Abstract
Background
Rotavirus is an important cause of severe diarrhoea in children. The aims of this study were to identify the rotavirus strains that cause diarrhoea in children in Yogyakarta and to determine the association between rotavirus positivity and its clinical manifestations.
Methods
Clinical data and stool samples were collected from children hospitalised at Kodya Yogyakarta Hospital, Indonesia. Rotavirus was detected in stool samples using an enzyme immunoassay (EIA), which was followed by genotyping using reverse transcriptase polymerase chain reaction (RT-PCR). Electropherotyping was performed for the rotavirus-positive samples.
Results
In total, 104 cases were included in the study, 57 (54.8%) of which were rotavirus-positive. Based on a multiple logistic regression analysis, age group, vomiting and stool mucous were associated with rotavirus positivity. Most of the 56 samples subjected to genotyping were classified as G1 (80.36%) and P[8] (69.64%) genotypes. The genotype combination G1P[8] was identified as the most prevalent strain (66.07%). Of the 19 samples subjected to electropherotyping, 17 G1 isolates and 1 G3 isolate had long patterns, and 1 G1 isolate had a short pattern.
Conclusion
G1P[8] was the most dominant strain of rotavirus causing diarrhoea in children in Yogyakarta. Age group, vomiting and stool mucous were associated with rotavirus positivity.
Keywords: rotavirus, diarrhoea, G-type, P-type, children
Introduction
Rotavirus is an RNA virus that mainly infects the gastrointestinal tract. It is the most prevalent agent causing severe diarrhoea among infants and young children in both developed and developing countries, including Indonesia (1–3). It was recently estimated that rotavirus is responsible for 7,200,000–11,500,000 severe diarrhoea episodes and 133,000–284,000 deaths, most of which occur in developing countries (2). The incidence might be lower in countries that have introduced rotavirus vaccination (2).
Since rotavirus diarrhoea is prevalent among children in developed and developing countries, it is unlikely that improvements in sanitation and hygiene will be able to lower the incidence of the disease. Therefore, one of the best strategies for decreasing the global burden of disease is development and implementation of effective vaccines (1). The current strategy for developing rotavirus vaccines involves oral administration of live attenuated viruses that protection is expected to be similar to natural rotavirus infection (4, 5).
Rotavirus has three essential antigenic specificities, determined by the viral proteins VP6, VP7 and VP4. Based on the VP6 protein, rotavirus is classified into seven groups (A to G), yet only groups A, B and C infect humans. VP7, a glycoprotein, determines G-genotype, while VP4, which is protease-sensitive, determines P-genotype. VP7 and VP4 form the outer layer of rotavirus and elicit neutralising antibody responses. Therefore, they are implicated in the dual classification of rotavirus group A as G- and P-type (6).
The effectiveness of rotavirus vaccine can be enhanced by identification of the patterns of circulating virus serotypes and the role of these serotypes in causing clinical illness (7). During vaccine implementation, strain surveillance is necessary to monitor antibody-escape variants and changes in the circulating rotavirus strains among humans (8).
Two rotavirus vaccines, Rotarix and RotaTeq, have been developed by GSK and Merck, respectively. RotaTeq is a live attenuated pentavalent vaccine containing G1, G2, G3, G4 and P[8] rotavirus derived from reassortants of human and bovine strains. Rotarix is a live attenuated monovalent vaccine derived from the most common human rotavirus strain, G1P[8]. These vaccines have been proven to be highly effective for the global prevention of severe diarrhoea (9, 10). Before implementing the rotavirus vaccination programme in Yogyakarta, it is crucial to perform a study to ensure that the vaccines used in the programme match with the most prevalent and clinically relevant rotavirus strain in Yogyakarta.
The aims of this study were to identify the rotavirus strains currently circulating in Yogyakarta and to determine the association between rotavirus positivity and the illness’s clinical manifestation. This study provides accurate information about the rotavirus strains circulating in Yogyakarta before and after the implementation of the vaccination programme.
Materials and Methods
Study population and case definition
From February to August 2009, we enrolled 104 children under five years of age that were hospitalised at Kodya Yogyakarta General Hospital, Indonesia due to acute diarrhoea. We defined acute diarrhoea as 3 or more instances of loose stool within 24 hours for a duration of fewer than 2 weeks (11). We excluded patients whose faecal samples could not be obtained within 48 hours of admission to the hospital.
Clinical data collection
The patients’ medical histories, including previous occurrences of blood and mucous in stool, fever, vomiting, dehydration and diarrhoea, were collected.
Sample collection and storage
Stool samples were collected within the first 48 hours after admission according to WHO protocol (12). Stool specimens were stored at 4 °C – 8 °C before they were transported to the Department of Microbiology, Faculty of Medicine, Universitas Gadjah Mada, Indonesia. The specimens were then aliquoted into several tubes and stored at −20 °C.
Detection of rotavirus in stool samples
All stool samples were examined for the presence of group A rotavirus by enzyme immunoassay (EIA) using the IDEIA™ Rotavirus (DakoCytomation) kit according to the manufacturer’s instructions. Other possible causes of diarrhoea were not examined.
RT-PCR genotyping
Rotavirus RNA was extracted from rotavirus-positive faecal specimens using Trizol (Invitrogen) according to the manufacturer’s instructions. Rotavirus RNA was analysed to determine the VP7 (G-type) and VP4 (P-type) genotypes using reverse transcriptase polymerase chain reaction (RT-PCR) as previously described (13–15). For G typing, the primers 9con1L (13) and VP7R (15) were used during the first round of RT-PCR (20 cycles) to amplify the 905-bp of the VP7 gene segment. Then, 9con1L was used in the second round of PCR (20 cycles) with the type-specific primers 9T1 (G1), 9T-2 (G2), 9T-3P (G3), 9T4 (G4) and 9T-9B (G9) (Table 1) (13). For P-typing, the primers con2 and con3 were used in the firstround of RT-PCR (20 cycles) to amplify the 877-bp fragment of the VP4 gene segment (14). Con3 was used in the second round of PCR (30 cycles) with the type-specific primers 1T-1D (P[8]), 2T-1 (P[4]), 3T-1(P[6]), 4T-1 (P[9]) and 5T-1 (P[10]) (Table 2) (14).
Table 1.
Primers used for VP7 genotyping of rotavirus strains
| Primer | Sequence ( 5′-3′) | Position | PCR prouct (bp) | References | |
|---|---|---|---|---|---|
| First amplification of G-typing as described by Das et al. (13) and Gomara et al. (15) | |||||
| 9con1L | TAG CTC CTT TTA ATG TAT GGT AT | 37–59 | 895 | Das BK et al. (13) | |
| VP7R | AAC TTG CCA CCA TTT TTT CC | 914–932 | Gomara MI et al. (15) | ||
| First amplification of G-typing as described by Gomara et al. (15) | |||||
| VP7F | ATG TAT GGT ATT GAA TAT ACC AC | 51–71 | 881 | Gomara MI et al. (15) | |
| VP7R | AAC TTG CCA CCA TTT TTT CC | 914–932 | Gomara MI et al. (15) | ||
| Second amplification of G-typing as described by Das et al. (13) | |||||
| G1 | 9T-1 | TCT TGT CAA AGC AAA TAA TG | 176–195 | 158 | Das BK et al. (13) |
| G2 | 9T-2 | GTT AGA AAT GAT TCT CCA CT | 262–281 | 244 | Das BK et al. (13) |
| G3 | 9T-3P | GTC CAG TTG CAG TGT AGC | 484–501 | 464 | Das BK et al. (13) |
| G4 | 9T-4 | GGG TCG ATG GAA AAT TCT | 423–440 | 403 | Das BK et al. (13) |
| G9 | 9T-9B | TAT AAA GTC CAT TGC AC | 131–147 | 110 | Das BK et al. (13) |
| Second amplification of G-typing as described by Gouvea et al. (16) | |||||
| G1 | aBT1 | CAA GTA CTC AAA TCA ATG ATG G | 314–335 | 618 | Gouvea et al. (16) |
| G2 | aCT2 | CAA TGA TAT TAA CAC ATT TTC TGT G | 411–435 | 521 | Gouvea et al. (16) |
| G3 | G3 | ACG AAC TCA ACA CGA GAG G | 250–259 | 682 | Gouvea et al. (16) |
| G4 | aDT4 | CGT TTC TGG TGA GGA GTT G | 480–498 | 452 | Gouvea et al. (16) |
| G8 | aAT8 | GTC ACA CCA TTT GTA AAT TCG | 178–198 | 754 | Gouvea et al. (16) |
| G9 | aFT9 | CTT GAT GTG ACT AYA AAT AC | 757–776 | 179 | Gouvea et al. (16) |
Table 2.
Primers used for VP4 genotyping of rotavirus strains
| Primer | Sequence (5′-3′) | Position | PCR product (bp) | References | |
|---|---|---|---|---|---|
| First amplification of P-typing as described by Gentsch et al. (14) | |||||
| Con-3 | TGG CTT CGC TCA TTT ATA GAC A | 11–32 | 876 | Gentsch et al. (14) | |
| Con-2 | ATT TCG GAC CAT TTA TAA CC | 868–887 | Gentsch et al. (14) | ||
| First amplification of P-typing as described by Simmonds et al. (8) | |||||
| VP4F | TAT GCT CCA GTN AAT TGG | 132–149 | 663 | Simmonds et al. (8) | |
| VP4R | ATT GCA TTT CTT TCC ATA ATG | 775–795 | Simmonds et al. (8) | ||
| Second amplification of P-typing as described by Gentsch et al. (14) | |||||
| P[4] | 2T-1 | CTA TTG TTA GAG GTT AGA GTC | 474–494 | 483 | Gentsch et al. (14) |
| P[6] | 3T-1 | TGT TGA TTA GTT GGA TTC AA | 259–278 | 267 | Gentsch et al. (14) |
| P[8] | 1T-1D | TCT ACT TGG ATA ACG TGC | 339–356 | 345 | Gentsch et al. (14) |
| P[9] | 4T-1 | TGA GAC ATG CAA TTG GAC | 385–402 | 391 | Gentsch et al. (14) |
| P[10] | 5T-1 | ATC ATA GTT AGT AGT CGG | 575–594 | 583 | Gentsch et al. (14) |
Samples that failed to yield a detectable PCR product from secondary amplification of the primer-specific PCR typing were re-typed with other primers using the methods described by Gouvea et al. (16) and Simmond et al. (8). For G typing, the consensus primers VP7F and VP7R were used in the first round of RT-PCR (30 cycles) to generate the 881-bp of the VP7 gene segment (15). VP7F was used in the second round of PCR (30 cycles) with the type-specific primers aBT1 (G1), aCT-2 (G2), G3 (G3), aDT4 (G4) and G9 (G9) (Table 1) (16). For P-typing, the consensus primers VP4F and VP4R were used in the first round of RT-PCR (30 cycles) to generate a 663-bp fragment of the VP4 gene segment (8). VP4F was used in the second round of PCR (40 cycles) with the type-specific primers 1T-1D (P[8]), 2T-1 (P[4]), 3T-1(P[6]), 4T-1 (P[9]) and 5T-1 (P[10]) (Table 2) (14). All PCR products were separated in 2% agarose gel and visualised under UV light after staining with ethidium bromide.
Electropherotyping
Rotavirus RNA was extracted using Minikit (Qiagen) according to the manufacturer’s instructions. RNA was subjected to polyacrylamide gel electrophoresis to separate the 11 segments of dsRNA, which were then visualised by silver staining.
Statistical analyses
The variables were described using frequencies and percentages. Chi-square tests and logistic regression analyses were used to determine the association between rotavirus infections and clinical manifestations using STATA 13. The variables that had P-values of less than 0.25 according to the chi-square tests were entered into a logistic regression analysis model. Age group and all the clinical symptoms analysed (vomiting, stool mucous, bloody stool and fever) had P-values of less than 0.25 (Table 3). Bloody stool was omitted because the chi-square test revealed cells with a count of zero. Therefore, age group, vomiting, stool mucous and fever were examined in the multiple logistic regression analysis. The fit of the model was checked using the Hosmer-Lemeshow test. The results were presented as crude and adjusted odds ratios with a 95% confidence interval. P-values of less than 0.05 were considered statistically significant.
Table 3.
Characteristics and clinical symptoms of children under five years of age hospitalised at Kodya Yogyakarta General Hospital (n = 104)
| Variable | n (%) of patients with rotavirus-positive diarrhoea (n = 57) | n (%) of patients with rotavirus-negative diarrhoea (n = 47) | n | X2-Statistica (df) | P-valuea | |
|---|---|---|---|---|---|---|
| Gender | Male | 41 (71.9) | 29 (61.7) | 70 | 1.22 (1) | 0.27 |
| Female | 16 (28.1) | 18 (38.3) | 34 | |||
| Age groups | 0–11 | 11 (19.3) | 25 (53.2) | 36 | 4.57 (1) | 0.03* |
| 12–23 | 29 (50.9) | 9 (19.1) | 38 | 2. 96 (1) | 0.08 | |
| 24–59 | 17 (29.8) | 13 (27.7) | 30 | |||
| Clinical manifestation | Vomiting | 46 (80.7) | 27 (57.4) | 73 | 6.66(1) | 0.01* |
| Stool mucous | 6 (10.5) | 17 (36.2) | 23 | 9.83 (1) | 0.00* | |
| Bloody stool | 0 (0) | 11 (23.4) | 11 | 14.92 (1) | 0.00* | |
| Fever | 18 (31.6) | 23 (48.9) | 41 | 3.25 (1) | 0.07 | |
chi-square test for independence
Ethical approval
The research protocol was approved by the Ethical Committee of the Faculty of Medicine, Universitas Gadjah Mada, Yogyakarta, Indonesia. Informed consent was provided by the parents or guardians of each child before the children were enrolled in the study.
Results
From February to August 2009, 104 children under five years of age who were hospitalised at Kodya Yogyakarta General Hospital, Indonesia due to acute diarrhoea were enrolled in the study. There were 70 (67.3%) male and 34 (32.7%) female patients (Table 3). Of the 104 samples collected, 57 (54.8%) were positive for rotavirus, as determined by EIA.
Rotavirus diarrhoea particularly affects young children; 70.2% of the cases involved children under two years of age. Most cases involved children aged 12–23 months, with the next largest group comprised of children aged 24–59 months (Table 3). Additionally, the chi-square tests revealed significant differences in vomiting, stool mucous and bloody stool between rotavirus-positive and rotavirus-negative patients (Table 3).
We present the final model of the multiple logistic regression analysis in Table 4. There are four variables associated with rotavirus positivity: age group, including the 0–11-month-old group (adjusted OR = 0.31; CI 95% = 0.10 to 0.93; P = 0.04) and 12–23-month-old group (adjusted OR = 3.46; CI 95% 1.03 to 11.60; P = 0.04), stool mucous (adjusted OR = 0.24; CI 95% = 0.07 to 0.79; P = 0.02), and vomiting (adjusted OR = 3.97; CI 95% = 1.32 to 11.97; P = 0.01). In our study, only one patient had mild dehydration, and all the others (n = 103) had no dehydration.
Table 4.
Association between age groups, clinical symptoms and rotavirus positivity revealed by multiple logistic regression
| Variable | b | Adjusted OR (95% CI) | P-valuea |
|---|---|---|---|
| Age group: 0–11 months vs. 24–59 monthsb | −1.17 | 0.31 (0.10–0.93) | 0.04 |
| Age group: 12–23 months vs. 24–59 monthsb | 1.24 | 3.46 (1.03–11.60) | 0.04 |
| Vomiting | 1.38 | 3.97 (1.32–11.97) | 0.01 |
| Stool mucous | −1.41 | 0.24 (0.07–0.79) | 0.02 |
OR = odds ratio.
Likelihood ratio test;
reference category.
Of the 57 rotavirus-positive samples, 56 faecal specimens were characterised as G or P genotypes. G1 was the most prevalent genotype (80.36%), followed by G2 (16.07%) and G3 (3.57%) (Table 5). Regarding P-typing, they were classified as P[8] (69.64%), P[4] (17.86%), P[6] (7.14%) or untypeable (5.36%) (Table 5). The identified G- and P-type combinations were G1P[8] (66.07%), G2P[4] (16.07%), G1P[6] (7.14%), G1P[untypeable] (5.36%), G3P[8] (3.57%) and G1P[4] (1.79%) (Table 5).
Table 5.
Rotavirus genotype distribution in Kodya Yogyakarta Hospital
| Rotavirus strains | Total (%) (n=56) |
|---|---|
| G genotype | |
| G1 | 45 (80.36) |
| G2 | 9 (16.07) |
| G3 | 2 (3.57) |
| P genotype | |
| P[8] | 39 (69.64) |
| P[4] | 10 (17.86) |
| P[6] | 4 (7.14) |
| P[untypeable] | 3 (5.36) |
| G- and P-type combinations | |
| G1P[8] | 37 (66.07) |
| G2P[4] | 9 (16.07) |
| G1P[6] | 4 (7.14) |
| G1P[untypeable] | 3 (5.36) |
| G3P[8] | 2 (3.57) |
| G1P[4] | 1 (1.79) |
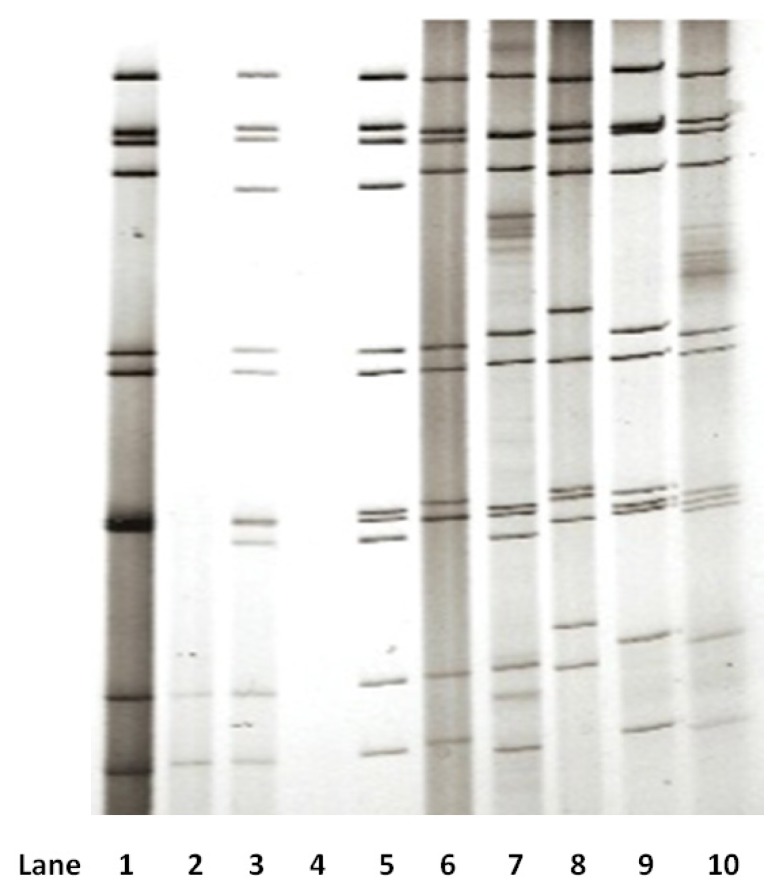
A number of rotavirus-positive stool samples did not have sufficient volume for polyacrylamide gel electrophoresis (PAGE) analysis. Only 24 samples were subjected to PAGE analysis, and 19 isolates had good results. Eighteen samples were identified as G1, and one sample was identified as G3. All of them showed the migration pattern of the genome of rotavirus group A: 4-2-3-2. The results of electropherotyping showed that 17 of 18 G1 isolates and 1 G3 isolate had long patterns, while 1 G1 isolate had a short pattern (Figure 1).
Figure 1.
Electropherotype profile of rotavirus RNA genome. Lanes 1–5: sample RW13 (G3P[8]) - long pattern; sample RW22 - RNA insufficient; sample RW23 (G1P[8]) - long pattern; sample RW26 - no RNA; sample RW30 (G1P[8]) - long pattern. Lanes 6–10 are control strains: RV3 (G3P[6]) - long pattern; RV4 (G1P[8]) - long pattern; RV5 (G2P[4]) - short pattern; F45 (G9P[8]) - long pattern; ST3 (G4P[6]) - long pattern.
Discussion
This hospital-based study described a high incidence of rotavirus infection in Yogyakarta, Indonesia. Rotavirus infection was identified in 54.8% of 104 children hospitalised with acute diarrhoea. Compared to previous hospital-based studies performed in Indonesia in the late 1970s and 1990s, the incidence rate was similarly high. Bishop et al. (17) reported rotavirus infection in 38% of children with severe acute diarrhoea in Yogyakarta. Subekti et al. (18), who conducted a study in Jakarta from 1997–1999, showed that 170 of 236 (72%) children with diarrhoea were infected with rotavirus. Putnam et al. (19), who conducted a study in Medan, Makassar, Jakarta, Yogyakarta, Surabaya, Bali and Nusa Tenggara, found that 748 of 1,660 (45.1%) children under five years of age suffered from diarrhoea due to rotavirus. Moreover, Soenarto et al. (20), who conducted a study of children under five years of age in Yogyakarta, Jakarta, Bandung, Mataram, Denpasar and Palembang in 2006, showed that 60% of the 2,240 children studied suffered from diarrhoea due to rotavirus. These findings support the need for continuous surveillance of the disease in various locations.
Our study found a high incidence of rotavirus infection in Yogyakarta, Indonesia, which is comparable to the findings of similar studies in surrounding countries, such as Malaysia (38%) (21), Thailand (43%) (22), Cambodia (56%) (23) and Myanmar (57%) (24). Our study provides evidence that rotavirus infection gives a substantial burden to the diarrhoeal diseases and it is critical to develop an effective approach to preventing infection.
Rotavirus infection in children clinically manifests with several symptoms, such as fever, vomiting and watery, bloodless diarrhoea (25, 26). These clinical manifestations are more severe than those of other causes of viral gastroenteritis (27). In our study, vomiting was associated with rotavirus positivity (Table 4), which aligns with the findings of other studies (20, 28). Those studies also reported that rotavirus-positive patients were less likely to have bloody diarrhoea (20, 28), in agreement with our study.
Severe complications of rotavirus infection, such as bloody diarrhoea associated with necrotising enterocolitis (NEC) and bowel perforation, have been described (25, 26, 29). Even though they have not yet been fully elucidated, several factors were associated with the pathogenesis of the disease (30). However, an enterotoxin protein, NSP4, and the ability of rotavirus to trigger the apoptosis of intestinal cells might be the factors that most affect the manifestation of the disease (31).
Our study demonstrated that the percentage of children aged 0–11 months who suffered from rotavirus diarrhoea was lower than that of children aged 12–23 and 24–59 months (Table 3). This phenomenon is probably due to the presence of anti-rotavirus immunoglobulin G (IgG) and IgA antibodies in breast milk (32). In their study of children in Yogyakarta, Chan et al. (33) found a high concentration of anti-rotavirus IgG and IgA in colostrum as well as in breast milk. Moreover, Clemens et al. (34) found that exclusive breastfeeding confers better protection against severe rotavirus infection than partial or no breastfeeding. However, further study is required to fully address this issue in our patient population.
The genetic diversity of circulating rotavirus strains may have a crucial role in the initiation of the vaccination programme and its clinical evaluation. Some studies have been conducted to identify the rotavirus strains circulating in Indonesia. Using samples collected in Yogyakarta from 1978–1979, Bishop et al. (17) identified G1 (2%); G2 (9%), G3 (53%) and G4 (36%) strains. However, Soenarto et al. (20) reported that the G1 and G9 genotypes were the most prevalent, accounting for 73 (30%) and 69 (29%) of the 240 samples, respectively, followed by G2, which accounted for 34 samples (14%). Putnam et al. (19) reported that the rotavirus strains causing diarrhoea in Indonesia were P[4] (16.2%), P[6] (11.3%), P[8] (24.7%), P[9] (0.7%), P[10] (1.3%), P[11] (0.5%), mixed infection (3.6%) and untypeable (1.6%). However, Soenarto et al. (20) found that the P-type rotavirus strains causing diarrhoea were distributed differently: P[4] (16.4%), P[6] (55.6%), P[8] (17.5%), mixed infection (9%) and untypeable (1.6%).
We found that G1P[8] was the most dominant strain of rotavirus causing diarrhoea in children under five years of age hospitalised at Kodya Yogyakarta Hospital. G1P[8] was also the predominant strain identified in Malaysia in a 1996 study (35). However, G9P[8] genotypes became the most common strains in Malaysia from 2001–2003 and in 2007 (21, 35).
In our study, G2P[4], G1P[6], G3P[8] and G1P[4] were also common causative agents. Since the majority of rotavirus strains circulating in Yogyakarta are similar to those circulating globally, we suggest that the current rotavirus vaccines can effectively reduce the prevalence of rotavirus diarrhoea in children under five years of age in Yogyakarta.
Rotavirus has several mechanisms to generate genetic diversity. One is accumulation of point mutation (genetic drift), which is due to the error-prone nature of RNA-dependent RNA polymerase (RdRp) enzyme (36). Such mutations could be introduced at the primer binding sites and result in typing failure (8, 15). Another important mechanism of rotavirus evolution is genetic shift due to reassortment of the segmented RNA genome. This event could lead to a new combination of G and P types (36) and introduce novel genotypes through zoonotic transmission (37). Both mechanisms may explain our finding that the P-type of about 5% of samples could not be identified. The rapid evolution of rotavirus may increase the number of strains that cannot be genotyped with currently available assays. Therefore, our study underscores the importance of regularly updating typing methods to successfully detect the currently circulating rotavirus strains.
Continuous monitoring is important for identifying circulating rotavirus strains before and after the introduction of the vaccine. It will provide information about the impact of the vaccine on genotype distribution and the possible emergence of less common rotavirus strains circulating in certain geographical areas. It is also crucial to assess the effectiveness of the vaccination programme against rotavirus diarrhoea.
According to the migration of its 11 RNA gene segments, rotavirus can also be classified as either long or short RNA electropherotype using polyacrylamide gels. The short electropherotype occurs as a result of partial duplication in gene segment 11. This gene runs more slowly than gene segment 10; because of its standard size, gene segment 11 in long-electropherotype strains migrates faster than segment 10 (38). In this study, we found that most G1 and G3 isolates had a long electropherotype. Long electropherotypes usually belong to the Wa genogroup, whereas short electropherotypes typically belong to the DS-1 genogroup (36).
Conclusion
We found that rotavirus caused acute diarrhoea in 54.8% of children under five years of aged hospitalised at Kodya Yogyakarta General Hospital. The majority (66.07%) was G1P[8], the most prevalent strain identified globally. Age group, vomiting and stool mucous were associated with rotavirus positivity.
Acknowledgement
This research was supported by a grant from the Indonesian Government (RISBINIPTEKDOK 2009/2010). Also supported by the Indonesia Endowment Fund for Education (LPDP) for funding PhD fellowship to Mohamad S. Hakim. We thank Murdoch Children Research Institute Melbourne, Australia, for providing facilities to conduct G- and P-genotyping; Abdul Wahab (Universitas Gadjah Mada) for providing assistance in data analysis; Linda Oktabriana (Universitas Gadjah Mada), Nada Bogdanovic-Sakran and Carl D. Kirkwood (Murdoch Children Research Institute) for their technical support; and the parents of the children involved in this study for their willingness to participate.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Authors’ Contributions
Conception and design: HN, ATA
Analysis and interpretation of the data: HN, MSH
Drafting of the article: HN, MSH
Critical revision of the article for important intellectual content: QP
Final approval of the article: QP, ATA
Provision of study materials or patients: SA
Obtaining of funding: ATA
Administrative, technical, or logistic supports: IBNPD
Collection and assembly of data: HN, MSH, IBNPD
References
- 1.Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD, et al. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12(2):136–141. doi: 10.1016/S1473-3099(11)70253-5. https://doi.org/10.1016/S1473-3099(11)70253-5. [DOI] [PubMed] [Google Scholar]
- 2.Walker CL, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, et al. Global burden of childhood pneumonia and diarrhoea. Lancet. 2013;381(9875):1405–1416. doi: 10.1016/S0140-6736(13)60222-6. https://doi.org/10.1016/S0140-6736(13)60222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parashar UD, Gibson CJ, Bresee JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis. 2006;12(2):304–306. doi: 10.3201/eid1202.050006. https://doi.org/10.3201/eid1202.050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bresee JS, Hummelman E, Nelson EA, Glass RI. Rotavirus in Asia: the value of surveillance for informing decisions about the introduction of new vaccines. J Infect Dis. 2005;192(Suppl 1):S1–S5. doi: 10.1086/431515. https://doi.org/10.1086/431515. [DOI] [PubMed] [Google Scholar]
- 5.Glass RI, Bresee JS, Turcios R, Fischer TK, Parashar UD, Steele AD. Rotavirus vaccines: targeting the developing world. J Infect Dis. 2005;192(Suppl 1):S160–S166. doi: 10.1086/431504. https://doi.org/10.1086/431504. [DOI] [PubMed] [Google Scholar]
- 6.Dennehy PH. Rotavirus vaccines: an overview. Clin Microbiol Rev. 2008;21(1):198–208. doi: 10.1128/CMR.00029-07. https://doi.org/10.1128/CMR.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bern C, Unicomb L, Gentsch JR, Banul N, Yunus M, Sack RB, et al. Rotavirus diarrhea in Bangladeshi children: correlation of disease severity with serotypes. J Clin Microbiol. 1992;30(12):3234–3238. doi: 10.1128/jcm.30.12.3234-3238.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmonds MK, Armah G, Asmah R, Banerjee I, Damanka S, Esona M, et al. New oligonucleotide primers for P-typing of rotavirus strains: Strategies for typing previously untypeable strains. J Clin Virol. 2008;42(4):368–373. doi: 10.1016/j.jcv.2008.02.011. https://doi.org/10.1016/j.jcv.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 9.Desai R, Oliveira LH, Parashar UD, Lopman B, Tate JE, Patel MM. Reduction in morbidity and mortality from childhood diarrhoeal disease after species A rotavirus vaccine introduction in Latin America - a review. Mem Inst Oswaldo Cruz. 2011;106(8):907–911. doi: 10.1590/s0074-02762011000800002. [DOI] [PubMed] [Google Scholar]
- 10.Parashar U, Steele D, Neuzil K, Quadros C, Tharmaphornpilas P, Serhan F, et al. Progress with rotavirus vaccines: summary of the Tenth International Rotavirus Symposium. Expert Rev Vaccines. 2013;12(2):113–117. doi: 10.1586/erv.12.148. https://doi.org/10.1586/erv.12.148. [DOI] [PubMed] [Google Scholar]
- 11.Neves MA, Pinheiro HH, Silva RS, Linhares AC, Silva LD, Gabbay YB, et al. High prevalence of G12P[8] rotavirus strains in Rio Branco, Acre, Western Amazon, in the post-rotavirus vaccine introduction period. J Med Virol. 2016;88(5):782–789. doi: 10.1002/jmv.24404. https://doi.org/10.1002/jmv.24404. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization. Generic protocol for (i) hospital-based surveillance to estimate the burden of rotavirus gastroenteritis in children and (ii) a community-based survey on utilization of health care services for gastroenteritis in children. Field test version. Geneva, Switzerland: Department of Vaccines and Biologicals, World Health Organization; 2002. [Google Scholar]
- 13.Das BK, Gentsch JR, Cicirello HG, Woods PA, Gupta A, Ramachandran M, et al. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994;32(7):1820–1822. doi: 10.1128/jcm.32.7.1820-1822.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, et al. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30(6):1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomara MI, Cubitt D, Desselberger U, Gray J. Amino acid substitution within the VP7 protein of G2 rotavirus strains associated with failure to serotype. J Clin Microbiol. 2001;39(10):3796–3798. doi: 10.1128/JCM.39.10.3796-3798.2001. https://doi.org/10.1128/JCM.39.10.3796-3798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, et al. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28(2):276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bishop RF, Unicomb LE, Soenarto Y, Suwardji H, Ristanto, Barnes GL. Rotavirus serotypes causing acute diarrhoea in hospitalized children in Yogyakarta, Indonesia during 1978–1979. Arch Virol. 1989;107(3–4):207–213. doi: 10.1007/BF01317917. [DOI] [PubMed] [Google Scholar]
- 18.Subekti D, Lesmana M, Tjaniadi P, Safari N, Frazier E, Simanjuntak C, et al. Incidence of Norwalk-like viruses, rotavirus and adenovirus infection in patients with acute gastroenteritis in Jakarta, Indonesia. FEMS Immunol Med Microbiol. 2002;33(1):27–33. doi: 10.1111/j.1574-695X.2002.tb00568.x. [DOI] [PubMed] [Google Scholar]
- 19.Putnam SD, Sedyaningsih ER, Listiyaningsih E, Pulungsih SP, Komalarini, Soenarto Y, et al. Group A rotavirus-associated diarrhea in children seeking treatment in Indonesia. J Clin Virol. 2007;40(4):289–294. doi: 10.1016/j.jcv.2007.09.005. https://doi.org/10.1016/j.jcv.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Soenarto Y, Aman AT, Bakri A, Waluya H, Firmansyah A, Kadim M, et al. Burden of severe rotavirus diarrhea in Indonesia. J Infect Dis. 2009;200(Suppl 1):S188–S194. doi: 10.1086/605338. https://doi.org/10.1086/605338. [DOI] [PubMed] [Google Scholar]
- 21.Hung LC, Wong SL, Chan LG, Rosli R, Ng AN, Bresee JS. Epidemiology and strain characterization of rotavirus diarrhea in Malaysia. Int J Infect Dis. 2006;10(6):470–474. doi: 10.1016/j.ijid.2006.05.008. https://doi.org/10.1016/j.ijid.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Jiraphongsa C, Bresee JS, Pongsuwanna Y, Kluabwang P, Poonawagul U, Arporntip P, et al. Epidemiology and burden of rotavirus diarrhea in Thailand: results of sentinel surveillance. J Infect Dis. 2005;192(Suppl 1):S87–S93. doi: 10.1086/431508. https://doi.org/10.1086/431508. [DOI] [PubMed] [Google Scholar]
- 23.Nyambat B, Meng CY, Vansith K, Vuthy U, Rin E, Kirkwood C, et al. Hospital-based surveillance for rotavirus diarrhoea in Phnom Penh, Cambodia, March 2005 through February 2007. Vaccine. 2009;27(Suppl 5):F81–F84. doi: 10.1016/j.vaccine.2009.08.085. https://doi.org/10.1016/j.vaccine.2009.08.085. [DOI] [PubMed] [Google Scholar]
- 24.Moe K, Thu HM, Oo WM, Aye KM, Shwe TT, Mar W, et al. Genotyping of rotavirus isolates collected from children less than 5 years of age admitted for diarrhoea at the Yangon Children’s Hospital, Myanmar. Vaccine. 2009;27(Suppl 5):F89–F92. doi: 10.1016/j.vaccine.2009.08.068. https://doi.org/10.1016/j.vaccine.2009.08.068. [DOI] [PubMed] [Google Scholar]
- 25.Koukou D, Chatzichristou P, Trimis G, Siahanidou T, Skiathitou AV, Koutouzis EI, et al. Rotavirus Gastroenteritis in a Neonatal Unit of a Greek Tertiary Hospital: Clinical Characteristics and Genotypes. PLoS One. 2015;10(7):1–13. doi: 10.1371/journal.pone.0133891. https://doi.org/10.1371/journal.pone.0133891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma R, Hudak ML, Premachandra BR, Stevens G, Monteiro CB, Bradshaw JA, et al. Clinical manifestations of rotavirus infection in the neonatal intensive care unit. Pediatr Infect Dis J. 2002;21(12):1099–1105. doi: 10.1097/00006454-200212000-00003. https://doi.org/10.1097/01.inf.0000040560.26682.88. [DOI] [PubMed] [Google Scholar]
- 27.Chen SY, Chang YC, Lee YS, Chao HC, Tsao KC, Lin TY, et al. Molecular epidemiology and clinical manifestations of viral gastroenteritis in hospitalized pediatric patients in Northern Taiwan. J Clin Microbiol. 2007;45(6):2054–2057. doi: 10.1128/JCM.01519-06. https://doi.org/10.1128/JCM.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salim H, Karyana IP, Sanjaya-Putra IG, Budiarsa S, Soenarto Y. Risk factors of rotavirus diarrhea in hospitalized children in Sanglah Hospital, Denpasar: a prospective cohort study. BMC Gastroenterol. 2014;14:54. doi: 10.1186/1471-230X-14-54. https://doi.org/10.1186/1471-230X-14-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dearlove J, Latham P, Dearlove B, Pearl K, Thomson A, Lewis IG. Clinical range of neonatal rotavirus gastroenteritis. Br Med J (Clin Res Ed) 1983;286(6376):1473–1475. doi: 10.1136/bmj.286.6376.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenberg HB, Estes MK. Rotaviruses: from pathogenesis to vaccination. Gastroenterology. 2009;136(6):1939–1951. doi: 10.1053/j.gastro.2009.02.076. https://doi.org/10.1053/j.gastro.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin Y, Metselaar HJ, Sprengers D, Peppelenbosch MP, Pan Q. Rotavirus in organ transplantation: drug-virus-host interactions. Am J Transplant. 2015;15(3):585–593. doi: 10.1111/ajt.13135. https://doi.org/10.1111/ajt.13135. [DOI] [PubMed] [Google Scholar]
- 32.Yolken RH, Peterson JA, Vonderfecht SL, Fouts ET, Midthun K, Newburg DS. Human milk mucin inhibits rotavirus replication and prevents experimental gastroenteritis. J Clin Invest. 1992;90(5):1984–1991. doi: 10.1172/JCI116078. https://doi.org/10.1172/JCI116078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan J, Nirwati H, Triasih R, Bogdanovic-Sakran N, Soenarto Y, Hakimi M, et al. Maternal antibodies to rotavirus: could they interfere with live rotavirus vaccines in developing countries? Vaccine. 2011;29(6):1242–1247. doi: 10.1016/j.vaccine.2010.11.087. https://doi.org/10.1016/j.vaccine.2010.11.087. [DOI] [PubMed] [Google Scholar]
- 34.Clemens J, Rao M, Ahmed F, Ward R, Huda S, Chakraborty J, et al. Breast-feeding and the risk of life-threatening rotavirus diarrhea: prevention or postponement? Pediatrics. 1993;92(5):680–685. [PubMed] [Google Scholar]
- 35.Zuridah H, Kirkwood CD, Bishop RF, Bogdanovic-Sakran N, Yap KL. Molecular characterization and epidemiology of rotavirus isolates obtained from children with diarrhoea in Malaysia. Med J Malaysia. 2009;64(3):193–196. [PubMed] [Google Scholar]
- 36.Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, et al. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J Infect Dis. 2005;192(Suppl 1):S146–S159. doi: 10.1086/431499. https://doi.org/10.1086/431499. [DOI] [PubMed] [Google Scholar]
- 37.Martella V, Banyai K, Matthijnssens J, Buonavoglia C, Ciarlet M. Zoonotic aspects of rotaviruses. Vet Microbiol. 2010;140(3–4):246–255. doi: 10.1016/j.vetmic.2009.08.028. https://doi.org/10.1016/j.vetmic.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 38.Matsui SM, Mackow ER, Matsuno S, Paul PS, Greenberg HB. Sequence analysis of gene 11 equivalents from “short” and “super short” strains of rotavirus. J Virol. 1990;64(1):120–124. doi: 10.1128/jvi.64.1.120-124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]