Abstract
Electronic reporting of genetic testing results is increasing, but they are often represented in diverse formats and naming conventions. Logical Observation Identifiers Names and Codes (LOINC) is a vocabulary standard that provides universal identifiers for laboratory tests and clinical observations. In genetics, LOINC provides codes to improve interoperability in the midst of reporting style transition, including codes for cytogenetic or mutation analysis tests, specific chromosomal alteration or mutation testing, and fully structured discrete genetic test reporting. LOINC terms follow the recommendations and nomenclature of other standards such as the Human Genome Organization Gene Nomenclature Committee’s terminology for gene names. In addition to the narrative text they report now, we recommend that laboratories always report as discrete variables chromosome analysis results, genetic variation(s) found, and genetic variation(s) tested for. By adopting and implementing data standards like LOINC, information systems can help care providers and researchers unlock the potential of genetic information for delivering more personalized care.
Keywords: Genetics, LOINC, Medical records systems, Clinical laboratory information systems, Vocabulary, controlled
INTRODUCTION
Strong arguments exist for delivering molecular genetic test results to electronic health records (EHRs) as standards-based, structured (computable) electronic reports for clinical and research purposes.1–4 The fact that most genetic tests apply for a lifetime and may have to be automatically reinterpreted as new knowledge becomes available3 only strengthens these arguments. Logical Observation Identifiers Names and Codes (LOINC; Regenstrief Institute Inc, Indianapolis, IN) and HL7 (Health Level Seven International, Ann Arbor, MI) are the mainstays for structured reporting of routine laboratory test results.5 These 2 key standards also provide a framework for structured reporting of genomic tests6 in conjunction with widely accepted vocabulary standards for naming genes and genetic variations.3,7–9 In this report, we will describe LOINC’s approach to naming/coding genetic tests (and their results), its scope, its mechanisms for gathering new content, and its current limitations.
LOINC
LOINC is a universal code system developed by the Regenstrief Institute for identifying laboratory tests and clinical observations in electronic messaging.10 The current release14 contains more than 73 000 terms, covering the full scope of laboratory testing (chemistry, microbiology, etc) and a broad range of clinical measurements (eg, vital signs, electrocardiograms, patient reported outcomes, clinical document titles,11,12 and radiology reports13,14). LOINC includes a data model for representing answer lists, panels of individual observations, other details like help text, and units of measure.15 New versions of the standard are published twice yearly. LOINC has been widely adopted as the standard for laboratory test result names in the United States, where it is a national standard,16,17 and internationally.18,19 Many genetic test reporting initiatives,20,21 including the HL7 Clinical Genomics Working Group,22,23 have adopted LOINC.
Here we focus on the 1400+ LOINC terms currently used in human genetic test reporting. Documentation and downloads of the entire LOINC database are available at http://loinc.org.
Naming conventions in LOINC
Each LOINC term has a fully specified name containing 6 main axes (Component/Analyte, Property, Timing, System/Specimen, and Method). The axes produce names detailed enough to distinguish among similar observations.18 For naming genetic tests that target specific genetic variations, LOINC uses the Human Genome Organization Gene Nomenclature Committee’s terminology24 to name the gene(s) and the Human Genome Variation Society’s (HGVS) syntax to name the variation(s)25 of interest. We are aware of the challenges due to misnaming and version changes in these nomenclatures26; however, the thousands of LOINC users tend to detect and report such problems. Updates to LOINC terms follow best practices.27 Where updates do not alter the meaning, we correct “wrong” portions of the name and keep the old names as synonyms. Alternatively, a new term can be created and the old term retired, adding a forward link to the new term.
Scope of LOINC coverage
LOINC provides codes for a broad range of human molecular genetic and cytogenetic tests. Here we provide a sampling of the LOINC content so that laboratories will be better able to understand and use LOINC terms for ordering and reporting. Many laboratories report the “simpler” genetic tests as individual and computer-understandable HL7 Version 2 observations in a way similar to the reporting of routine chemistry and hematology tests. These simpler kinds of genetic tests are the ones most commonly ordered in primary care,28 and LOINC has observation codes available for most of them.
Tests for specific gene mutations
As of July 2014, LOINC carries 192 terms for reporting single mutations (variants) qualitatively as present or absent, eg, HFE gene.p.C282Y [21695-2]—the variation most commonly associated with hereditary hemochromatosis in Caucasians. It also carries a quantitative measure of single mutations, eg, JAK2 p.V617F mutant/control [53761-3]—a report on the prevalence of white cells with the variation associated with polycythemia vera. Both of these examples use HGVS nomenclature as part of the test name. The National Center for Biotechnology Information’s (NCBI) Single Nucleotide Polymorphism database (dbSNP) codes29 may also be embedded in LOINC test names, especially when the variation is outside of the coding region, eg, IL28B gene associated variant rs12979860 [60279-7]. LOINC also contains nearly 50 codes for trinucleotide repeats, including both qualitative tests with answers like not expanded, intermediate, or expanded, and quantitative tests that report the actual number of repeats, eg, HTT gene.CAG repeats [53782-9].
Tests for chromosomal alterations
Many chromosomal alteration tests can also be reported as computer-understandable observations using one or more LOINC codes. For example, LOINC contains codes for qualitative studies for trisomies and uniparental disomies, eg, Chromosome 21 trisomy [Presence] [21771-1]. Today, aneuploidy risk can also be estimated by quantifying the dosage of chromosome-specific circulating cell-free DNA in maternal plasma [see 73970-6], and such tests are revolutionizing prenatal testing.
A variety of LOINC codes are available for reporting structural chromosome rearrangements. For example, BCR-ABL1 testing for diagnosis and management of blood cell malignancies is reported qualitatively as t(9;22)(q34.1;q11)(ABL1,BCR) b2a2+b3a2 fusion transcript [42714-6]. The following 2 approaches exist for reporting the ratio of a BCR-ABL1 fusion transcript to a control transcript: the raw transcript to control ratio [e.g., 55147-3], and a transformed ratio based on an international standard [e.g., 69380-4]. To increase computability, tests that report a chromosomal alteration, such as BCR-ABL1, should also report the results using the International System for Human Cytogenetic Nomenclature (ISCN)30,31 when possible.
Fully structured reporting of molecular genetic and cytogenetic tests
LOINC collaborated with HL7 to provide codes for all of the variables contained in two HL7 Clinical Genomics Working Group implementation guides for structured reporting of genetic tests—one for reporting genetic variations23 detected via sequencing, gene chips, and other methods, and another for reporting the results of cytogenetic studies.22 All of the LOINC codes in these 2 implementation guides include guidance on how they are to be used, and the categorical variables are linked to specific sets of allowed answers.
The genetic variation collection consists of a number of LOINC panels, including one that reports the overall results of the study, another that reports the details about each variation found, and yet another that describes the region of interest and the gene chip as applicable (see figure 1). To improve the integration of cytogenetic test results into EHRs, Heras et al18 examined a range of sample cytogenetic test names and result reports and, from this empirical effort, created a LOINC-enabled HL7 specification for reporting cytogenetic tests in a fully structured form.22 The LOINC codes for this specification are organized under the Chromosome analysis master panel [62389-2], which includes a choice of subpanels for discrete reporting of the 3 major testing approaches—chromosome banding, fluorescence in situ hybridization, and microarrays. Variables in this collection include the genomic source class (eg, germline, somatic), genetic disease assessed, chromosome analysis in ISCN nomenclature, and overall interpretation (for microarray example, see figure 2).
Figure 1:
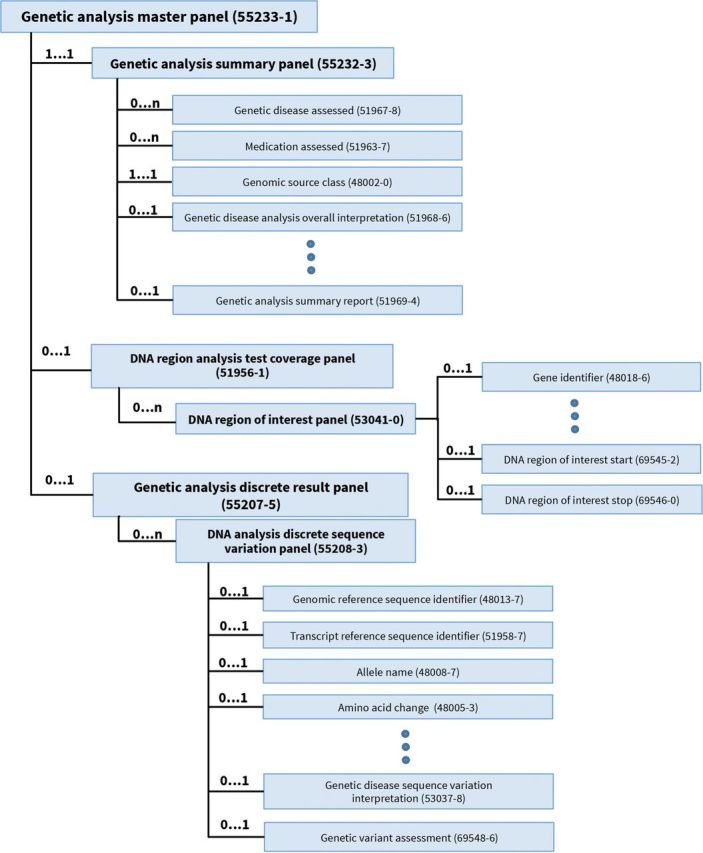
Genetic analysis master panel.
Figure 2:
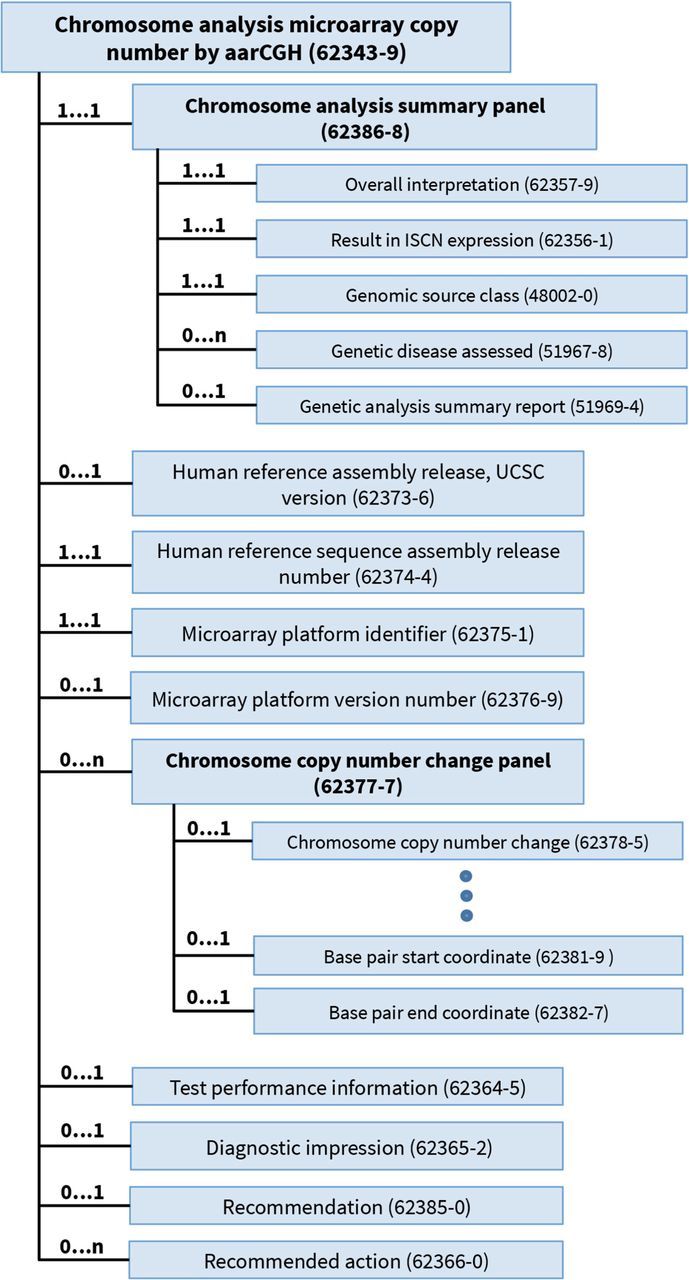
Chromosome analysis microarray copy number change panel, included within the Chromosome analysis master panel (62389-2) (not shown here).
Mutation analysis tests
These HL7 definitions have not yet been adopted widely, however; and molecular genetic tests, especially those for large numbers of variations, tend still to be reported as free-text narrative.28 To accommodate this reality and provide a standard way to order and report such tests, LOINC provides terms that include the gene name(s) and phrase mutation analysis. Currently LOINC distinguishes among the following different types of mutation analyses:
Mutation analysis for a specific gene or genes that target a fixed set of important mutations. Example: APC gene mutation analysis [20990-8].
Mutation analysis limited to one or more known mutations previously found in a family member. The mutations tested vary by patient. Example: ACADVL gene mutation analysis limited to known familial mutations [73736-1].
Full mutation analysis targeting the entire coding region in the targeted part of the genome. Example: ACADVL full gene mutation analysis … by sequencing [73735-3].
Mutation analyses for deletions/duplications. Example: ACVRL1 gene + ENG gene deletion and duplication mutation analysis [69481-9].
Augmenting narrative reports
Because we anticipate the transition to a fully structured molecular genomic reporting will take time, we encourage services that report their content as narrative reports to also include LOINC terms to deliver the core results in a computer-understandable form. Specifically, for mutation analysis studies, we encourage the inclusion of one LOINC term that reports the mutations found (eg, APC gene mutations found [20990-8]) and another that reports the mutations that could have been found. For tests that report only a fixed set of mutations, this can be done with 1 additional observation—APC gene mutations tested for [21618-4]. For sequencing and other tests to deliver the same information about what could have been found, terms from the DNA region of interest panel [53041-0] can be included as additional observations. All genetic studies should also include an observation that reports the reference sequence with a LOINC term such as Reference sequence Identifier [48012-9].
Cytogenetic tests that are delivered as narrative should include an additional observation that provides the findings in ISCN nomenclature to provide computability,30,31 eg, Chromosome analysis result in ISCN expression [62356-1]. However, it is better to provide the report in Heras’s full structured cytogenetic specification.18,22 Cytogenetic tests should also include an observation for the reference assembly build identifier such as Human reference sequence assembly release number [62374-4].
Result values for LOINC genetic codes
The use of LOINC codes to identify genetic tests is 1 ingredient for interoperability. Standards on the result values used to report the variations detected are equally important. Full details about the recommended result values for each code, ranging from Systematized Nomenclature of Medicine–Clinical Terms (International Health Standards Development Organization, Copenhagen, Denmark) to HGVS syntax, are provided in the LOINC database and in the HL7 implementation guides. In general, LOINC recommends the use of HGVS syntax for reporting variations and encourages the use of a second identifier, such as NCBI’s dbSNP29 or ClinVar32 IDs or, in the case of somatic mutations on cancer specimens, COSMIC33 (Genome Research Limited, Hinxton, UK) IDs. Since an HL7 Version 2 OBX-5 result field can carry 2 independent coding systems, both the HGVS notation and an alternative identifier can be reported together. For reporting pharmacogenomics genetic test results, LOINC encourages the use of the star-allele nomenclature34–36 as an additional way to report results because it is so widely used. For cytogenetic studies, ISCN is the standard nomenclature used to report genomic rearrangements identified either by standard karyotyping or molecular methodologies.30
Mechanisms for contributing to LOINC content
Almost all of LOINC’s content is based on external requests from laboratories, instrument vendors, test kit vendors, and public health departments around the world. Regenstrief welcomes requests for new LOINC terms. Submitters are asked to provide information for each variable requested, including a proposed LOINC name, a definition for the new analyte or method, units of measure for quantitative variables, answer lists for categorical variables, package inserts when a test is marketed, and deidentified sample reports when needed to understand the structure of test panels (see LOINC Users’ Guide37 for full details). Even with this amount of information, the LOINC team invests time in further research to enrich the definitions, verifying that existing LOINC terms do not satisfy the request and ensuring consistency across the database. The naming conventions in LOINC are informed by the input we receive from requestors and volunteer experts and are guided by the Laboratory LOINC Committee that meets twice a year in open meetings.
LIMITATIONS AND CHALLENGES
LOINC has substantial coverage of existing laboratory tests, but it is never complete. We hope to reach the point where we create LOINC codes for all tests in their premarketing stage. We are at that point for the test kits that the Centers for Disease Control distributes externally and may reach the same point for at least one large in vitro diagnostics vendor. LOINC lacks tests for whole exome and whole genome testing, including some of the newest whole genome microarray and high-resolution comparative genomic hybridization tests,38 because we have not yet received requests for them.
Variation in the styles for reporting genetic test results poses a substantial challenge to standardization. For example, some laboratories report variants identified on each allele as separate variables (eg, Allele 1 = TPMT*2, Allele 2 = TPMT*3C). Others report the value of both alleles as a single variable with a 2-part answer (eg, TPMT*2/*3C). In both cases, the allele naming style also varies considerably. One laboratory may include both the gene name and the allele name in the answer, another may report only the allele name, and yet another may report wild type and the letter code for the nucleotide result for the non–wild type. Further, some laboratories may only report presence or absence of a specific variant. We had discussions with the Food and Drug Administration about determining 1 standard approach to report the value of 2 alleles and providing guidance to the laboratory kit and instrument industry accordingly. They were open to this idea but have not yet acted.
CONCLUSION
The need to integrate genetic data with EHRs is compelling. LOINC is a widely adopted standard that provides codes to improve interoperability of genetic test reporting, including fully structured discrete reporting, tests for specific chromosomal alterations or gene mutations, and narrative cytogenetic or mutation analysis reports. By adopting and implementing data standards like LOINC, information systems can help clinicians unlock the potential of genetic information for delivering more personalized care.
Acknowledgments
The authors thank the many contributors to LOINC molecular pathology content, particularly the HL7 Clinical Genomics Work Group. This work was performed at the Regenstrief Institute, Indianapolis, IN, USA.
CONTRIBUTORS
JD and DJV conceived, wrote, and edited the manuscript. They are both guarantors. CJM provided critical revisions to the manuscript. All of the authors read and approved this manuscript for publication.
FUNDING
This work was supported by the National Library of Medicine grant numbers HSN2762008000006C, HHSN276201400138P, and HHSN276201400239P.
COMPETING INTERESTS
None.
REFERENCES
- 1.Kannry JL, Williams MS. Integration of genomics into the electronic health record: mapping terra incognita. Genet Med. 2013;15(10):757–760. doi:10.1038/gim.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470(7333):204–213. doi:10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 3.Kho AN, Rasmussen LV, Connolly JJ, et al. Practical challenges in integrating genomic data into the electronic health record. Genet Med. 2013;15(10):772–778. doi:10.1038/gim.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ullman-Cullere MH, Mathew JP. Emerging landscape of genomics in the Electronic Health Record for personalized medicine. Hum Mutat. 2011;32(5):512–516. doi:10.1002/humu.21456. [DOI] [PubMed] [Google Scholar]
- 5.US Department of Health and Human Services, Office of the National Coordinator for Health Information Technology. Clinical lab-test results. http://www.healthit.gov/providers-professionals/achieve-meaningful-use/core-measures-2/clinical-lab-test-results/. Accessed September 1, 2014. [Google Scholar]
- 6. Integrating pharmacogenetics with clinical care. Translational Software: Personalized Medicine in Practice Website. http://translationalsoftware.com/platform.html. Accessed September 1, 2014.
- 7.Hoffman MA. The genome-enabled electronic medical record. J Biomed Inform. 2007;40(1):44–46. doi:10.1016/j.jbi.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Chute CG, Ullman-Cullere M, Wood GM, et al. Some experiences and opportunities for big data in translational research. Genet Med. 2013;15(10):802–809. doi:10.1038/gim.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoenbill K, Fost N, Tachinardi U, et al. Genetic data and electronic health records: a discussion of ethical, logistical and technological considerations . J Am Med Inform Assoc. 2014;21(1):171–180. doi:10.1136/amiajnl-2013-001694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem. 2003;49(4):624–633. [DOI] [PubMed] [Google Scholar]
- 11.Frazier P, Rossi-Mori A, Dolin RH, et al. The creation of an ontology of clinical document names. Stud Health Technol Inform. 2001;84(Pt 1):94–98. [PubMed] [Google Scholar]
- 12.Hyun S, Shapiro JS, Melton G, et al. Iterative evaluation of the Health Level 7–Logical Observation Identifiers Names and Codes Clinical Document Ontology for representing clinical document names: a case report. J Am Med Inform Assoc. 2009;16(3):395–399. doi:10.1197/jamia.M2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vreeman DJ, McDonald CJ. Automated mapping of local radiology terms to LOINC. AMIA Annu Symp Proc. 2005;2005:769–773. [PMC free article] [PubMed] [Google Scholar]
- 14.LOINC. Version 2.48. Indianapolis, IN: The Regenstrief Institute; 2014. [Google Scholar]
- 15.Vreeman DJ, McDonald CJ, Huff SM. LOINC® - A universal catalog of individual clinical observations and uniform representation of enumerated collections. Int J Funct Inform Personal Med. 2010;3(4):273–291. doi:10.1504/IJFIPM.2010.040211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.US Department of Health, Human Services, Centers for Medicare & Medicaid Services. Medicare and Medicaid programs; electronic health record incentive program—stage 2. Fed Regist. 2012;77(171):53968 To be codified at 42 CFR §412, 413, and 495. [PubMed] [Google Scholar]
- 17.US Department of Health and Human Services, Office of the National Coordinator for Health Information Technology. Health information technology: standards, implementation specifications, and certification criteria for electronic health record technology, 2014 edition; revisions to the permanent certification program for health information technology. Fed Regist. 2012;77(171):53972. [PubMed] [Google Scholar]
- 18.Heras YZ, Mitchell JA, Williams MS, et al. Evaluation of LOINC for representing constitutional cytogenetic test result reports. AMIA Annu Symp Proc. 2009;2009:239–243. [PMC free article] [PubMed] [Google Scholar]
- 19.Vreeman DJ, Chiaravalloti MT, Hook J, et al. Enabling international adoption of LOINC through translation. J Biomed Inform. 2012;45(4):667–673. doi:10.1016/j.jbi.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011;3(79):79re1 doi:10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubinstein WS, Maglott DR, Lee JM, et al. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency . Nucleic Acids Res. 2013;41(D1):D925–D935. doi:10.1093/nar/gks1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Health Level Seven International (HL7). HL7 Version 2 Implementation Guide: Clinical Genomics; fully LOINC-Qualified Cytogenetics Model, Release 1 - US Realm . Ann Arbor, MI: Health Level Seven International; 2014. [Google Scholar]
- 23.Health Level Seven International (HL7). HL7 Version 2 Implementation Guide: Clinical Genomics; fully LOINC-Qualified Genetic Variation Model, Release 2. Ann Arbor, MI: Health Level Seven International: 2013. [Google Scholar]
- 24.HUGO Nomenclature. Human Genome Organisation (HUGO) International Website. http://hugo-international.org/comm_genenomenclaturecommittee.php. Accessed March 7, 2014. [Google Scholar]
- 25.Den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15(1):7–12. doi:10.1002/(SICI)1098-1004(200001)15:1<7:AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 26.Abhyankar S, Zuckerman A, McDonald CJ. A critical window of opportunity to standardize genetic testing results. AMIA 2012 Annual Symposium. Informatics: Transforming Health and Healthcare; 2012 Nov 7–9; Chicago, IL. [Google Scholar]
- 27.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37(4-5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 28.Ronquillo JG, Li C, Lester WT. Genetic testing behavior and reporting patterns in electronic medical records for physicians trained in a primary care specialty or subspecialty. J Am Med Inform Assoc. 2012;19(4):570–574. doi:10.1136/amiajnl-2011-000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaffer LG, McGowan-Jordan J, Schmid M, eds. ISCN (2013): An International System for Human Cytogenetic Nomenclature. Basel, Switzerland: S. Karger; 2013. [Google Scholar]
- 31.Simons A, Shaffer LG, Hastings RJ. Cytogenetic nomenclature: changes in the ISCN 2013 compared to the 2009 edition [published online ahead of print June 28, 2013]. Cytogenet Genome Res. doi:10.1159/000353118. [DOI] [PubMed] [Google Scholar]
- 32.Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(D1):D980–D985. doi:10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forbes SA, Bhamra G, Bamford S, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr Protoc Hum Genet. 2008;Chapter 10:Unit 10.11. doi:10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nebert DW. Suggestions for the nomenclature of human alleles: relevance to ecogenetics, pharmacogenetics and molecular epidemiology. Pharmacogenetics. 2000;10(4):279–290. [DOI] [PubMed] [Google Scholar]
- 35.Robarge JD, Li L, Desta Z, et al. The Star-Allele nomenclature: retooling for translational genomics. 2007; 82(3):244–248. [DOI] [PubMed] [Google Scholar]
- 36.Sim SC, Ingelman-Sundberg M. The Human Cytochrome P450 (CYP) Allele Nomenclature website: a peer-reviewed database of CYP variants and their associated effects. Hum Genomics. 2010;4(4):278–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald CJ, Huff SM, Deckard J, Holck K, Vreeman DJ, eds. Logical Observation Identifiers Names and Codes (LOINC®) Users’ Guide. Indianapolis: Regenstrief Institute; 2014. http://loinc.org/downloads. Accessed June 30, 2014. [Google Scholar]
- 38.Beaudet AL. The utility of chromosomal microarray analysis in developmental and behavioral pediatrics. Child Dev. 2013;84(1):121–132. doi:10.1111/cdev.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]


