Abstract
In eukaryotic cells DNA replication occurs in specific nuclear compartments, called replication factories, that undergo complex rearrangements during S-phase. The molecular mechanisms underlying the dynamics of replication factories are still poorly defined. Here we show that etoposide, an anticancer drug that induces double-strand breaks, triggers the redistribution of DNA ligase I and proliferating cell nuclear antigen from replicative patterns and the ensuing dephosphorylation of DNA ligase I. Moreover, etoposide triggers the formation of RPA foci, distinct from replication factories. The effect of etoposide on DNA ligase I localization is prevented by aphidicolin, an inhibitor of DNA replication, and by staurosporine, a protein kinase inhibitor and checkpoints' abrogator. We suggest that dispersal of DNA ligase I is triggered by an intra-S-phase checkpoint activated when replicative forks meet topoisomerase II-DNA–cleavable complexes. However, etoposide treatment of ataxia telangiectasia cells demonstrated that ataxia-telangiectasia-mutated activity is not required for the disassembly of replication factories and the formation of replication protein A foci.
INTRODUCTION
Eukaryotic chromosomes consist of a large number of replication units, or replicons, that are duplicated after a precise temporal order during S-phase. It is commonly accepted that replicons located in euchromatic, transcriptionally active, portions of the genome are replicated earlier than those embedded in heterochromatic, transcriptionally silent, regions. However, the molecular mechanisms underlying this program are still poorly understood. In addition, other control mechanisms operate during S-phase to define the nuclear regions where DNA replication takes place. DNA replication occurs in specific nuclear compartments, termed replication factories, that are composed of the numerous enzymes and factors involved in this process (Cardoso et al., 1993; Hozak et al., 1993, 1994; Montecucco et al., 1995b). Immunostaining of exponentially growing cells with antibodies directed either against 5-bromodeoxyuridine (BrdU)-labeled nascent DNA or toward replicative enzymes reveals that the distribution of replication factories varies during S-phase according to a precise program. In early S-phase replication takes place within many foci distributed throughout the cell nucleus (type I and II patterns). In mid-S-phase replication occurs at the nuclear periphery and in nucleolar regions (type III). In late S-phase several replication foci disperse in the nuclear volume, and finally very few large internal or peripheral foci (type IV and V) are detectable (O'Keefe et al., 1992).
Each replication factory undergoes an assembly/disassembly cycle that entails the recruitment of replicative factors to clusters of replicons and the ensuing release of the same factors upon the completion of DNA synthesis (Dimitrova and Gilbert, 2000; Leonhardt et al., 2000). How this dynamic process is controlled is still unknown; however, posttranslational modifications of replicative factors are likely to be involved.
It has been recently shown that the recruitment to replication factories of a few factors, among which DNA ligase I (hLigI; Montecucco et al., 1998), replication factor-C (Montecucco et al., 1998), DNA methylase (Chuang et al., 1997), and flap endonuclease-1 (FEN1) endonuclease (Chen et al., 1996), is directed by a short protein motif termed “replication factory-targeting sequence” (Montecucco et al., 1998; Rossi et al., 1999). The very same motif mediates the interaction with the sliding clamp PCNA, an essential protein of the replication apparatus (for review see Cox, 1997; Warbrick, 2000). Thus, in addition to coordinate the synthesis of leading and lagging strands, PCNA would act as the recruiter of other components of the replication machinery, increasing their local concentration to the biochemical optimum necessary for DNA synthesis to occur.
In response to DNA-damaging agents, biochemical pathways, commonly termed checkpoints, are activated to prevent the cell from entering a successive phase of the cell cycle and to allow DNA repair (for review see Dasika et al., 1999; Zhou and Elledge, 2000). Checkpoint activation leads to a delay of the cell cycle progression, inhibits replication and segregation of damaged DNA molecules, and induces transcription of several repair genes. Studies of different organisms, including Saccharomyces cerevisiae, Schizosaccharomyces pombe, and mammalian cells, have shown that DNA damage checkpoints act at three stages of the cell cycle, namely, at the G1/S transition, during S-phase and at the G2/M boundary. A subfamily of phosphoinositide kinase-related proteins, which comprises Mec1 of budding yeast, Rad3 of fission yeast, and mammalian ATM, ATM-Rad3-related and DNA-dependent protein kinase kinases, plays a central role in the DNA damage checkpoint. These kinases, through a complex and still poorly defined pathway, induce posttranslational modifications of replicative factors, such as RPA2 (Wold, 1997) or the budding yeast DNA polymerase α-primase complex (Pellicioli et al., 1999), and slow down or completely inhibit DNA replication.
In this study we have investigated the influence of etoposide (VP-16) on the dynamics of replication factories. VP-16 is a specific inhibitor of topoisomerase II (topo II), which induces replication-mediated double-strand DNA breaks (DSBs) and checkpoint activation (for review see Kaufmann, 1998). We observed that VP-16 triggers the dispersal of replication proteins, such as hLigI and PCNA, from the replication factories throughout the nucleus. This phenomenon temporally follows the formation of RPA2 foci distinct from replication factories. Finally, the redistribution of PCNA and hLigI is prevented by staurosporine, a checkpoint inhibitor, but does not require the ATM function.
MATERIALS AND METHODS
Drugs, Cell Lines, and Cell Treatments
AT1 BR (European Collection of Cell Cultures catalogue number BM0020), AT3 BR (ECACC catalogue number BM0026), and HeLa cells were grown as monolayers in complete DMEM supplemented with 10% fetal calf serum, 4 mM glutamine, and 50 μg/ml gentamicin. All reagents were from Sigma (St. Louis, MO). Cells were grown at 37°C in a humidified atmosphere containing 5% CO2, and trypsinized when confluent. Cells were treated for the time periods indicated in the text with different concentrations of etoposide (Vepesid: Bristol-Myers Squibb, New York, NY). Redistribution of hLigI and PCNA was studied in HeLa cells incubated for 3 h in etoposide-containing medium to better appreciate dispersal of replication factories. Formation of RPA2 foci and the relationship between RPA2 foci and replication factories was studied in cells treated for 1 h. Finally, the effect of staurosporine on the disassembly of replication factories was studied after a 2-h incubation to reduce toxic effects of staurosporine. Because of the high sensitivity of AT cells to etoposide, the effect on replication factories was determined after 2 h of incubation in the presence of 20 μM VP-16, the minimal conditions able to give the complete disassembly of replication factories in HeLa cells. Aphidicolin and staurosporine (Sigma) were stored in dimethyl sulfoxide at concentrations of 2 and 1 mM, respectively. Aliquots were diluted in DMEM immediately before use. In irradiation experiments, cells were exposed to UV-C radiation with the use of a Philips TUV 15-W lamp as previously described (Montecucco et al., 1995b). After removal of culture medium, cells were irradiated with a single dose (20 J/m2) of 254-nm light, and then fresh medium was added to the cells. Flow cytometry analysis was conducted on both untreated and VP-16-treated cells by propidium iodide staining, and cells were analyzed with an Epics XL flow cytometer (Coulter Pharmaceutical, Palo Alto, CA). Ten thousand cells were measured for each sample.
Analysis of Apoptotic Morphology
Cells grown on glass coverslips were fixed for 10 min with ice-cold 70% ethanol and washed several times with ice-cold phosphate-buffered saline (PBS). For the fraction of cells detached at the end of the treatments with VP-16, a further step was applied in which cells resuspended at a density of 106 cells/ml in PBS containing 10% fetal calf serum were cytocentrifuged on glass coverslips at 500 rpm for 3 min. After fixation, DNA was stained with 0.1 μg/ml Hoechst 33258 (Sigma) for 10 min at room temperature. The number of apoptotic cells was evaluated through fluorescence observation with the use of a Orthoplan microscope (Leitz) equipped with a 50× objective. The occurrence of apoptosis was measured as the percentage of cells with condensed or fragmented DNA over the total cell number. For each sample, 500 cells were counted.
Western Blot Analysis
Western blot analysis was performed on total cell extracts prepared from control and treated cells. Cells were harvested, washed twice with PBS, resuspended in 2× Laemmli sample buffer (Laemmli, 1970), and boiled for 10 min. Proteins were separated by electrophoresis in SDS-PAGE gels.
To evaluate the fraction of RPA bound to the nuclear structures, HeLa cells were trypsinized and washed first in PBS and then in cytoskeleton buffer (CSK: 10 mM HEPES-KOH, pH 7.4, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2; Dimitrova et al., 1999). Cells were then resuspended at 2 × 107 cells/ml in CSK buffer containing 0.5% Triton X-100, 0.2 mM hydrochloride 4-[2-aminoethyl]benzensulfonylfluoride HCl (Calbiochem, La Jolla, CA), 1 μg/ml pepstatin A, and 1 μg/ml leupeptin (Sigma). After 5 min on ice, cell lysate was separated into a soluble fraction and a nuclear pellet by centrifugation for 3 min at 1500 g at 4°C. The nuclear pellet was washed with CSK buffer and resuspended in Laemmli buffer. Proteins were fractionated by SDS-PAGE and electroblotted to a nitrocellulose transfer membrane (Protran, Schleicher & Schuell, Keene, NH) with the use of the Mini-Protean II (Bio-Rad, Hercules, CA). Membranes were blocked for 1 h with 2% skim milk (Difco, Detroit, MI) in TBS-T buffer (20 mM Tris-HCl, pH 7.5, 137 mM NaCl, and 0.1% Tween-20) and probed with the following primary antibodies: 1A4 monoclonal antibody (mAb) directed against a phosphoepitope of hLigI (Rossi et al., 1999), anti-hLigI polyclonal rabbit antiserum (Rossi et al., 1999), anti-RPA2 9H8 mAb (NeoMarkers, Fremont, CA). Primary antibodies were revealed with horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit antibodies and the enhanced chemiluminescence system (ECL, Amersham, Arlington Heights, IL).
Poly (ADP-ribose) polymicion (PARP) degradation during apoptosis was followed by Western blotting on cellular extracts prepared from control and apoptotic cells according to the method of Shah et al. (1995). Briefly, aliquots of cells (attached to the plastic dish or floating in the medium) were resuspended at a density of 106 cells/ml in 4 M urea, 62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, and 0.003% bromophenol blue. Cells were then disrupted by sonication on ice (two pulses of 20 s each at 50 W), and extracts were heated at 65°C for 15 min. Aliquots of extracts corresponding to the same number of cells were electrophoresed on a 7.5% SDS-PAGE gel. After an overnight incubation in PTN (PBS containing 0.1% Tween-20 and 10% newborn calf serum), the membrane was incubated for 3 h with C-2–10 mAb (Lamarre et al., 1988). Visualization of immunoreactive peptides was performed with horseradish peroxidase-conjugated goat anti-mouse IgG with the use of the ECL system (Amersham).
Immunofluorescence Microscopy
For immunostaining of protein antigens, HeLa cells grown on coverslips were washed with cold PBS and fixed for 4 min in cold methanol as previously described (Montecucco et al., 1995b). The following primary antibodies were used for detection of protein antigens: 9H8 mAb to RPA2 (NeoMarkers); PC10 mAb and FL-261 polyclonal antibody to PCNA (Santa Cruz Biotechnology, Santa Cruz, CA); 2B1 mAb and rabbit polyclonal antibody to hLigI (Rossi et al., 1999); fluorescein isothiocyanate (FITC)-conjugated anti-BrdU mAb (clone BMC 9318, Chemicon, Temecula, CA). The secondary antibodies used were FITC-conjugated goat anti-mouse IgG (Jackson Immunoresearch Laboratories, West Grove, PA) and cyanine dyes-conjugated goat anti-rabbit IgG (Jackson Immunoresearch). For costaining experiments, cells were incubated simultaneously with the anti-RPA2 mAb and the polyclonal rabbit antibodies against hLigI or PCNA. To detect sites of DNA synthesis, cells were grown in 50 μM BrdU (Sigma) for 30 min immediately before methanol fixation and treated as previously described (Montecucco et al., 1995b). Conventional epifluorescence microscopy was performed with a Leitz Orthoplan microscope equipped with a 63× objective. Pictures were taken with the use of 400 ASA film (Kodak, Rochester, NY). Confocal microscopy was performed with a TCS-NT digital scanning confocal microscope (Leica, Deerfield, IL) equipped with a 63×/NA = 1.32 oil immersion objective. We used the 488-nm laser line for excitation of FITC (detected at 500 nm <λFITC < 550 nm) and the 633-nm laser line for the Cy5 fluorescence (detected at 650 nm < λCy5 < 700 nm). The pinhole diameter was kept at 1 μm. Images were exported to Photoshop (Adobe Systems, Mountain View, CA).
RESULTS
Dephosphorylation of hLigI in Response to DNA Damage
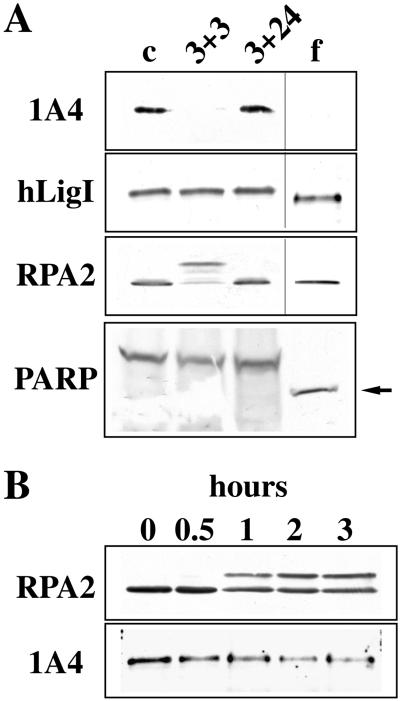
We have previously shown that hLigI is phosphorylated in a cell cycle-dependent manner. After dephosphorylation in early G1, the enzyme is progressively phosphorylated on Ser66 during S-phase and a hyperphosphorylated form of hLigI, with a lower electrophoretic mobility, is detectable in M-phase (Rossi et al., 1999). In this work we studied whether the phosphorylation status of hLigI could be modulated not only during the cell cycle but also in response to DNA damage. To this, exponentially growing HeLa cells were treated with VP-16, a drug known to induce replication-mediated DSBs. DSBs are, in fact, generated when replication forks encounter topo II-DNA–cleavable complexes trapped by the drug (Kaufmann, 1998). After a 3-h treatment with 100 μM VP-16, cells were allowed to recover for 3 or 24 h in normal medium before being analyzed by Western blotting to assess the phosphorylation status of hLigI. Because VP-16 is a well known apoptotic agent (Negri et al., 1993; Kaufmann, 1998), in parallel we determined the occurrence of apoptosis by following different morphological parameters such as nuclear condensation and fragmentation. A very low level of apoptosis (3.5 ± 0.1%) was detectable at the end of the 3-h treatment with VP-16. The apoptotic index increased during the recovery period, reaching 10 ± 0.6 or 49 ± 1.9% after 3 and 24 h of recovery, respectively. After a 24-h recovery most of the apoptotic cells were floating in the medium. Extracts prepared from untreated cells, from adherent cells collected after 3 or 24 h of recovery and from floating cells collected at 24 h, were analyzed by Western blotting with a rabbit polyclonal antiserum directed against hLigI and with 1A4 mAb that specifically recognized the enzyme phosphorylated on Ser66. As a control, we verified the occurrence of etoposide-induced phosphorylation of the p34 subunit of RPA (RPA2) detectable as a shift in the electrophoretic mobility of the protein (Shao et al., 1999). Western blot analysis with 9H8 mAb showed a transient phosphorylation of RPA2 at 3 h of recovery that was no longer detectable at 24 h either in adherent or in floating cells (Figure 1A). Concerning hLigI, although the level of the protein was fairly constant (Figure 1A), its phosphorylation status changed significantly during the experiment. Indeed, staining with 1A4 mAb showed that in adherent cells the enzyme was dephosphorylated at 3 h of recovery and rephosphorylated at later times (24 h). A form of hLigI with a higher electrophoretic mobility, most likely generated through extensive dephosphorylation, was instead detectable in floating cells collected at 24 h (Figure 1A). The presence of the 85-kDa proteolytic fragment of PARP (Figure 1A), recognized by the C-2–10 mAb (Lamarre et al., 1988), confirmed the apoptotic nature of these floating cells. A time course experiment showed that phosphorylation of RPA2 occurred in a short time (between 30 and 60 min of treatment with VP-16), whereas Ser66 of hLigI was gradually dephosphorylated over 3 h (Figure 1B).
Figure 1.
VP-16 affects the phosphorylation status of hLigI and RPA2. (A) Exponentially growing HeLa cells were incubated for 3 h with 100 μM VP-16. After the drug was removed, cells were grown for an additional 3 h (3 + 3) or 24 h in complete medium. After 24 h of recovery, approximately half of the cells were still adherent to the plastic dish (3 + 24), whereas the others were floating in the medium (f). Total cell extracts were analyzed by Western blotting with the 1A4 mAb directed to phosphorylated Ser66 of hLigI, a rabbit polyclonal antibody to hLigI, the 9H8 mAb to RPA2, and the C-2–10 mAb to PARP. We also analyzed a total cell extract prepared from untreated HeLa cells (c). The 85-kDa proteolytic fragment of PARP detectable in apoptotic cells is indicated (arrow). (B) Exponentially growing HeLa cells were treated with 100 μM VP-16. At the indicated times (hours) total cell extracts were analyzed by Western blotting with 9H8 mAb to RPA2 and 1A4 mAb to hLigI.
Altogether these data indicate that VP-16 triggers a reversible change of the phosphorylation status of hLigI and RPA2 that takes place in a relatively short time interval before the occurrence of apoptosis.
VP-16 Affects the Subnuclear Distribution of Replicative Factors
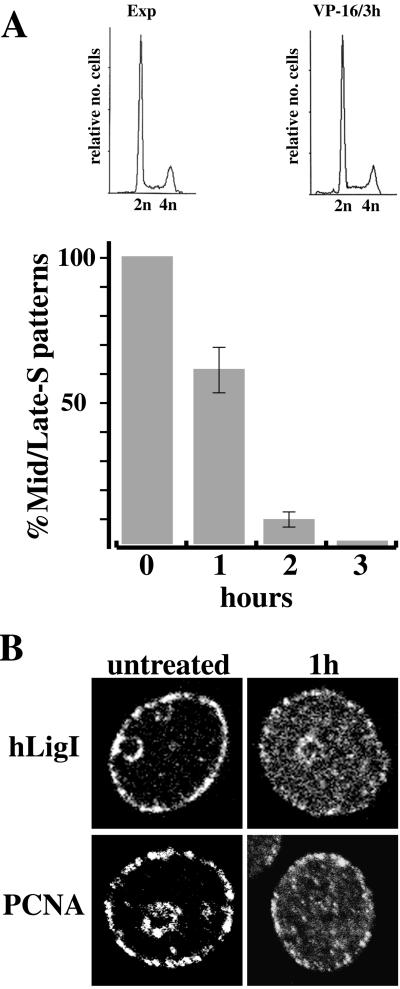
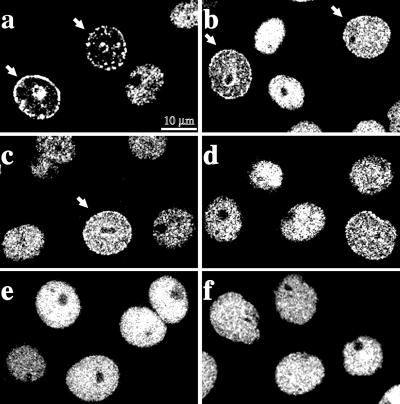
We have previously shown that during S-phase hLigI has a punctated nuclear distribution that reflects its association with replication factories (Montecucco et al., 1995b). To understand whether VP-16 could affect the subnuclear distribution of hLigI, HeLa cells were grown for increasing times in the presence of 100 μM VP-16 and then immunostained with anti-hLigI antibodies. As shown in Figure 2A, VP-16 caused the progressive reduction of the number of cells in which hLigI displayed the typical S-phase patterns observed in untreated cells. Indeed, after 3 h in the presence of the drug, the fraction of cells showing mid- and late S-phase patterns was reduced to <2% of the control value (Figure 2A). Flow cytometry analysis showed that the fraction of cells with an S-phase DNA content was not appreciably affected at the end of the 3-h incubation with etoposide, ruling out that the disappearance of mid- and late S-phase patterns was due to the completion of S-phase and accumulation of cells in G2-phase. A high fraction of G2 cells was instead detectable at later times during the recovery period (Scovassi and Prosperi, unpublished results). After 1 h of treatment, replication factories were still detectable in a significant fraction of cells (60 ± 9% of the control value, see Figure 2A). However, in the same cells most of hLigI was already dispersed throughout the nuclear volume (Figure 2B). Notably, VP-16 had a similar effect on the distribution of PCNA (Figure 2B). To determine the minimal dose able to elicit redistribution of hLigI, HeLa cells were incubated for 3 h with drug concentrations ranging from 2–100 μM (i.e., from 1.2–59 μg/ml). As shown in Figure 2A, replicative patterns were no longer detectable after 3 h of treatment with 100 μM VP-16. Immunostaining analysis with the anti-hLigI 2B1 mAb showed that a significant redistribution of the enzyme already occurred at 2 μM (Figure 3b) and mid- and late S-phase patterns were no longer detectable at 20 μM VP-16 (Figure 3d).
Figure 2.
VP-16 induces the disassembly of replication factories. (A) Exponentially growing HeLa cells were grown for the indicated time periods (hours) in the presence of 100 μM VP-16 and then immediately fixed. The distribution of hLigI was revealed with 2B1 mAb and with FITC-conjugated sheep anti-mouse IgG secondary antibody. At each time point we counted, in a pool of 500 randomly selected cells, the number of cells displaying hLigI in patterns typical of mid- and late S-phase (patterns III, IV, and V defined by O'Keefe et al. [1992]. The value measured at 1, 2, or 3 h of growth in the presence of VP-16 was expressed as the percentage of the number of cells with mid- and late S-phase patterns detected in control cells (0 h). Error bars indicate the average error of the mean as determined from three separate experiments. Flow cytometry analysis of the DNA content of asynchronously growing cells (Exp) and cells incubated for 3 h in the presence of 100 μM VP-16 is shown in the top. (B) Immunofluorescence analysis of HeLa cells untreated or incubated for 1 h in 100 μM VP-16 (1 h). Cells were stained either with 2B1 mAb to hLigI or with PC10 mAb to PCNA. FITC-conjugated goat anti-mouse IgG was used as a secondary antibody. For each point the confocal laser image of a single cell nucleus, displaying a mid-S-phase pattern, is shown to better visualize the dispersion of PCNA and hLigI from the replication factories triggered by VP-16.
Figure 3.
The effect of VP-16 on replication factories is dose dependent. Exponentially growing HeLa cells were treated for 3 h with different concentrations of VP-16 and analyzed in indirect immunofluorescence with 2B1 mAb and with FITC-conjugated goat anti-mouse IgG secondary antibody. Confocal laser images were taken. a to f correspond, respectively, to cells treated with 0, 2, 10, 20, 50 and 100 μM VP-16. The arrows point to cells in which hLigI has a typical mid-S-phase pattern. Note the progressive disassembly of the replication factories.
Altogether these results indicate that VP-16 causes hLigI to leave replication factories, probably before dephosphorylation of Ser66. The fact that PCNA also undergoes a similar redistribution suggests that VP-16 could trigger the complete disassembly of replication factories, possibly through the activation of an intra-S-phase checkpoint.
RPA2 Is Recruited to Nuclear Foci Distinct from Replication Factories in Response to VP-16 Treatment
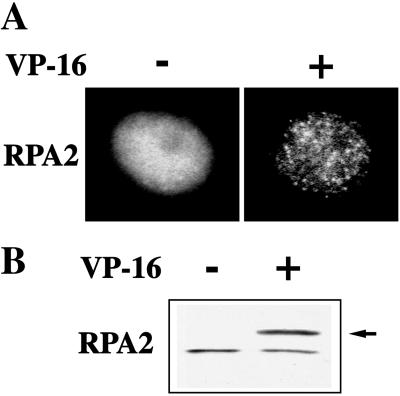
We asked whether, similarly to PCNA and hLigI, RPA2 could relocalize in response to VP-16 treatment. Immunostaining with 9H8 mAb showed that in untreated HeLa cells RPA2 displayed an almost homogeneous nuclear distribution (Figure 4A). In accord with the findings of Dimitrova et al. (1999), the association with replication factories was detectable only when most of RPA2 was extracted from the cells with 0.5% Triton X-100 before fixation, indicating that only a small fraction of the protein was stably bound to nuclear structures. In contrast, RPA2 foci were clearly detectable in 49 ± 2% of HeLa cells grown for 1 h in the presence of 100 μM VP-16. The fact that these foci were visible even if Triton X-100 extraction was omitted (Figure 4A) suggested a massive association of RPA2 to nuclear structures in response to VP-16 treatment. To test this hypothesis, untreated and etoposide-treated cells were subjected to Triton X-100 extraction and analyzed by Western blotting with 9H8 mAb. As shown in Figure 4B, the fraction of RPA2 resistant to Triton X-100 extraction significantly increased after VP-16 treatment. Indeed, while comparable levels of nonphosphorylated protein were present in control and treated cells, hyperphosphorylated RPA2 accounted for most of the signal detectable in VP-16 treated cells. Although nonconclusive, these results indicate a higher affinity of phosphorylated RPA2 for Triton X-100–insoluble structures probably corresponding to the RPA2 foci.
Figure 4.
VP-16 induces the formation of RPA2 foci and the concomitant phosphorylation of the protein. (A) Exponentially growing HeLa cells, either untreated (−) or held for 1 h in 100 μM VP-16 (+) were stained with 9H8 mAb to RPA2 and with FITC-conjugated goat anti-mouse IgG secondary antibody. Conventional epifluorescence microscopy images of representative cell nuclei are shown. (B) Triton X-100–insoluble cell extract (0.5% ) was prepared from cells treated as above. Extracts were analyzed by Western blotting with 9H8 mAb. The arrow points to the hyperphosphorylated form of RPA2 detectable after VP-16 treatment.
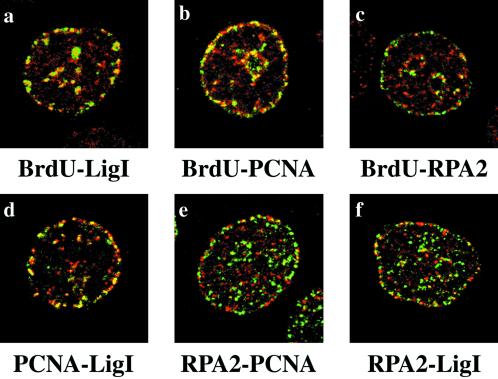
In a fraction of cells treated for 1 h with 100 μM VP-16, RPA2 displayed a subnuclear distribution that was reminiscent of replication patterns observed in mid-S-phase. We have shown (Figure 2A) that under these conditions hLigI and PCNA were still detectable in mid- and late S-phase patterns, although a significant fraction of these proteins was already dispersed throughout the nuclear volume (Figure 2B). Thus, the possibility existed that hLigI and PCNA could colocalize with VP-16–induced RPA2 foci. However, immunostaining with antibodies specific for the three proteins (Figure 5, d–f) showed that hLigI colocalized with PCNA but not with RPA2 foci. To investigate whether hLigI/PCNA foci corresponded to sites of DNA synthesis, HeLa cells were pulse labeled with 50 μM BrdU during the last 30 min of treatment with etoposide. Although BrdU incorporation was drastically reduced by the drug, sites of DNA synthesis were still detectable and colocalized with hLigI/PCNA foci (Figure 5, a and b). Notably, under the same conditions most of the etoposide-induced RPA2 foci did not colocalize with replication factories stained with anti-BrdU, anti-PCNA, or anti-hLigI antibodies (Figure 5, c, e, and f). Thus, VP-16–induced RPA foci are distinct from replication factories.
Figure 5.
Etoposide-induced RPA2 foci do not colocalize with replication factories. HeLa cells were treated for 1 h with 100 μM VP-16. Cells were stained with the polyclonal antibody to hLigI (a), with the polyclonal antibody to PCNA (b), and with the 9H8 mAb to RPA2 (c). Antibodies were revealed with a Cy5-conjugated secondary antibody (red). In the same panels sites of BrdU incorporation were revealed by the FITC-conjugated anti- BrdU mAb (green). Confocal laser images of the same cell were taken and merged. Yellow spots indicate the extent of protein-BrdU colocalization. d, cells were costained with the PC10 mAb to PCNA (green) and the polyclonal antibody to hLigI (red). e, cells were costained with the 9H8 mAb to RPA2 (green) and the polyclonal antibody to PCNA (red). f, cells were costained with the 9H8 mAb to RPA2 (green) and with the polyclonal antibody to hLigI (red). mAbs were revealed with the FITC-conjugated anti-mouse secondary antibody, and polyclonal antibodies were revealed with the Cy5-conjugated anti-rabbit secondary antibody. Confocal laser images of the same cell were taken and merged. Yellow spots indicate the extent of colocalization between the different proteins.
UV Irradiation and Aphidicolin Do Not Trigger the Disassembly of Replication Factories
We asked whether other DNA-damaging agents, such as UV light, could trigger the dispersal of hLigI from replication factories. HeLa cells were treated at a 20-J/m2 dose and after increasing recovery periods (0–3 h) were immunostained with 2B1 mAb directed toward hLigI. As shown in Figure 6, a and b, there was no difference in the distribution of hLigI in UV-irradiated compared with untreated cells, indicating that the dispersal of the enzyme was not part of a general response of the cell to DNA damage.
Figure 6.
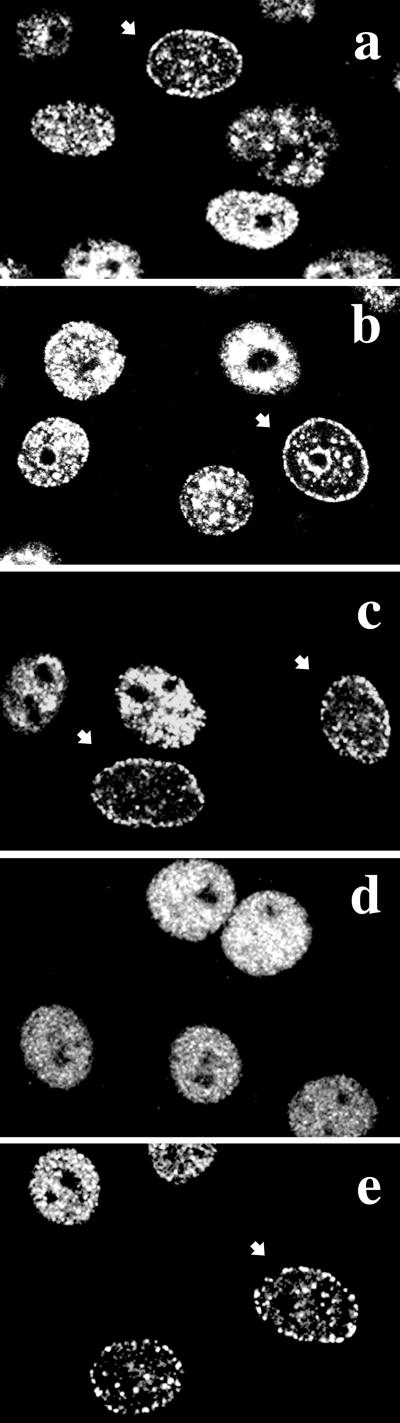
Effect of aphidicolin and UV light on the subnuclear distribution of hLigI. Exponentially growing HeLa cells were stained after the indicated treatments with 2B1 mAb and with FITC-conjugated goat anti-mouse IgG secondary antibody. a, untreated cells; b, cells stained after 3 h from UV irradiation at 20 J/m2; c, cells kept for 3 h in 2 μg/ml aphidicolin; d, cells treated for 3 h with 100 μM VP-16; e, cells grown for 3 h in 2 μg/ml aphidicolin and 100 μM VP-16. Arrows point to cells in which hLigI has a typical mid-S-phase pattern.
We next investigated whether stalled replication forks could trigger redistribution of hLigI. To explore this possibility, we used aphidicolin, a drug known to block DNA replication by specifically inhibiting replicative DNA polymerases. HeLa cells were grown for 3 h in the presence of 2 μg/ml aphidicolin and then immunostained with 2B1 mAb. As shown in Figure 6c, aphidicolin failed to induce relocalization of hLigI, indicating that stalling of replicative forks, per se, was not sufficient for the dispersal of the enzyme and more in general for the disassembly of replication factories.
Cytotoxicity induced by etoposide is critically linked to DNA replication and is reduced by aphidicolin, which does not affect the formation of cleavable complexes (Holm et al., 1989; Kaufmann, 1998). We observed that aphidicolin also prevented the dispersal of hLigI (Figure 6e), suggesting that ongoing DNA replication was critical for this event.
Staurosporine Prevents the Disassembly of Replication Factories
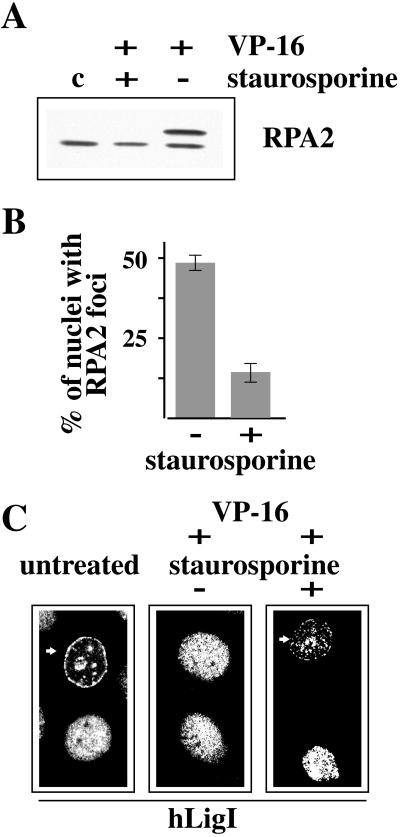
The results described in the previous section suggested that the signals that elicited the disassembly of replication factories originated from the encounter of replication forks with topo II-DNA–cleavable complexes trapped by etoposide. It was conceivable that these signals led to the activation of checkpoint kinases. Therefore, we investigated whether the inhibition of checkpoint kinases could prevent the dispersal of hLigI. It has been recently shown that 7-hydroxystaurosporine, a protein kinase C inhibitor and a cell-cycle checkpoint abrogator, prevents RPA2 phosphorylation in human colon carcinoma cells treated either with camptothecin or with VP-16 (Shao et al., 1999). Similarly, we found that preincubation of HeLa cells with 10 μM staurosporine prevented phosphorylation of RPA2 induced by VP-16 (Figure 7A). In addition, we observed that staurosporine inhibited the formation of etoposide-induced RPA2 foci. Indeed, the analysis of a large number of cells (n = 500) treated for 1 h with VP-16 showed that the fraction of nuclei with RPA2 foci decreased from 49 ± 2 to 15 ± 5% in cells preincubated with staurosporine (Figure 7B). The effect of staurosporine on the phosphorylation status and on the subnuclear distribution of RPA2 in VP-16–treated cells prompted us to investigate whether this inhibitor could affect the disassembly of replication factories as well. We found that 15 ± 3% of the cells incubated for 2 h in the presence of both VP-16 and staurosporine still displayed hLigI in mid- and late S-phase patterns compared with a value of <2% observed when staurosporine was omitted. Figure 7C exemplifies the distribution patterns of hLigI in HeLa cells untreated or treated with etoposide either in the absence (−) or in the presence (+) of staurosporine. No effect of the sole staurosporine on the distribution of hLigI was detected. Moreover, we did not observe any effect of staurosporine on the phosphorylation status of Ser66 of hLigI (Montecucco and Biamonti, unpublished results).
Figure 7.
Staurosporine prevents the phosphorylation of RPA2 and the redistribution of RPA2 and hLigI. (A) Western blot analysis of 0.5% Triton X-100–insoluble cell extracts prepared from HeLa cells treated for 1 h with 100 μM VP-16 either in the presence (+) or in the absence (−) of 10 μM staurosporine. Staurosporine was added to the medium 15 min before VP-16. Untreated cells (c) were also analyzed. RPA2 was revealed with 9H8 mAb. (B) The histogram indicates the percentage of cells with RPA2 foci after a treatment of 1 h with VP-16 (100 μM), either in the absence (−) or in the presence (+) of 10 μM staurosporine. Error bars indicate the average error of the mean as determined from three separate experiments. (C) Immunofluorescence analysis of the subnuclear distribution of hLigI in untreated HeLa cells or in cells grown for 2 h in 100 μM VP-16–containing medium either in the absence (−) or in the presence (+) of 10 μM staurosporine. Cells were stained with 2B1 mAb and with FITC-conjugated goat anti-mouse IgG secondary antibody. Arrows point to cells in which hLigI has a typical mid-S-phase pattern.
ATM Is Not Required for the Etoposide-induced Disassembly of Replication Factories
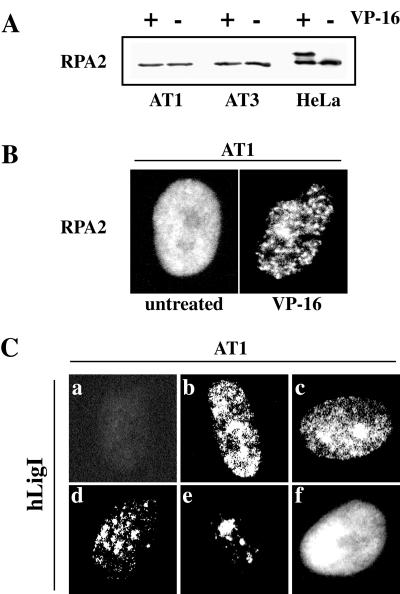
The results in the previous section suggested that checkpoint kinases, including ATM, could have a role in the disassembly of replication factories induced by VP-16. To explore whether ATM were involved in this process we studied the effect of etoposide on the distribution of RPA2 and hLigI in two ataxia telangiectasia (AT) cell lines, AT1 BR and AT3 BR. In both cell lines, a 1-h treatment with 100 μM VP-16 triggered the formation of RPA2 foci similar in size and number to those detected, under the same conditions, in HeLa cells (Figure 8B). However, Western blot analysis showed that etoposide-induced phosphorylation of RPA2 did not occur in AT cells, at least after 1 h of treatment (Figure 8A). This result was consistent with a previous report by Liu and Weaver (1993) showing that phosphorylation of RPA2 in response to DNA damage was delayed in AT cells. Thus, our findings suggest an involvement of ATM in phosphorylation but not in relocalization of RPA2 occurring in etoposide-treated cells. We next analyzed the distribution of hLigI in AT cells before and after VP-16 treatment. In untreated cells, replication factories were easily recognizable with anti-hLigI 2B1 mAb (as exemplified in Figure 8C) and coincided with sites of BrdU incorporation (Montecucco and Biamonti, unpublished results). Moreover, in 15–20% of the AT cells the distribution patterns of hLigI was similar to that observed in HeLa cells during mid- and late S-phase (Figure 8C, c–e). After a 2-h incubation in 20 μM VP-16, namely, the minimal dose able to elicit dispersal of hLigI in HeLa cells, mid- and late S-phase patterns were no longer visible. On the basis of these data we concluded that the redistribution of hLigI and RPA triggered by VP-16 did not require the ATM function.
Figure 8.
ATM is not necessary for the redistribution of RPA2 and hLigI induced by VP-16. (A) Total cell extracts were prepared from AT1, AT3, and HeLa cells, untreated (−) or grown for 1 h in 100 μM VP-16. Extracts were analyzed in Western blotting with 9H8 mAb to RPA2. (B) AT1 cells, untreated or grown for 1 h in 100 μM VP-16, were analyzed by indirect immunofluorescence with 9H8 mAb and with FITC-conjugated goat anti-mouse IgG secondary antibody. Conventional epifluorescence microscopy images of representative cell nuclei are shown. Identical results were observed with AT3 cells. (C) Exponentially growing AT1 cells were stained with 2B1 mAb to hLigI and with FITC-conjugated goat anti-mouse IgG secondary antibody. a, nucleus of a cell in G1; b–e, different patterns observed in nuclei of S-phase cells; f, nucleus of a cell in G2.
DISCUSSION
In this paper we show that the antitumor drug VP-16 causes a drastic redistribution of hLigI, PCNA, and RPA2, three replicative factors recruited to replication factories in S-phase. Indeed, after a 3-h incubation in the presence of VP-16, hLigI and PCNA are no longer found in replicative patterns, although the fraction of S-phase cells, as determined by flow cytometry analysis, is identical to that of untreated cells. It has been previously described that the recruitment of some replicative factors, including hLigI, replication factor-C, FEN1, and DNA methylase, to replication factories depends on a binding site to PCNA that would act as a recruiter (Chen et al., 1996; Chuang et al., 1997; Montecucco et al., 1998). Therefore redistribution of PCNA suggests that etoposide would affect the subnuclear distribution of PCNA-interacting factors leading to the disassembly of replication factories.
Etoposide-induced redistribution of hLigI is followed by dephosphorylation of Ser66. We have previously proposed that phosphorylation of Ser66 could mark hLigI molecules used during DNA replication. Dephosphorylation of Ser66 requires the interaction with PCNA and seems to occur even during S-phase. We proposed that after a replicon has been completely replicated, dephosphorylation of Ser66 could be required for the recycling of hLigI to another replication unit (Rossi et al., 1999). The dephosphorylation of Ser66 after etoposide-induced disassembly of replication factories is consistent with this model.
Redistribution of hLigI and PCNA is preceded by the formation of RPA2 foci. After 1 h of growth in the presence of VP-16, in a subset of cells with RPA2 foci, a fraction of hLigI was still associated with replication factories. We observed that in these cells the RPA2 and hLigI lay in close proximity without overlapping. We propose that proximity between RPA2 foci and replication factories could reflect the preferential accumulation of RPA in postreplicative chromatin, as expected from a role of this protein in postreplicative DNA repair. This hypothesis is in accord with the fact that Rad51, which colocalizes with RPA2 at sites of recombinational repair (Raderschall et al., 1999), also has been recently found associated with postreplicative chromatin, close to the replication foci (Tashiro et al., 2000). In this scenario the disassembly of replication factories induced by etoposide treatment would originate when replication forks encounter topo II-DNA–cleavable complexes trapped by VP-16. This event would be accompanied by the formation of DNA repair foci in nearby regions.
We found that aphidicolin, which produces the stalling of replication forks, prevents the disassembly of replication factories. Because aphidicolin does not affect the formation of topo II-DNA–cleavable complexes (Holm et al., 1989; Kaufmann, 1998), our observation suggests that the disassembly of replication factories is due neither to the stalling of replication forks nor to topo II-DNA–cleavable complexes. Instead, it is conceivable that the signal that triggers dispersal of hLigI and PCNA originates when replication forks meet topo II-DNA–cleavable complexes leading to the activation of an intra-S-phase checkpoint. The involvement of checkpoint kinases is suggested by the ability of staurosporine, an inhibitor of phospholipid/calcium-dependent protein kinases and a cell-cycle checkpoint abrogator, to prevent 1) the formation of RPA2 foci, 2) the hyperphosphorylation of RPA2, and 3) the disassembly of replication factories. The analysis of AT cells indicates that the redistribution of PCNA, hLigI, and RPA occurs even in the absence of an active ATM protein kinase. ATM is instead involved in RPA2 phosphorylation that is not detectable in AT cells grown for 1 h in the presence of VP-16. This finding is consistent with a previous report by Liu and Weaver (1993) who showed that phosphorylation of RPA2 in response to DNA damage is delayed in AT cells. Altogether these results indicate that the formation of RPA2 foci does not require phosphorylation of RPA2 and suggest that phosphorylation of RPA2 can occur within the foci.
In yeast an intra-S-phase checkpoint, mediated by the Rad53/Mec1 protein kinases, is activated both by DSBs and by stalled replication forks (Santocanale and Diffley, 1998). This seems to be different in mammalian cells in which independent checkpoint pathways appear to be activated in response to DNA damage and to the block of DNA replication. We have observed that etoposide-induced DNA damage triggers the disassembly of replication factories and that inhibition of checkpoints by staurosporine efficiently prevents this process. On the other hand, Dimitrova and Gilbert (2000) recently reported that stalled replication forks stabilize replication factories. Inhibition of checkpoints in aphidicolin-arrested cells results in redistribution of PCNA and RPA2 from early to late replicating domains in the absence of DNA replication. The checkpoint activation by stalled replication forks could serve to maintain genome integrity. Indeed, the disintegration of the stalled forks, coupled with the displacement of MCM proteins triggered by initiation of DNA replication, would completely prevent cells from replicating large portions of the genome (Dimitrova and Gilbert, 2000). We suggest that the disassembly of replication factories occurring after VP-16 treatment could unveil a strategy devised by the cells to cope with spontaneous DSBs during DNA replication. In S-phase, a fraction of DSBs are repaired by the homologous recombination pathway that entails the production of extended single-stranded regions in the postreplicative chromatin and the successive pairing of the two sister chromatids. This strand invasion would create a modified replication fork involving leading and lagging strand synthesis from the donor template as it has been recently proposed for the MAT locus in S. cerevisiae (Holmes and Haber, 1999). It is conceivable that this process is accompanied by the disassembly of single replication factories. Etoposide could amplify this process leading to the massive, intra-S-phase disassembly of replication factories and preventing large portion of the genome to be duplicated by the replication apparatus. In this perspective our results suggest an additional mechanism by which etoposide can exert its cytotoxic effect.
ACKNOWLEDGMENTS
The authors thank Centro Grandi Strumenti of the University of Pavia for the confocal microscopy facility. This work was supported by a grant from Associazione Italiana per la Ricerca sul Cancro to A.M. and from Ministero dell'Universitá e della Ricerca Scientifica e Technologica-Consiglio Nazionale delle Ricerche “Biomolecole per la salute umana” L. 95/95 to G.B. R.R. was supported by a fellowship from Consiglio Nazionale delle Ricerche. G.F. was supported by a fellowship from the Fondazione Adriano Buzzati Traverso.
REFERENCES
- Cardoso MC, Leonhardt H, Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin A and cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- Chen J, Chen S, Saha P, Dutta A. p21Cip1/Waf1disrupts the recruitment of human Fen1 by proliferating-cell nuclear antigen into the DNA replication complex. Proc Natl Acad Sci USA. 1996;93:11597–11602. doi: 10.1073/pnas.93.21.11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LS-H, Ian H-I, Koh T-W, Ng H-H, Xu G, Li BFL. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- Cox LS. Who binds wins: competition for PCNA rings out cell-cycle changes. Trends Biochem Sci. 1997;7:493–498. doi: 10.1016/S0962-8924(97)01170-7. [DOI] [PubMed] [Google Scholar]
- Dasika GK, Lin SC, Zhao S, Sung P, Tomkinson A, Lee EY. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat Cell Biol. 2000;2:686–694. doi: 10.1038/35036309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C, Covey JM, Kerrigan D, Pommier Y. Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res. 1989;49:6365–6368. [PubMed] [Google Scholar]
- Holmes AM, Haber JE. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Hozak P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to a nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- Hozak P, Jackson DA, Cook PR. Replication factories and nuclear bodies: the ultrastructural characterization of replication sites during the cell cycle. Cell. 1994;107:2191–2202. doi: 10.1242/jcs.107.8.2191. [DOI] [PubMed] [Google Scholar]
- Kaufmann SH. Cell death induced by topoisomerase-targeted drugs: more questions than answers. Biochim Biophys Acta. 1998;1400:195–211. doi: 10.1016/s0167-4781(98)00136-5. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamarre D, Talbot B, de Murcia G, Laplante C, Leduc Y, Mazen A, Poirier GG. Monoclonal antibodies against poly(ADP-ribose)polymerase: epitope mapping, inhibition of activity, inter-species immunoreactivity and cellular distribution of enzyme. Biochim Biophys Acta. 1988;950:147–160. doi: 10.1016/0167-4781(88)90007-3. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Rahn H-P, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC. Dynamics of DNA replication factories in living cells. J Cell Biol. 2000;149:271–279. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu V, Weaver D. The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol Cell Biol. 1993;13:7222–7231. doi: 10.1128/mcb.13.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A, Rossi R, Levin DS, Gary R, Park MS, Motycka TA, Ciarrocchi G, Villa A, Biamonti G, Tomkinson AE. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A, Savini E, Biamonti G, Stefanini M, Focher F, Ciarrocchi G. Late induction of human DNA ligase I after UV-C irradiation. Nucleic Acids Res. 1995a;23:962–966. doi: 10.1093/nar/23.6.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A, Savini E, Weighardt F, Rossi R, Ciarrocchi G, Villa A, Biamonti G. The N-terminal domain of human DNA ligase I contains the nuclear localization signal and directs the enzyme to sites of DNA replication. EMBO J. 1995b;14:5379–5386. doi: 10.1002/j.1460-2075.1995.tb00222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri C, Bernardi R, Braghetti A, Ricotti GC, Scovassi AI. The effect of the chemotherapeutic drug VP-16 on poly(ADP-ribosylation) in apoptotic HeLa cells. Carcinogenesis. 1993;14:2559–2564. doi: 10.1093/carcin/14.12.2559. [DOI] [PubMed] [Google Scholar]
- O'Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raderschall E, Golub EI, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci USA. 1999;96:1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi R, Villa A, Negri C, Scovassi I, Ciarrocchi G, Biamonti G, Montecucco A. The replication factory targeting sequence/PCNA-binding site is required in G(1) to control the phosphorylation status of DNA ligase I. EMBO J. 1999;18:5745–5754. doi: 10.1093/emboj/18.20.5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- Shah GM, Poirier D, Duchaine C, Brochu G, Desnoyers S, Lagueux J, Verreault A, Hoflack J-C, Kirkland JB, Poirier GG. Detection of poly(ADP-ribose) polymerase and its apoptosis-specific fragment by a nonisotopic activity-western blot technique. Anal Biochem. 1995;227:1–13. [Google Scholar]
- Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro S, Walter J, Shinohara A, Kamada N, Cremer T. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J Cell Biol. 2000;150:283–291. doi: 10.1083/jcb.150.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Zhou B-BS, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]