Abstract
Objective Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) is the emergent international health terminology standard for encoding clinical information in electronic health records. The CORE Problem List Subset was created to facilitate the terminology’s implementation. This study evaluates the CORE Subset’s coverage and examines its growth pattern as source datasets are being incorporated.
Methods Coverage of frequently used terms and the corresponding usage of the covered terms were assessed by “leave-one-out” analysis of the eight datasets constituting the current CORE Subset. The growth pattern was studied using a retrospective experiment, growing the Subset one dataset at a time and examining the relationship between the size of the starting subset and the coverage of frequently used terms in the incoming dataset. Linear regression was used to model that relationship.
Results On average, the CORE Subset covered 80.3% of the frequently used terms of the left-out dataset, and the covered terms accounted for 83.7% of term usage. There was a significant positive correlation between the CORE Subset’s size and the coverage of the frequently used terms in an incoming dataset. This implies that the CORE Subset will grow at a progressively slower pace as it gets bigger.
Conclusion The CORE Problem List Subset is a useful resource for the implementation of Systematized Nomenclature of Medicine Clinical Terms in electronic health records. It offers good coverage of frequently used terms, which account for a high proportion of term usage. If future datasets are incorporated into the CORE Subset, it is likely that its size will remain small and manageable.
Keywords: problem-oriented medical record, problem list, electronic health record, SNOMED Clinical Terms, controlled medical terminology, medical vocabulary
INTRODUCTION
The Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) is descended from a long lineage of terminological artifacts spanning almost 50 years. Its origin dates back to 1965, when the College of American Pathologists published the Systematized Nomenclature of Pathology, which was later broadened to cover other fields of medicine. The merger of SNOMED RT (Reference Terminology) and the United Kingdom’s Clinical Terms Version 3 (also known as Read Codes) in 1999 gave rise to SNOMED CT. Efforts to promote SNOMED CT as the international standard clinical terminology began in earnest in 2007, when the SNOMED CT intellectual property rights were transferred from the College of American Pathologists to the newly formed International Health Terminology Standards Development Organisation (IHTSDO). From the original nine countries, the IHTSDO membership has tripled in the last7 years and now includes, among others, the United States, Canada, the United Kingdom, the Netherlands, Denmark (the country of registration), India, and Australia.1,2
Not unlike other standardization activities, SNOMED CT’s adoption has been slow. However, there is evidence that SNOMED CT-related research and implementation activities are on the rise.3–5 In the United States, the “Meaningful Use” incentive program for electronic health record (EHR) usage now requires the use of SNOMED CT for encoding data elements such as clinical problems, encounter diagnosis, and procedures.6,7 While, in the previous phase of the Meaningful Use program, either ICD-9-CM or SNOMED CT could be used for encoding clinical problems, going forward only SNOMED CT can be used. This change is in line with the general opinion that SNOMED CT is a better choice for encoding clinical information in the EHR.
SNOMED CT vs. ICD for the EHR
The International Statistical Classification of Diseases and Related Health Problems (ICD) is endorsed by the World Health Organization as the international standard diagnostic classification for epidemiology and health management, as well as for some clinical purposes. Its clinical modification (CM) has been used in the United States for classifying morbidity and mortality, reimbursement, creation of diagnosis-related groups, analysis of healthcare delivery, and epidemiological and clinical research. In order to generate ICD codes to support various administrative activities, many existing EHRs use ICD as a clinical terminology to directly encode clinical information. While ICD serves many important functions (which cannot be replaced by SNOMED CT), the fact that it is a statistical classification poses some limitations on its use in EHRs. Being a clinical terminology by purpose and design, SNOMED CT is inherently more suitable for capturing clinical information and, thus, for supporting patient care.
Content coverage
Numerous studies have concluded that SNOMED CT provides better clinical coverage than the ICD classifications.8–16 There are over 100 000 SNOMED CT concepts covering clinical findings, symptoms, and diagnoses, compared to only 14 000 codes in ICD-9-CM. ICD-10-CM will include significantly more codes (68 000). However, the increase in granularity is not uniform, but, instead, is concentrated in specific chapters (eg, injury and external cause), and a big portion of the new codes are created by formulaic expansion (eg, laterality, episode of care).17Table 1 shows examples of the loss of specificity encountered when encoding diseases in ICD-9-CM or ICD-10-CM, compared to SNOMED CT. Many rare congenital skin anomalies are encompassed by a single code in ICD-9-CM and ICD-10-CM and cannot be distinguished from each other. Lumping groups of diagnoses together occurs even for more common conditions like acidosis and brachial plexus disorders. The lack of specificity in ICD-9-CM and ICD-10-CM will affect the ability of the EHR to deliver disease-specific clinical decision support, because diseases with very different etiologies and management strategies are lumped together in these terminologies.
Table 1:
Comparison of the Granularity of ICD-9-CM, ICD-10-CM, and SNOMED CT
| Condition | SNOMED CT disease codes | Corresponding ICD-9-CM code | Corresponding ICD-10-CM code |
|---|---|---|---|
| Congenital skin anomalies | 205573006 Focal dermal hypoplasia 79468000 Familial benign pemphigus 5132005 Keratosis pilaris … (total 21 codes) | 757.39 Other specified congenital anomalies of skin | Q82.8 Other specified congenital malformations of skin |
| Acidosis | 59455009 Metabolic acidosis 12326000 Respiratory acidosis 91273001 Lactic acidosis … (total 60 codes) | 276.2 Acidosis | E87.2 Acidosis |
| Brachial plexus disorders | 72893007 Brachial neuritis 278065000 Pancoast’s syndrome 78141002 Erb-Duchenne paralysis … (total 33 codes) | 353.0 Brachial plexus lesions | G54.0 Brachial plexus disorders |
The clinical coverage of SNOMED CT is not limited by the scope of its existing concepts. One unique feature of SNOMED CT is extensibility by post-coordination – the creation of new meaning by modifying or refining existing concepts. The only way to extend ICD is to add local extension codes, which (as the name implies) are only interpretable locally and are meaningless to an outside reader. With post-coordination, computability and interoperability are preserved. It is possible to determine equivalence and subsumption between existing concepts and post-coordinated expressions, so that post-coordinated expressions can be shared and integrated. Liu et al.18 found that post-coordination could potentially increase SNOMED CT’s coverage of summary-level clinical concepts from 80 to 96%. According to Campbell et al.,19 only 1.5% of problem list terms could not be represented in SNOMED CT with post-coordination. However, from an implementation perspective, post-coordination is nontrivial. Challenges include data entry interface design, data storage and retrieval, and computational requirements. In the study by Lee et al.,4 6 of the 13 healthcare organizations interviewed were able to use post-coordination in some way.
Clinical orientation
ICD descriptions are often criticized by clinical users as being awkward. This is because ICD codes are closely related to statistical groupings for epidemiological and other purposes, and the descriptions need to reflect the inclusion and exclusion criteria for classification. For example, an unsuccessful tendon grafting operation will be coded in SNOMED CT simply as 281430007 Failure of tendon graft. In ICD-9-CM, the corresponding code is E878.2 Surgical operation with anastomosis, bypass, or graft, with natural or artificial tissues used as implant causing abnormal patient reaction, or later complication, without mention of misadventure at time of operation. Since such descriptions cannot be used directly in data entry, the ICD implementers often need to create interface terms that clinical users can readily recognize. There is no such need in SNOMED CT, because the terms in SNOMED CT are words and phrases directly used in clinical discourse.
Additionally, in some areas of ICD (eg, causes of injury), the emphasis seems to be more on public health than the individual patient. Extreme examples (eg, burning water-skis, turtle bite) have been used as jokes about ICD-10-CM.20 Given that the primary purpose of the original ICD classifications was to collect global epidemiological data, rather than patient clinical data, it is understandable why mechanisms of injury are given such attention. To implement effective preventive measures, one would need detailed statistics. However, exposing these detailed injury codes to clinicians and requiring their use is likely to cause confusion and opposition.
Data entry and retrieval
As a statistical classification, ICD needs to ensure that codes are mutually exclusive (ie, that there is only one way to code a condition) and jointly exhaustive (ie, that there is always a code for any condition). To facilitate counting and ensure data comparability, only codes at the lowest level (the leaf codes) can be used. To satisfy these requirements, two special types of codes are necessary: the “unspecified” (also known as “not otherwise specified” or NOS) and “not elsewhere classified” (or NEC) codes. The unspecified codes are used when specific information is missing. For example, 480.0 Viral pneumonia, unspecified is used when the medical record does not contain the specific virus causing pneumonia. The unspecified code is necessary, since the parent code 480 Viral pneumonia (which essentially conveys the same meaning as Viral pneumonia, unspecified) cannot be used. NEC codes are used when there is additional specific information in the medical record, but no corresponding specific code. Pneumonia caused by Human metapneumovirus is coded as 480.8 Pneumonia due to other virus not elsewhere classified, since there is no specific code for this condition. In data entry, clinical users may be confused by the NOS and NEC codes. In SNOMED CT, there are no NOS and NEC codes. The clinician can use codes at any level of specificity, as warranted by the clinical situation.
In data retrieval, it is important for a code to maintain the same meaning over time. In SNOMED CT, a concept code always represents the same meaning. This is not necessarily true in ICD. Codes can change across versions. For example, the code for Meconium aspiration syndrome changed from 770.1 to 770.12 in 2006. The subtle change in the meaning of NEC codes is even more problematic, because it is difficult to detect (since the code and its description remain the same). This is called semantic drift, in Cimino’s desiderata paper.21 For example, the code 480.8 Pneumonia due to other virus not elsewhere classified included SARS Pneumonia before 2003, but not afterwards, because 480.3 Pneumonia due to SARS-associated coronavirus was added.
Two unique characteristics of SNOMED CT facilitate data retrieval. Firstly, SNOMED CT is a poly-hierarchy (one concept can have multiple parents), while ICD is a strict hierarchy (one parent per code). A strict hierarchy is necessary in a statistical classification, to avoid double counting. ICD codes for similar diseases can be assigned (somewhat arbitrarily) to different sub-branches or chapters, making it a challenge to find them all. To identify all hypertensive patients, one might be tempted to use the codes under 401–405 Hypertensive disease, but will find that set is missing codes like 410.9 Myocardial infarction with hypertension and 642 Hypertension complicating pregnancy, childbirth, and the puerperium. In SNOMED CT, one can use a simple query to get all descendants of 38341003 Hypertensive disorder.
Secondly, logical definitions in SNOMED CT make it possible to retrieve concepts using relationships and attributes. For example, to find diseases caused by blockage of arteries anywhere except in the intestine or kidneys, a researcher can retrieve descendants of 2929001 Occlusion of artery (183 concepts), excluding those whose finding site is mesenteric artery or renal artery (11 concepts). However, in ICD-9-CM, she will need to manually search for a list of codes like the following:
414.0 Coronary atherosclerosis
416.0 Idiopathic pulmonary arteriosclerosis
437.0 Cerebral atherosclerosis
440 Atherosclerosis (and descendants, except 440.1 Of renal artery)
362.3 Retinal vascular occlusion and descendants
etc …
Furthermore, the search has to be repeated with any new release of ICD-9-CM. In SNOMED CT, she can simply re-run the query to pick up the changes.
The CORE Problem List Subset of SNOMED CT
The CORE Problem List Subset of SNOMED CT (the CORE Subset) was first published in 2009. To facilitate the implementation of a SNOMED CT-based problem list, we identified a subset of SNOMED CT concepts commonly used in actual problem list data. CORE stands for “clinical observations recording and encoding” and refers to the use of controlled terminologies to encode clinical information at a summary level, such as the problem list, discharge diagnosis, or reason for encounter sections of an EHR.22 The first CORE Subset was based on datasets from seven large-scale healthcare institutions (Kaiser Permanente, KA; Mayo Clinic, MA; University of Nebraska Medical Center, NU; Hong Kong Hospital Authority, HA; Intermountain Healthcare, IH; Regenstrief Institute, RI; and Beth Israel Deaconess Medical Center, BI). The dataset from the US Veterans Administration (VA) was added in 2012.
The CORE Subset was created empirically by identifying the most commonly used problem list terms that accounted for 95% of total usage in each institution and mapping them to SNOMED CT.22 The main reason for adopting a usage-based cut-off was that all the datasets had very long tails of infrequently used terms. By focusing on the frequently used terms, we made the mapping effort more manageable. The intended use of the CORE Subset is as a starter set to build a local SNOMED CT-based problem list terminology. The CORE Subset is not expected to be exhaustive or able to provide every concept the user needs. It is anticipated that some concepts outside the subset will need to be added.
Based on our analysis of the pattern of overlap between the source datasets, we believe that the CORE Subset will provide good coverage for frequently used problem list terms and total usage in most institutions. Also, by filtering out rarely used terms, the subset’s size is more likely to remain manageable when more datasets are incorporated. In this study, we examined two properties of the CORE Subset. Firstly, we assessed the coverage of the CORE Subset at the term- and usage-level. Secondly, we studied the CORE Subset’s pattern of growth to date, to project how it will grow in future.
METHODS
To compare the term and usage coverage of problem list data by SNOMED CT, ICD-9-CM, and ICD-10-CM, we first calculated the CORE Subset’s coverage of the frequently used terms and their corresponding usage in the source datasets. Using the same mapping method (lexical matching with synonym substitution), we mapped the local terms to ICD-9-CM and ICD-10-CM to estimate their coverage in the datasets in a similar manner.22
To estimate the coverage of the CORE Subset when applied to a new dataset that is not used to build the subset, we did “leave-one-out” analysis of our source datasets. We constructed new CORE Subsets using any seven of the eight datasets and calculated their coverage of frequently used SNOMED CT concepts and total usage for the “left-out” dataset. We only considered terms within the 95% usage cut-off that are mappable to SNOMED CT. For SNOMED CT concepts in the left-out dataset that are not covered by the CORE Subset, we identified those that are directly related to a CORE concept, either as a direct child or a parent.
To estimate the future growth of the CORE Subset, we did a retrospective “growth experiment” by “growing” the subset one dataset at a time. We started with an initial base CORE Subset built from the two biggest and two smallest datasets (based on number of local terms), then added other datasets one by one, in all possible orders. For each addition, we noted the size of the starting CORE Subset and the coverage of the frequently used SNOMED CT concepts in the incoming dataset. Our hypothesis is that, as the CORE Subset gets bigger, the term coverage of the incoming dataset will increase and fewer new concepts will be added. The CORE Subset will grow at a progressively slower rate and eventually level off or only change very slowly. We examined the relationship between CORE Subset size and term coverage by scatter plot and by calculating the correlation coefficients (Pearson, Kendall, and Spearman). We used linear regression to estimate the potential ceiling of the CORE Subset. We used IBM SPSS © for Windows (version 21) for statistical analysis.
RESULTS
Coverage
Table 2 shows the characteristics of the eight datasets and their mappings to SNOMED CT, ICD-9-CM, and ICD-10-CM. The problem list vocabularies varied considerably in the number of unique terms, but all had a long tail of infrequently used terms. Across all datasets, 22.8% of unique terms accounted for 95% of usage. On average, 93.1% of the frequently used local terms within the 95% usage range could be mapped to SNOMED CT. These SNOMED CT-mapped terms corresponded to an average usage of 90.5%. An average of 43.4 and 49.9% of the frequently used terms could be mapped to ICD-9-CM and ICD-10-CM, respectively, corresponding to usage coverage of 48.4 and 59.3%.
Table 2:
Characteristics of the Source Datasets and Coverage of the Frequently Used Terms and Usages by SNOMED CT, ICD-9-CM, and ICD-10-CM
| Institution | Unique local terms | Frequently used termsa (percentage of unique terms) | SNOMED CT | ICD-9-CM | ICD-10-CM | |||
|---|---|---|---|---|---|---|---|---|
| Frequently used terms coverage (%) | Usage coverage % | Frequently used terms, coverage (%) | Usage coverage % | Frequently used terms, coverage (%) | Usage coverage % | |||
| KP | 26 890 | 2961 (11.0) | 2657 (89.7) | 85.3 | 1000 (33.8) | 41.8 | 1139 (38.5) | 54.8 |
| VA | 21 221 | 2069 (9.7) | 1983 (95.8) | 93.8 | 1292 (62.5) | 60.6 | 1089 (52.6) | 64.8 |
| MA | 14 921 | 3610 (24.2) | 3165 (87.7) | 86.4 | 1037 (28.7) | 38.4 | 1396 (38.7) | 48.4 |
| NU | 13 126 | 3320 (25.3) | 3154 (95.0) | 91.3 | 1199 (36.1) | 44.7 | 1448 (43.6) | 56.7 |
| HA | 12 449 | 2635 (21.2) | 2279 (86.5) | 87.2 | 1214 (46.1) | 47.9 | 1103 (41.9) | 46.1 |
| IH | 5685 | 1077 (18.9) | 1051 (97.6) | 94.4 | 510 (47.4) | 54.4 | 622 (57.8) | 70.5 |
| RI | 3166 | 792 (25.0) | 762 (96.2) | 91.8 | 336 (42.4) | 45.5 | 447 (56.4) | 60.6 |
| BI | 879 | 410 (46.6) | 396 (96.6) | 93.6 | 206 (50.2) | 53.7 | 287 (70.0) | 72.9 |
| Mean | 12 292 | 2109 (22.8) | 1930 (93.1) | 90.5 | 849 (43.4) | 48.4 | 941 (49.9) | 59.3 |
aMost heavily used terms that, collectively, accounted for 95% of term usage.
KP, Kaiser Permanente; VA, Veterans Administration; MA, Mayo Clinic; NU, University of Nebraska Medical Center; HA, Hong Kong Hospital Authority; IH, Intermountain Healthcare; RI, Regenstrief Institute; BI, Beth Israel Deaconess Medical Center.
Table 3 summarizes the results of the “leave-one-out” analysis. The average size of the CORE Subsets based on seven datasets was 5758, and this covered, on average, 80.3% of the frequently used SNOMED CT concepts in the left-out dataset. The average usage coverage was 83.7%. Among the frequently used SNOMED CT concepts not covered by the CORE Subset, 55% were direct parents or children of CORE concepts. It was more common to find a CORE parent (45.3%) than a child (20.7%), meaning that the missing concepts were generally more specific than the CORE concepts.
Table 3:
Coverage of the CORE Subset for a New Dataset Estimated by “Leave-One-Out” Analysis
| Dataset left out | CORE Subset size (based on the other seven datasets) | Coverage of frequently used concepts in dataset left out (%) | Usage coverage % | Frequently used concepts in dataset left out that are missing from CORE Subset |
||
|---|---|---|---|---|---|---|
| with CORE parent (percentage of missing concepts) | with CORE child (percentage of missing concepts) | with either CORE parent or child (percentage of missing concepts) | ||||
| KP | 5733 | 1745 (79.8) | 79.6 | 246 (55.5) | 106 (23.9) | 287 (64.8) |
| VA | 5856 | 1222 (79.2) | 87.6 | 170 (53.1) | 67 (20.9) | 195 (60.9) |
| MA | 5286 | 1977 (69.0) | 77.8 | 449 (50.4) | 82 (9.2) | 487 (54.7) |
| NU | 5508 | 2050 (75.4) | 85.4 | 290 (43.4) | 127 (19.0) | 351 (52.5) |
| HA | 5313 | 1232 (58.8) | 66.8 | 419 (48.6) | 87 (10.1) | 456 (52.8) |
| IH | 6091 | 899 (91.4) | 92.6 | 37 (43.5) | 23 (27.1) | 52 (61.2) |
| RI | 6112 | 660 (91.2) | 86.8 | 29 (45.3) | 21 (32.8) | 38 (59.4) |
| BI | 6167 | 374 (97.7) | 93.1 | 2 (22.2) | 2 (22.2) | 3 (33.3) |
| Mean | 5758 | 1270 (80.3) | 83.7 | 205 (45.3) | 64 (20.7) | 234 (55.0) |
KP, Kaiser Permanente; VA, Veterans Administration; MA, Mayo Clinic; NU, University of Nebraska Medical Center; HA, Hong Kong Hospital Authority; IH, Intermountain Healthcare; RI, Regenstrief Institute; BI, Beth Israel Deaconess Medical Center.
Growth pattern and convergence
We started with an initial base CORE Subset built from the two largest (KP and VA) and two smallest (RI and BI) datasets, then sequentially built bigger CORE Subsets by adding the other datasets one by one, in all possible orders. There were 32 unique paths to build the full CORE Subset from the initial base Subset (Table 4).
Table 4:
Examining the Relationship Between the Size of the CORE Subset and Coverage of the Frequently Used Terms in an Incoming Dataset by a Retrospective “Growth Experiment,” Starting With an Initial Subset Based on Four Datasets
| Datasets constituting the starting CORE Subset | Incoming data set | Size of starting CORE Subset | Coverage of frequently used SNOMED CT concepts in incoming dataset % |
|---|---|---|---|
| KP, VA, RI, BI | MA | 3052 | 52.7 |
| KP, VA, RI, BI | NU | 3052 | 57.7 |
| KP, VA, RI, BI | HA | 3052 | 43.5 |
| KP, VA, RI, BI | IH | 3052 | 81.7 |
| KP, VA, RI, BI, MA | NU | 4408 | 70.2 |
| KP, VA, RI, BI, MA | HA | 4408 | 53.4 |
| KP, VA, RI, BI, MA | IH | 4408 | 85.7 |
| KP, VA, RI, BI, NU | MA | 4201 | 64.6 |
| KP, VA, RI, BI, NU | HA | 4201 | 52.6 |
| KP, VA, RI, BI, NU | IH | 4201 | 89.3 |
| KP, VA, RI, BI, HA | MA | 4235 | 59.9 |
| KP, VA, RI, BI, HA | NU | 4235 | 64.7 |
| KP, VA, RI, BI, HA | IH | 4235 | 84.6 |
| KP, VA, RI, BI, IH | MA | 3232 | 54.1 |
| KP, VA, RI, BI, IH | NU | 3232 | 60.5 |
| KP, VA, RI, BI, IH | HA | 3232 | 44.9 |
| KP, VA, RI, BI, MA, NU | HA | 5217 | 58.3 |
| KP, VA, RI, BI, MA, NU | IH | 5217 | 90.2 |
| KP, VA, RI, BI, MA, HA | NU | 5384 | 74.0 |
| KP, VA, RI, BI, MA, HA | IH | 5384 | 87.4 |
| KP, VA, RI, BI, MA, IH | NU | 4549 | 71.9 |
| KP, VA, RI, BI, MA, IH | HA | 4549 | 54.2 |
| KP, VA, RI, BI, NU, HA | MA | 5194 | 68.7 |
| KP, VA, RI, BI, NU, HA | IH | 5194 | 90.7 |
| KP, VA, RI, BI, NU, IH | MA | 4306 | 64.9 |
| KP, VA, RI, BI, NU, IH | HA | 4306 | 53.2 |
| KP, VA, RI, BI, HA, IH | MA | 4387 | 60.9 |
| KP, VA, RI, BI, HA, IH | NU | 4387 | 66.9 |
| KP, VA, RI, BI, MA, NU, HA | IH | 6091 | 91.4 |
| KP, VA, RI, BI, MA, NU, IH | HA | 5313 | 58.8 |
| KP, VA, RI, BI, MA, HA, IH | NU | 5508 | 75.4 |
| KP, VA, RI, BI, NU, HA, IH | MA | 5286 | 69.0 |
KP, Kaiser Permanente; VA, Veterans Administration; MA, Mayo Clinic; NU, University of Nebraska Medical Center; HA, Hong Kong Hospital Authority; IH, Intermountain Healthcare; RI, Regenstrief Institute; BI, Beth Israel Deaconess Medical Center.
Figure 1 is a scatter plot of the 32 data points of starting subset size and term coverage. There was significant positive correlation between subset size and term coverage (Pearson correlation coefficient = 0.528, 2-tailed P = 0.002; Kendall’s τ = 0.413, 2-tailed P = 0.001; Spearman’s ρ = 0.523, 2-tailed P = 0.002). A linear regression equation could be fitted:
Figure 1.
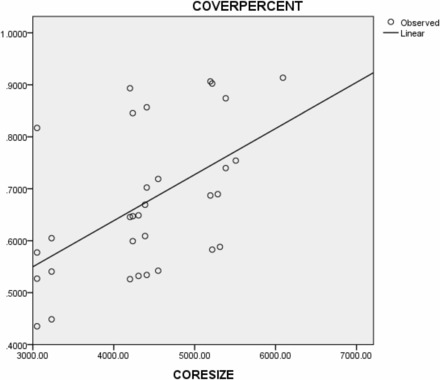
Scatter plot of starting CORE Subset size vs. coverage of frequently used SNOMED CT concepts in the incoming dataset (Observed – 32 data points, Linear – fitted linear regression line).
Assuming the equation holds for bigger subsets (which may or may not be true), the coverage of frequently used SNOMED CT concepts in the incoming dataset will get very close to 100% when the subset reaches 8000 concepts.
DISCUSSION
SNOMED CT is the most comprehensive, multilingual clinical terminology in the world. It has 300 000 active concepts, about 100 000 of which are suitable for use in the problem list. Most institutions use a local problem list terminology of under 30 000 terms and, so, require some term selection. Starting with the CORE Subset saves time and effort in term selection. Moreover, starting from a common subset reduces variability between local terminologies and improves interoperability. Detailed statistics about SNOMED CT implementation are difficult to find in the literature.4 Based on the 2011 Unified Medical Language System (UMLS) user annual reports, 823 users have used the CORE Subset, about 40% of them in relation to the EHR.
In the leave-one-out analysis, the CORE Subset covered, on average, 80.3% of the frequently used SNOMED CT concepts in a new dataset, corresponding to a total usage of 83.7%. Note that we only looked at the coverage of the terms that were within the 95% usage cut-off and were mappable to SNOMED CT. Wright et al. analyzed the CORE Subset’s coverage of problem list data in a large healthcare network and found coverage of 71.1% of all unique problem list terms and 94.8% of problem list entries.23 The high coverage of local terms is a bit surprising, but can be explained, because almost all of their problem list terms are mappable to SNOMED CT (only 15 out of 1494 terms are not mapped to it). It is likely that one would see a lower overall coverage of local terms in a dataset where not all the terms are mappable to SNOMED CT. On the other hand, the high usage coverage is consistent with our results. For the three smallest datasets (IH, RI, and BI), which are comparable to the Wright dataset, the average usage coverage was 90.8%.
In our study, we only considered pre-coordinated SNOMED CT concepts. The use of post-coordination has been shown to significantly increase the coverage of SNOMED CT.18,19 In our own analysis of the source datasets, we found that, of 348 frequently used local terms not mappable to SNOMED CT, 260 terms (74.7%) can be represented by post-coordination. Moreover, 68.8% of the post-coordinated expressions involve a focal concept that is already part of the CORE Subset, which means that these expressions can be directly linked to the CORE concepts.
As far as we know, the CORE Subset is the first of its kind using the Pareto distribution analysis to identify frequently used concepts that are generalizable to other datasets. Whether this will result in a convergent, stable subset if we incorporate more datasets is an open question. One prerequisite for convergence is that frequently used concepts are clustered and not randomly distributed in SNOMED CT. There is evidence for such clustering. A CORE Subset of approximately 5000 concepts, corresponding to only 5% of SNOMED CT clinical concepts, already covers 80% of the frequently used concepts in any dataset. Furthermore, over half of the frequently used concepts not covered are direct parents or children of CORE concepts. As we showed in our previous study, the most heavily used terms are also the ones most likely to be shared among institutions.22
In this study, we found a significant positive correlation between the size of the CORE Subset and the coverage of frequently used terms in an incoming dataset. If this relationship holds for future datasets, fewer and fewer new terms will need to be added, and the CORE Subset will plateau, resulting in a relatively stable subset. According to the regression formula, term coverage will approach 100% with a subset size of 8000 concepts. However, since this number was derived by extrapolation outside the range of our data points, it should be regarded as speculative.
Apart from good term and usage coverage, we have previously studied the utility of the CORE Subset in data capture. We compared the term-finding efficiency of the CORE Subset, a clinical subset of SNOMED CT (100 000 concepts), and the problem list terminology of a hospital (24 000 concepts).24 Despite its small size, the CORE Subset was able to provide a level of partial and exact matches comparable to the clinical SNOMED CT subset. The CORE Subset provided the fastest way to find a term, because a search of the subset returned the fewest terms to choose from.
There are other uses for the CORE Subset, outside of EHRs. The CORE Subset identifies a relatively small collection of about 6000 clinical concepts whose importance is substantiated by actual usage data. Compared to the whole SNOMED CT terminology, the CORE Subset is a more manageable target and stands as a proxy for the study of SNOMED CT. In 2010, the IHTSDO did a comprehensive review of 100 CORE concepts as a quality assurance exercise. The CORE Subset has also been the focus of other SNOMED CT quality assurance,25–27 inter-terminology mapping,28,29 and terminology research30–35 activities.
Our study has the following limitations. The analysis is based on the eight problem list datasets that we obtained for the CORE Project. These are mostly US institutions (except one from Hong Kong) that provide care in all major medical specialties. The datasets together cover about 18 million patients. Only the most frequently used local terms accounting for 95% of term usage that can be mapped to SNOMED CT are considered. In mapping to SNOMED CT, we only mapped to pre-coordinated SNOMED CT concepts and do not use post-coordination. The mappings from local terms to SNOMED CT are mostly done by lexical matching supplemented by manual review and have not been independently verified.
CONCLUSION
SNOMED CT is inherently more suitable for capturing clinical information in EHRs than the ICD classifications because of its better content coverage, clinical orientation, and more flexible data entry and retrieval. The CORE Problem List Subset of SNOMED CT is a useful resource for the implementation of SNOMED CT in EHRs, providing over 80% coverage of frequently used terms and total usage. In the future, if the CORE Subset grows in size with the addition of new source datasets, the rate of growth will gradually slow, and it is likely that the CORE Subset will remain a manageable size.
Supplementary Material
Acknowledgments
The authors would like to thank the following institutions for providing their datasets for this project: Beth Israel Deaconess Medical Center, Hong Kong Hospital Authority, Intermountain Healthcare, Kaiser Permanente, Mayo Clinic, Nebraska University Medical Center, Regenstrief Institute, and Veterans Administration.
CONTRIBUTORS
K.W.F. conceived and designed the study. K.W.F. and J.X. performed the data analysis. K.W.F. drafted the manuscript, and both K.W.H. and J.X. contributed substantially to its revision
FUNDING
This research was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
COMPETING INTERESTS
None
REFERENCES
- 1. Cornet R, de Keizer N. Forty Years of SNOMED: a Literature Review. BMC Med Inform Decis Mak 2008;8(Suppl 1):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. IHTSDO. History of SNOMED CT. [cited January 14, 2015]. http://www.ihtsdo.org/snomed-ct/what-is-snomed-ct/history-of-snomed-ct. [Google Scholar]
- 3. Elhanan G, Perl Y, Geller J. A Survey of Direct Users and Uses of SNOMED CT: 2010 Status. AMIA Annu Symp Proc. 2010;2010:207–211. [PMC free article] [PubMed] [Google Scholar]
- 4. Lee D, Cornet R, Lau F, de Keizer N. A survey of SNOMED CT implementations. J Biomed Inform. 2013;46(1):87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee D, de Keizer N, Lau F, Cornet R. Literature review of SNOMED CT use .J Am Med Inform Assoc. 2014;21(e1):e11–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records .N Engl J Med. 2010;363(6):501–504. [DOI] [PubMed] [Google Scholar]
- 7. US Office of the National Coordinator for Health Information Technology, Department of Health and Human Services. Health Information Technology: Standards, Implementation Specifications, and Certification Criteria for Electronic Health Record Technology, 2014 Edition. 2012 [cited January 14, 2015]. http://www.gpo.gov/fdsys/pkg/FR-2012-09-04/pdf/2012-20982.pdf.
- 8. Yasini M, Ebrahiminia V, Duclos C, Venot A, Lamy JB. Comparing the use of SNOMED CT and ICD10 for coding clinical conditions to implement laboratory guidelines. Stud Health Technol Inform. 2013;186:200–204. [PubMed] [Google Scholar]
- 9. Tvede I, Bredegaard K, Andersen JS. Quality improvements based on detailed and precise terminology. Stud Health Technol Inform. 2010;155:71–77. [PubMed] [Google Scholar]
- 10. Chiang MF, Casper DS, Cimino JJ, Starren J. Representation of ophthalmology concepts by electronic systems: adequacy of controlled medical terminologies .Ophthalmology. 2005;112(2):175–183. [DOI] [PubMed] [Google Scholar]
- 11. Chen JW, Flaitz C, Johnson T. Comparison of accuracy captured by different controlled languages in oral pathology diagnoses. AMIA Annu Symp Proc. 2005;2005:918. [PMC free article] [PubMed] [Google Scholar]
- 12. McClay JC, Campbell J. Improved coding of the primary reason for visit to the emergency department using SNOMED. Proc AMIA Symp. 2002;2002:499–503. [PMC free article] [PubMed] [Google Scholar]
- 13. Elkin PL, Ruggieri AP, Brown SH, Buntrock J, Bauer BA, Wahner-Roedler D, Litin SC, Beinborn J, Bailey KR, Bergstrom L. A randomized controlled trial of the accuracy of clinical record retrieval using SNOMED-RT as compared with ICD9-CM. Proc AMIA Symp. 2001;2001:159–163. [PMC free article] [PubMed] [Google Scholar]
- 14. Vardy DA, Gill RP, Israeli A. Coding medical information: classification versus nomenclature and implications to the Israeli medical system. J Med Syst. 1998;22(4):203–210. [DOI] [PubMed] [Google Scholar]
- 15. Chute CG, Cohn SP, Campbell KE, Oliver DE, Campbell JR. The content coverage of clinical classifications. For The Computer-Based Patient Record Institute's Work Group on Codes & Structures. J Am Med Inform Assoc. 1996;3(3):224–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Campbell JR, Payne TH. A comparison of four schemes for codification of problem lists. Proc Annu Symp Comput Appl Med Care. 1994;1994:201–205. [PMC free article] [PubMed] [Google Scholar]
- 17. Steindel SJ. International classification of diseases, 10th edition, clinical modification and procedure coding system: descriptive overview of the next generation HIPAA code sets. J Am Med Inform Assoc. 2010;17(3):274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu H, Wagholikar K, Wu ST. Using SNOMED-CT to encode summary level data - a corpus analysis .AMIA Summits Transl Sci Proc. 2012;2012:30–37. [PMC free article] [PubMed] [Google Scholar]
- 19. Campbell JR, Xu J, Fung KW. Can SNOMED CT fulfill the vision of a compositional terminology? Analyzing the use case for problem list .AMIA Annu Symp Proc. 2011;2011:181–188. [PMC free article] [PubMed] [Google Scholar]
- 20. Walked Into a Lamppost? Hurt While Crocheting? Help Is on the Way. The Wall Street J. 2011 [cited January 14, 2015]. http://online.wsj.com/article/SB10001424053111904103404576560742746021106.html?mod=dist_smartbrief.
- 21. Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37(4–5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 22. Fung KW, McDonald C, Srinivasan S. The UMLS-CORE project: a study of the problem list terminologies used in large healthcare institutions. J Am Med Inform Assoc. 2010;17(6):675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wright A, Feblowitz J, McCoy AB, Sittig DF. Comparative analysis of the VA/Kaiser and NLM CORE problem subsets: an empirical study based on problem frequency. AMIA Annu Symp Proc. 2011;2011:1532–1540. [PMC free article] [PubMed] [Google Scholar]
- 24. Fung KW, Xu J, Rosenbloom ST, Mohr D, Maram N, Suther T. Testing three problem list terminologies in a simulated data entry environment. AMIA Annu Symp Proc. 2011;2011:445–454. [PMC free article] [PubMed] [Google Scholar]
- 25. Rector A, Iannone L. Lexically suggest, logically define: quality assurance of the use of qualifiers and expected results of post-coordination in SNOMED CT. J Biomed Inform. 2012;45(2):199–209. [DOI] [PubMed] [Google Scholar]
- 26. Rector AL, Brandt S, Schneider T. Getting the foot out of the pelvis: modeling problems affecting use of SNOMED CT hierarchies in practical applications .J Am Med Inform Assoc. 2011;18(4):432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Agrawal A, He Z, Perl Y, Wei D, Halper M, Elhanan G, Chen Y. The readiness of SNOMED problem list concepts for meaningful use of electronic health records. Artif Intell Med. 2013;58(2):73–80. [DOI] [PubMed] [Google Scholar]
- 28. US National Library of Medicine. SNOMED CT to ICD-10-CM Map. 2014 [cited January 14, 2015]. http://www.nlm.nih.gov/research/umls/mapping_projects/snomedct_to_icd10cm.html.
- 29. Campbell JR, Brear H, Scichilone R, White S, Giannangelo K, Carlsen B, Solbrig H, Fung KW. Semantic Interoperation and Electronic Health Records: Context Sensitive Mapping from SNOMED CT to ICD-10. Stud Health Technol Inform. 2013;192:603–607. [PubMed] [Google Scholar]
- 30. Nadkarni PM, Darer JA. Migrating existing clinical content from ICD-9 to SNOMED. J Am Med Inform Assoc. 2010;17(5):602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Steindel SJ. A comparison between a SNOMED CT problem list and the ICD-10-CM/PCS HIPAA code sets. Perspect Health Inf Manag. 2012;9:1b. [PMC free article] [PubMed] [Google Scholar]
- 32. Lopez-Garcia P, Boeker M, Illarramendi A, Schulz S. Usability-driven pruning of large ontologies: the case of SNOMED CT. J Am Med Inform Assoc. 2012;19(e1):e102–e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abdoune H, Merabti T, Darmoni SJ, Joubert M. Assisting the translation of the CORE subset of SNOMED CT into French. Stud Health Technol Inform. 2011;169:819–823. [PubMed] [Google Scholar]
- 34. Lamy JB, Tsopra R, Venot A, Duclos C. A Semi-automatic Semantic Method for Mapping SNOMED CT Concepts to VCM Icons. Stud Health Technol Inform. 2013;192:42–46. [PMC free article] [PubMed] [Google Scholar]
- 35. Hogan WR, Slee VN. Measuring the information gain of diagnosis vs. diagnosis category coding. AMIA Annu Symp Proc. 2010;2010:306–310. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


