typhimurium lpf fimbrial operon by phase variation. Mol Microbiol 29:311–320. doi: 10.1046/j.1365-2958.1998.00934.x. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Escobedo G, Gunn JS. 2013. Gallbladder epithelium as a niche for chronic Salmonella carriage. Infect Immun 81:2920–2930. doi: 10.1128/IAI.00258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez-Escobedo G, Gunn JS. 2013. Identification of Salmonella enterica serovar Typhimurium genes regulated during biofilm formation on cholesterol gallstone surfaces. Infect Immun 81:3770–3780. doi: 10.1128/IAI.00647-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forest C, Faucher SP, Poirier K, Houle S, Dozois CM, Daigle F. 2007. Contribution of the stg fimbrial operon of Salmonella enterica serovar Typhi during interaction with human cells. Infect Immun 75:5264–5271. doi: 10.1128/IAI.00674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahan MJ, Slauch JM, Mekalanos JJ. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Wang S, Scarpellini G, Gunn B, Xin W, Wanda SY, Roland KL, Curtiss R III. 2009. Evaluation of new generation Salmonella enterica serovar Typhimurium vaccines with regulated delayed attenuation to induce immune responses against PspA. Proc Natl Acad Sci U S A 106:593–598. doi: 10.1073/pnas.0811697106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baek CH, Wang S, Roland KL, Curtiss R III. 2009. Leucine-responsive regulatory protein (Lrp) acts as a virulence repressor in Salmonella enterica serovar Typhimurium. J Bacteriol 191:1278–1292. doi: 10.1128/JB.01142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang HY, Dozois CM, Tinge SA, Lee TH, Curtiss R III. 2002. Transduction-mediated transfer of unmarked deletion and point mutations through use of counterselectable suicide vectors. J Bacteriol 184:307–312. doi: 10.1128/JB.184.1.307-312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xin W, Wanda SY, Li Y, Wang S, Mo H, Curtiss R III. 2008. Analysis of type II secretion of recombinant pneumococcal PspA and PspC in a Salmonella enterica serovar Typhimurium vaccine with regulated delayed antigen synthesis. Infect Immun 76:3241–3254. doi: 10.1128/IAI.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S, Curtiss R III. 2014. Development of Streptococcus pneumoniae vaccines using live vectors. Vaccines 2:49–88. doi: 10.3390/vaccines2010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briles DE, Hollingshead SK, Swiatlo E, Brooks-Walter A, Szalai A, Virolainen A, McDaniel LS, Benton KA, White P, Prellner K, Hermansson A, Aerts PC, Van Dijk H, Crain MJ. 1997. PspA and PspC: their potential for use as pneumococcal vaccines. Microb Drug Resist 3:401–408. doi: 10.1089/mdr.1997.3.401. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto M, McDaniel LS, Kawabata K, Briles DE, Jackson RJ, McGhee JR, Kiyono H. 1997. Oral immunization with PspA elicits protective humoral immunity against Streptococcus pneumoniae infection. Infect Immun 65:640–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folkesson A, Advani A, Sukupolvi S, Pfeifer JD, Normark S, Löfdahl S. 1999. Multiple insertions of fimbrial operons correlate with the evolution of Salmonella serovars responsible for human disease. Mol Microbiol 33:612–622. doi: 10.1046/j.1365-2958.1999.01508.x. [DOI] [PubMed] [Google Scholar]
- 30.Strindelius L, Folkesson A, Normark S, Sjöholm I. 2004. Immunogenic properties of the Salmonella atypical fimbriae in BALB/c mice. Vaccine 22:1448–1456. doi: 10.1016/j.vaccine.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Bhuiyan S, Sayeed A, Khanam F, Leung DT, Rahman Bhuiyan T, Sheikh A, Salma U, LaRocque RC, Harris JB, Pacek M, Calderwood SB, LaBaer J, Ryan ET, Qadri F, Charles RC. 2014. Cellular and cytokine responses to Salmonella enterica serotype Typhi proteins in patients with typhoid fever in Bangladesh. Am J Trop Med Hyg 90:1024–1030. doi: 10.4269/ajtmh.13-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheikh A, Charles RC, Rollins SM, Harris JB, Bhuiyan MS, Khanam F, Bukka A, Kalsy A, Porwollik S, Brooks WA, LaRocque RC, Hohmann EL, Cravioto A, Logvinenko T, Calderwood SB, McClelland M, Graham JE, Qadri F, Ryan ET. 2010. Analysis of Salmonella enterica serotype Paratyphi A gene expression in the blood of bacteremic patients in Bangladesh. PLoS Negl Trop Dis 4:e908. doi: 10.1371/journal.pntd.0000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salih O, Remaut H, Waksman G, Orlova EV. 2008. Structural analysis of the Saf pilus by electron microscopy and image processing. J Mol Biol 379:174–187. doi: 10.1016/j.jmb.2008.03.056. [DOI] [PubMed] [Google Scholar]
- 34.Barondess JJ, Beckwith J. 1990. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature 346:871–874. doi: 10.1038/346871a0. [DOI] [PubMed] [Google Scholar]
- 35.White AP, Gibson DL, Grassl GA, Kay WW, Finlay BB, Vallance BA, Surette MG. 2008. Aggregation via the red, dry, and rough morphotype is not a virulence adaptation in Salmonella enterica serovar Typhimurium. Infect Immun 76:1048–1058. doi: 10.1128/IAI.01383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meli R, Bentivoglio C, Nuzzo I, Mattace Raso G, Galdiero E, Galdiero M, Di Carlo R, Carratelli CR. 2003. Th1-Th2 response in hyperprolactinemic mice infected with Salmonella enterica serovar Typhimurium. Eur Cytokine Netw 14:186–191. [PubMed] [Google Scholar]
- 37.O’Garra A, Arai N. 2000. The molecular basis of T helper 1 and T helper 2 cell differentiation. Trends Cell Biol 10:542–550. doi: 10.1016/S0962-8924(00)01856-0. [DOI] [PubMed] [Google Scholar]
- 38.Ferreira DM, Darrieux M, Oliveira ML, Leite LC, Miyaji EN. 2008. Optimized immune response elicited by a DNA vaccine expressing pneumococcal surface protein A is characterized by a balanced immunoglobulin G1 (IgG1)/IgG2a ratio and proinflammatory cytokine production. Clin Vaccine Immunol 15:499–505. doi: 10.1128/CVI.00400-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtis SR., III 1965. Chromosomal aberrations associated with mutations to bacteriophage resistance in Escherichia coli. J Bacteriol 89:28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Briles DE, King JD, Gray MA, McDaniel LS, Swiatlo E, Benton KA. 1996. PspA, a protection-eliciting pneumococcal protein: immunogenicity of isolated native PspA in mice. Vaccine 14:858–867. doi: 10.1016/0264-410X(96)82948-3. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook JE, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 43.Baek CH, Kim KS. 2003. lacZ- and aph-based reporter vectors for in vivo expression technology. J Microbiol Biotechnol 13:872–880. [Google Scholar]
- 44.Sun W, Olinzock J, Wang S, Sanapala S, Curtiss R III. 2014. Evaluation of YadC protein delivered by live attenuated Salmonella as a vaccine against plague. Pathog Dis 70:119–131. doi: 10.1111/2049-632X.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curtiss R III, Porter SB, Munson M, Tinge SA, Hassan JO, Gentry-Weeks C, Kelly SM. 1991. Nonrecombinant and recombinant avirulent Salmonella live vaccines for poultry, p 169–198. In Blankenship L (ed), Colonization control of human bacterial enteropathogens in poultry. Academic Press, New York, NY. [Google Scholar]
- 46.Roland K, Curtiss R III, Sizemore D. 1999. Construction and evaluation of a Δcya Δcrp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis 43:429–441. [PubMed] [Google Scholar]
- 47.Hitchcock PJ, Brown TM. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol 154:269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 49.Wang S, Li Y, Scarpellini G, Kong W, Shi H, Baek CH, Gunn B, Wanda SY, Roland KL, Zhang X, Senechal-Willis P, Curtiss R III. 2010. Salmonella vaccine vectors displaying delayed antigen synthesis in vivo to enhance immunogenicity. Infect Immun 78:3969–3980. doi: 10.1128/IAI.00444-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang HY, Srinivasan J, Curtiss R III. 2002. Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar Typhimurium vaccine. Infect Immun 70:1739–1749. doi: 10.1128/IAI.70.4.1739-1749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 52.Nakayama K, Kelly SM, Curtiss R III. 1988. Construction of an Asd+ expression-cloning vector: stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Nat Biotechnol 6:693–697. doi: 10.1038/nbt0688-693. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Production of PspA antigen in S. Typhimurium RASV strains. Western blot showing whole-cell lysates obtained from mid-log-phase cultures, electrophoresed on a 12% SDS-polyacrylamide gel, transferred onto nitrocellulose, and probed with anti-rPspA serum. Download FIG S1, TIF file, 0.2 MB (258.4KB, tif) .
Copyright © 2017 Łaniewski et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Additional strains and plasmids used in this study. Download TABLE S1, DOCX file, 0.03 MB (30.8KB, docx) .
Copyright © 2017 Łaniewski et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S2, DOCX file, 0.02 MB (20.1KB, docx) .
Copyright © 2017 Łaniewski et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.