Abstract
Introduction The semantic interoperability of electronic healthcare records (EHRs) systems is a major challenge in the medical informatics area. International initiatives pursue the use of semantically interoperable clinical models, and ontologies have frequently been used in semantic interoperability efforts. The objective of this paper is to propose a generic, ontology-based, flexible approach for supporting the automatic transformation of clinical models, which is illustrated for the transformation of Clinical Element Models (CEMs) into openEHR archetypes.
Methods Our transformation method exploits the fact that the information models of the most relevant EHR specifications are available in the Web Ontology Language (OWL). The transformation approach is based on defining mappings between those ontological structures. We propose a way in which CEM entities can be transformed into openEHR by using transformation templates and OWL as common representation formalism. The transformation architecture exploits the reasoning and inferencing capabilities of OWL technologies.
Results We have devised a generic, flexible approach for the transformation of clinical models, implemented for the unidirectional transformation from CEM to openEHR, a series of reusable transformation templates, a proof-of-concept implementation, and a set of openEHR archetypes that validate the methodological approach.
Conclusions We have been able to transform CEM into archetypes in an automatic, flexible, reusable transformation approach that could be extended to other clinical model specifications. We exploit the potential of OWL technologies for supporting the transformation process. We believe that our approach could be useful for international efforts in the area of semantic interoperability of EHR systems.
Keywords: electronic health records, semantic interoperability, Clinical Element Model, archetype, OWL
INTRODUCTION
Objective
Our main goal is facilitating the reuse of existing clinical models in different electronic health records (EHRs) standards by providing methods that allow the transformation of clinical models between such standards. Demonstrating that ontologies and the Web Ontology Language (OWL) can be a technological backbone for such transformation processes is another goal of this work. The approach will be applied in this paper to the transformation of Clinical Element Models (CEMs) into openEHR archetypes.
Background and significance
The lack of semantic interoperability is cited as a reason for inefficiencies within the healthcare system in the United States, contributing to the waste of billions of dollars annually.1–3 Therefore, it is essential to share EHR across different organizations and allow healthcare professionals to transparently access the complete EHR of patients regardless of the institutions that may have participated in patient record creation. In the last decades, many efforts have addressed the development of EHR standards4 based on dual model architectures; that is, separating the information and knowledge levels. Examples of such standards are ISO 13606,5 the openEHR specification,6 HL7 CDA,7 and the CEM.8 In the information level, the information model defines the entities that form the building blocks of the EHR. The knowledge level enables definition of clinical concepts in the form of structured and constrained combinations of the entities contained in the information model. Each definition of clinical concept is usually referred to as clinical model.
Clinical models are built in a similar way in the different EHR specifications and standards, basically, by constraining the entities of the information model. Despite the use of similar processes in each community, there are three main differences between the clinical models obtained by those communities: (1) the formalism used to specify the models; (2) the content and structure of the information model; and (3) the types of constraints used. Consequently, the same clinical concepts are represented in the different formalisms using different constructs and, most times, the same clinical concept can be represented in different ways in the same formalism. Differences (2) and (3) are due to the availability of different EHR standards, but (1) could be solved by using a common formalism.
Current international efforts assume the co-existence of different EHR standards and propose complementary solutions for semantic interoperability. SemanticHealthNet (SHN)9 pursues the development of an organizational and governance process for reaching the semantic interoperability of clinical information across Europe and solutions for the semantic harmonization of clinical models, information models, and clinical terminologies. SHN asserts that ontologies should play a fundamental role in the achievement of semantic interoperability. SHN proposals include making clinical models interoperable through the development and application of semantic patterns that could bridge the structural difference of such models while capturing the semantic equivalence of the constructs.10,11 The Clinical Information Modeling Initiative (CIMI)12 pursues the development of interoperable clinical models, which requires research on the alignment, mapping, and transformation of clinical model formalisms. CIMI has decided to use archetypes in their Clinical Information Model specification.
Conceptually speaking, SHN and CIMI pursue the development of semantically equivalent clinical models expressed in different EHR standards (i.e., CEM, openEHR, HL7, ISO 13606). Clinical model transformation methods would permit existing models to be made available to other communities, sharing the clinical knowledge and facilitating the semantic interoperability of clinical information. In cases in which the same clinical concepts have been rendered in different existing models, representing them in the same formalism would facilitate their comparison and alignment. This would be important for achieving semantic interoperability at the data level, since clinical models transformation methods may also guide the transformation of clinical data instances. The transformation of clinical models requires understanding the meaning of the different modeling primitives and constructs provided by the information model. By such conceptual analysis we can generate conceptual mappings between the information models that guide our transformation process.
In recent years, semantic web technologies have been used in different approaches for managing EHR information and knowledge. The reason for this is the potential of technologies like OWL,13 which enables a formal representation of the domain information entities and knowledge that can be exploited by automated means. The research community has developed ontologies for representing some information models14–16 and ontological frameworks for the interoperability of clinical data, models, and applications have been proposed.17–22 Moreover, SHN promotes the use of ontology patterns for the achievement of semantic interoperability. Ontology patterns can be viewed as templates for the creation of semantic content, and those templates are based on formal ontologies.23
The current state of the art supports transforming clinical models by aligning and mapping clinical model formalisms from an ontological perspective.24 However, we are not aware of any approach whose transformation engine is implemented using OWL technologies and patterns in the transformation process. To provide methods for the transformation of clinical models between different formalisms, we propose a transformation strategy that will be based on ontologies, OWL technologies, and transformation templates, which will describe how particular types of models from the source formalism are represented in the target formalism. We propose an extensible, flexible transformation architecture able to deal with the definition of transformation templates that provide syntactic, structural transformation, and templates driven by the meaning of the clinical concepts. The transformation architecture will exploit the reasoning and inferencing capabilities of OWL technologies, which will permit the definition of generic transformation templates and ensure that only logically consistent content is transformed and generated. In addition, OWL may help the integration of the clinical models with terminologies available in this format. Given the large amount of CEMs available and the commitment of CIMI to use archetypes, we believe that the transformation of CEMs into archetypes is an interesting case study because of the large amount of CEMs available and the commitment of CIMI to use archetypes.
The contributions of this paper are: (1) an extensible, flexible architecture for the transformation of clinical models between EHR standards based on ontologies and OWL technologies; (2) promotion of EHR (i.e., CEM, openEHR) and semantic web (i.e., OWL) standards as key players for the achievement of semantic interoperability; and (3) the creation of a base of openEHR archetypes from CEMs, which can be used for purposes such as comparison with pre-existing openEHR archetypes or transformation of clinical data between both standards.
MATERIALS AND METHODS
Clinical models: CEM and openEHR archetypes
CEM specifications describe the representation of detailed clinical data models and the instances of data that conform to these models. CEM includes the Abstract Instance Model, which provides the structure to represent instances of medical data, and the Abstract Constraint Model, which defines the constraints on values in the Abstract Instance Model. Each CEM is classified into a basic structural category, which captures the common attributes assignable to a common class model. Figure 1 shows part of the CEM used for recording the blood pressure of a patient. This CEM belongs to the structural category of “Panel,” which represents a grouping of clinical observations. This Panel contains two “Statements,” one for recording systolic blood pressure and the other for recording diastolic blood pressure. CEM Statements represent complete assertions about a particular aspect or condition of a panel. Each CEM model may be complemented using “Components.” Unlike Panels or Statements, CEM Components do not have a clinical meaning by themselves but have meaning only in the context of other CEMs. In our example, the Component Method Device captures the device used to perform the measurement of the blood pressure.
Figure 1:
Part of Blood Pressure Panel.
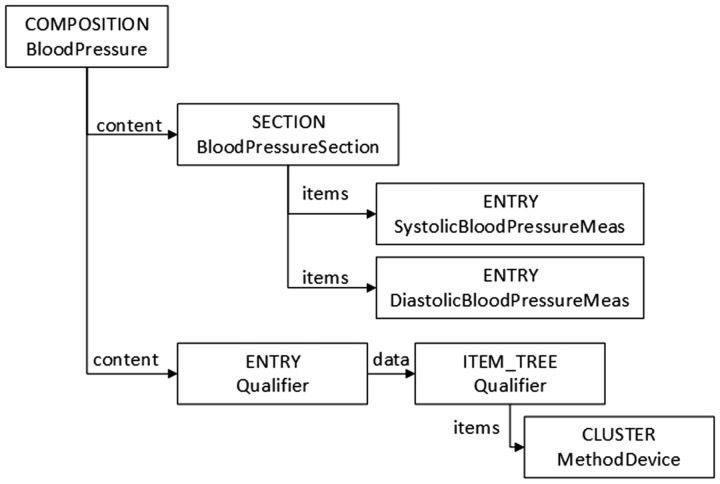
The openEHR Foundation has produced specifications for representing and exchanging EHR content in a meaningful way. OpenEHR specifications distinguish information and knowledge levels. In this case, the information model provides the static entities of the EHR domain and archetypes are used for modeling the clinical models. Archetypes are also constraint-based models of domain entities and are specified through constraints on the entities of the information model. In this way, an archetype describes configurations of clinical data using the information model classes. Figure 2 shows part of an archetype equivalent to the CEM model shown in Figure 1. The openEHR class “Entry” is defined as a clinical statement, so the measures of systolic and diastolic blood pressure are both Entrys. The openEHR class “Composition” groups EHR content related to the same clinical event. The model shown in Figure 2 uses a “Section” to arrange Entrys and separate the ones for the recording of blood pressure from those providing additional information (i.e., capturing the device used to perform the measure).
Figure 2:
Part of the Blood Pressure archetype.
Therefore, CEM and openEHR information models have different entities and structures, but clinical models are built in both specifications by constraining the entities of the information model. Such constrained entity represents a specialization of that entity of the information model. The constraints are applied to the attributes defined for each entity: range, cardinality, and so on.
Representation of clinical models in OWL
OWL has been proposed as a common formalism for the representation of clinical models because it provides a formalism in which each construct has a concrete meaning and in which information models, clinical models, and terminologies can be smoothly combined. OWL also provides a formalism that supports automated reasoning, ensuring, for instance, that only logically consistent content is transformed. Hence, our starting point is OWL as common formalism for representing CEM and openEHR archetypes, and OWL will be used for studying and proposing solutions for the alignment and transformation of clinical models between these specifications.
In recent years, OWL representations have been developed for both the CEM and openEHR information model classes and clinical models. In this work, the OWL representations for CEM developed in the context of the Strategic Health IT Advanced Research Project, secondary use of EHR,16,25 and the OWL representation for openEHR developed in the context of the Archeck project26 will be used.
Regarding information model entities, both representations have been developed using similar methodological principles representing the main concepts of the information models as classes and representing the clinical models; that is, CEMs and archetypes, as subclasses of the information model classes. Regarding constraints, each constrained entity is defined by means of an OWL class in which the corresponding constraints are defined. Note that OWL allows combining the information model and the clinical models into the same modeling framework, in which clinical models definitions are based on information model classes. Therefore, there is no modeling boundary between the information model and the clinical models beyond the modular organization of ontologies: the clinical models’ ontologies import the information model ontology.
Transformation of clinical models
Our starting point is the availability of OWL ontologies for each information model and the premise that clinical models are built by constraining the entities of such an information model. Our transformation process is guided by the mappings between the entities of CEM and openEHR. Our approach uses mappings between ontological entities. Once such mappings are defined, the transformation is executed with the support of OWL technologies, thus reducing the implementation effort. Finally, the resulting OWL content can be validated according to the target clinical model specification using automated reasoning, exploited in OWL or transformed into other formats such as the openEHR ADL language.
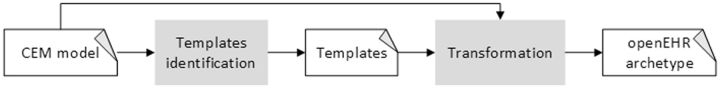
The basic goal is to transform the entities and constraints defined in the CEMs into openEHR archetypes. It should be noted that clinical models of a particular type have a similar structure, which means that many structural transformations will be shared by different clinical models. For example, a common set of transformations can be applied to every CEM Panel model to obtain the corresponding openEHR representation. We propose to use generic openEHR archetypes as templates for the transformation process because this approach facilitates obtaining structurally homogeneous openEHR archetypes. Consequently, the transformation of a CEM model, shown in Figure 3, requires (1) identifying its corresponding template and (2) applying it to obtain the corresponding openEHR archetype.
Figure 3:
Overview of the transformation approach.
Figure 4 provides a partial, schematic representation of a CEM Panel and its corresponding representation in openEHR. Panels contain a set of items and might contain qualifiers, modifiers, and attributions. For simplicity, we focus our example on modeling the items. The left side represents a Panel according to CEM; the right side represents the corresponding openEHR archetype, where gray concepts correspond to the openEHR information model, and white concepts correspond to our transformation template. Hence, the transformations depicted in the figure can be interpreted as the generic transformation of a CEM Panel into openEHR.
Figure 4:
Transformation template to openEHR archetype for a CEM Panel.
CEM Panel, Simple Statement, and Data Component are mapped, respectively, onto PANEL, STATEMENT_ENTRY and DATA_ELEMENT in openEHR, which are not entities of the openEHR information model. We define them as subclasses of the openEHR classes COMPOSITION, GENERIC_ENTRY and ELEMENT, respectively. Consequently, direct mappings between such CEM and openEHR classes can be defined.
The scenario is different for the transformation of the constrained properties. In Figure 4, the constraint 1..* on the CEM property item, which means that a Panel must have at least one Simple Statement, cannot be mapped onto a similar property in openEHR. A constrained CEM property must be mapped to a combination of openEHR properties and classes. In this example, the openEHR representation of this property is a Section (ITEM_SECTION), because SECTION and COMPOSITION have no property with the semantics of the CEM item. This is an additional, intermediate class needed to bridge the gap between the information models.
Given that both CEM and openEHR information models are available in OWL, such mappings can be expressed using OWL axioms. More concretely, given that we intend to perform a unidirectional transformation from CEM to openEHR, we use the subClassOf (⊑) axiom to define the mapping from CEM to openEHR. We will use the Manchester OWL Syntax27 to represent the OWL axioms. Two types of mapping are defined in our approach (see Table 1).
Table 1:
Types of mappings for the transformation of clinical models
| Mapping | Description and example |
|---|---|
| Class2Class | This mapping links two classes. cem:A⊑openehr:B means that a CEM A is a logical subclass of an openEHR B, which would be used by the transformation engine to transform the A model into a B Archetype. |
| Example: cem:Panel ⊑openehr:PANEL ⊑openehr:COMPOSITION Meaning: A CEM Panel would be transformed into a PANEL archetype, which is a COMPOSITION. | |
| Property2Structure | This mapping links a property axiom from CEM to an ontological structure in openEHR. One axiom of such type is defined for every property associated with the classes of the CEM information model. |
| Example: (cem:Panel SubClassOf cem:item min 1 cem:SimpleStatement) ⊑ (openehr:PANEL SubClassOf openehr:content exactly 1 (openehr:ITEM_SECTION and openehr:items min 1 openehr:STATEMENT_ENTRY)) Meaning: Representation of the CEM property item in openEHR. |
Execution
The transformation engine takes a CEM model and obtains the corresponding openEHR archetype by applying the corresponding mappings and template archetypes. The mappings define which OWL axioms have to be created to represent the CEM model as an openEHR archetype. The transformation process is generative, since new axioms are added to a base openEHR archetype.
We use the Ontology Pre-Processing Language version 2 (OPPL2)28 for generating the corresponding axioms. OPPL2 is a scripting language for OWL that can be used to modify the axioms of an ontology using a patterns approach.29,30 OPPL2 offers an API that permits the execution of the patterns while controlling the transformation processes. OPPL2 works in conjunction with reasoners, which has advantages for our purpose: (1) defining patterns that exploit inferencing (i.e., a pattern that affects CEM Statements will also affect Simple and Compound Statements unless we explicitly mention that the rule only applies to asserted axioms) and (2) ensuring the transformation of only logically consistent content of the clinical models (i.e., only CEM Panels with at least one Simple Statement would be transformed).
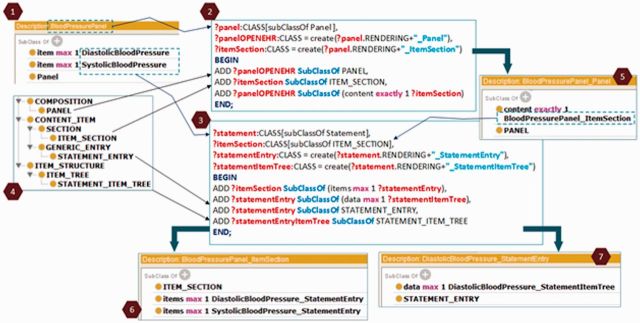
Our transformation method creates the openEHR archetype in OWL format by executing OPPL2 patterns. Figure 5 illustrates the transformation of the BloodPressurePanel (1), which consists of two Simple Statements, namely, DiastolicBloodPressure and SystolicBloodPressure. This transformation uses two OPPL patterns: (2) transforms the Panel and (3) transforms the CEM property item and the Simple Statements. The axioms to be generated appear between the BEGIN and END keywords of the OPPL patterns. The first OPPL pattern has the parameter ?panel, which represents the CEM BloodPressurePanel in this example. The second one has two parameters: (i)?statement, which stands for DiastolicBloodPressure and SystolicBloodPressure CEM Simple Statements and (ii)?item- Section, which stands for BloodPressurePanel_ItemSection openEHR ITEM_SECTION, created by the first pattern. The axioms are added to the OWL content that represents the template archetype for Panels (4). PANEL, ITEM_SECTION, STATEMENT_ENTRY, and STATEMENT_ITEM_TREE are classes of such ontology. Figure 5 also contains the OWL openEHR representation of: (5) the description of the Panel; (6) the transformation of the CEM property item; and (7) the DiastolicBloodPressure Simple Statement. Finally, once the openEHR clinical model is obtained in OWL format, we can obtain the corresponding openEHR archetype using the Archeck tool.26
Figure 5.
OPPL2 patterns for the transformation of the CEM BloodPressurePanel into an openEHR archetype.
RESULTS
Four major results have been obtained in this research work. The transformation approach is the first result, since it provides the method for the transformation of CEMs into openEHR archetypes. The second result is the set of transformation templates for CEM, which were not available to date for the research community. We have used the tools developed by our research groups to create the OWL version of CEM, since the input to our transformation pipeline has been the OWL representation of CEM models. The third result is the implementation of the transformation engine from CEM OWL to openEHR OWL as described in subsection Transformation of clinical models, which has been used to validate our approach. The interface of an online version receives a CEM OWL and returns (1) the openEHR OWL version and (2) the instantiation of the OPPL patterns used to transform it. Such patterns can be used to analyze what has been transformed. The fourth result is the set of transformed CEMs into openEHR archetypes, which includes 5 Panels, 57 Statements, and 153 Components. All the templates, patterns, archetypes, and the web interface of the transformation engine are available at http://sele.inf.um.es/CEM2Archetypes.
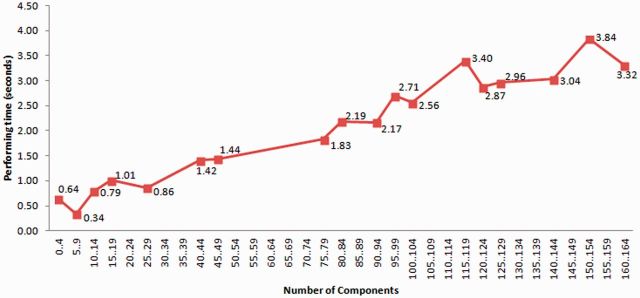
We have performed a technical evaluation of our results. We evaluated the execution of the transformations for a series of CEM models, verifying that the obtained results matched the expected outcomes according to our openEHR archetype ontology and transformation templates design. This ensures that the transformations are correctly executed from a technical perspective. The mean time for transforming a CEM model to openEHR archetype in OWL is 1.587 s on a server with 2.13 GHz, 8 cores processor, using a 6 GB Java Virtual Machine, Hermit 1.3.5 as reasoner, OWLAPI 3.4.5, and OPPL2. There is a positive correlation between the total time and the number of Components in the source CEM (Pearson’s correlation coefficient = 0.964). A linear regression model could explain the relation (see Figure 6) between the number of Components in the CEM models and time (R = 0.996; P = 0.0 for both constant and number of components), which is good in terms of scalability. Components transformed in the parent model are not transformed again due to the CEM inheritance mechanism. Nevertheless, Components may include other Components in the form of qualifiers, modifiers, or attributions, which may have more Components included.
Figure 6.
Time performance of the transformation process with respect to the number of Components of the CEM.
DISCUSSION AND CONCLUSION
We have presented an approach for obtaining openEHR archetypes from CEM models. The methodological approach supposes a shift with respect to our previous work with ISO 13606 and openEHR.18 The new method is based on OWL technologies instead of on model-driven engineering technologies. OWL technologies reduce the number of steps in the transformation process. This therefore becomes simpler and technologically more homogeneous. Our transformation process is based on flexible templates instead of requiring a predefined mapping between the entities of the corresponding information models. However, both efforts can be linked through the use of OWL ontologies.
Mappings could have been defined and implemented using a different technology, i.e., XML. OWL provides better support for this process because (1) OWL representations for both CEM and openEHR are available; (2) OWL supports automated reasoning, which permits to ensure that only logically consistent content is transformed and generated; and (3) OPPL2 supports the enrichment and analysis of ontologies and permits defining templates that exploit the taxonomic structure of the CEM and openEHR ontologies. OPPL2 permits the transformation process to be generative, adding axioms to the corresponding archetype template. OPPL2 patterns are also easy to extend and reuse.
The definition of the transformation templates is manual and requires expertise in clinical model specifications and OWL. We would expect these patterns to be defined by communities having experts with such profiles and systems developers to transform clinical models with the support of software tools. Our approach is not limited to our templates, but would work with those defined by a particular community, meaning that particular transformations could be achieved by defining the corresponding archetype templates.
The technological artifacts presented here are a proof-of-concept implementation that validates the methodological approach, showing the potential of OWL technologies to support the process and studying aspects like scalability. To the best of our knowledge, this is the first effort for algorithmically generating archetypes from CEM. Our development team has extensive experience with CEM, ISO 13606, and openEHR archetypes and has evaluated the mappings to the templates, the archetypes generated, as well as of its structural semantics to ensure the semantic and syntactic correctness. Our future plans include the comparison of the transformed archetypes with those already existing in the openEHR community, from which interesting information could be obtained, such as the existence of similar or different modeling principles in the CEM and openEHR communities.
Further work is required to complete the transformation templates and to develop robust end-user tools that support the process and permit users to define their own transformation templates. A technical upgrade of the process might permit the transformation of each component as a separate archetype and to include it as a slot in the archetype of its corresponding CEM, thus facilitating the reuse and modularity of the content of the clinical models. Our method generates new openEHR archetypes. This makes our approach of special interest for those clinical concepts without available archetypes. If such archetypes exist, our method provides a representation of both clinical models in OWL for comparing them. Such comparison could reveal important relations like equivalency between archetypes or suggesting specialization relations between the archetype generated from CEM and the existing openEHR archetype. OWL technologies are appropriate for this purpose because of the possibility of automated reasoning, which has already been used by our research group for evaluating specialization in archetypes.26
Our transformation approach generates OWL for the clinical model by applying patterns based on openEHR archetypes. We use structural ontology patterns, which permit the expression of the source clinical model content according to the target information model. These patterns are different from the ontology content patterns proposed by SHN. SHN patterns are meant to provide a formal definition of the meaning of the clinical content, but not to transform content from one EHR standard to another. SHN patterns, which can be play a similar role to terminological bindings, can be combined with our structural transformation patterns. The additional formalization of the meaning of the information model and clinical model entities could be exploited in the mapping rules and in the transformation templates.
We think that such integration with SHN patterns would be the most effective way to include terminological content in the transformation process. In our current method, the CEM terminological bindings are transformed into term bindings in archetypes. Once the CEM is transformed into an openEHR archetype with the corresponding term binding, our method can semantically exploit such binding for different purposes (i.e., checking the correctness of specializations by applying our Archeck method26).
We believe that these results can be useful for efforts like CIMI. Expressing the clinical models in OWL would permit CIMI to benefit from the aforementioned OWL good properties and our framework could be applied to get the corresponding models for the different EHR standards, thereby facilitating the sharing of semantics.
In this work, only the transformation of CEM into openEHR archetypes has been addressed. A similar approach could be applied for generating ISO 13606 archetypes or HL7 clinical models. In recent years, OWL representations for HL7 standards have been proposed.15 We are currently studying the suitability of such representations for our approach. We are also examining the application of the transformation patterns to clinical data transformation.
In summary, we have presented an approach for facilitating the interoperability of clinical models between EHR standards, illustrated for CEM and openEHR. This work helps demonstrate how ontologies and OWL technologies can support the transformation of clinical models and in turn, how EHR standards and specifications can be effectively used and integrated into interoperability efforts powered by semantic technologies.
CONTRIBUTORS
M.C.L.G. participated in the design of the study and the experiments, implemented the software tools, participated in the data analysis and evaluation of the results, and drafted and revised the paper. C.T., M.M.T., J.T.F.B., and C.G.C. participated in the design of the study and the experiments, participated in the data analysis and evaluation of the results, and drafted and revised the paper.
FUNDING
This work was supported by the Ministerio de Economía y Competitividad and the FEDER program through grant TIN2010-21388-C02, the Fundación Séneca through grants 15295/PI/10 and 15555/FPI/2010, the Office of the National Coordinator for Health Information Technology through the Strategic Health IT Advanced Research Projects Program (grant 90TR002), and the National Library Of Medicine of the National Institutes of Health under Award Number R01LM011829.
COMPETING INTERESTS
None.
REFERENCES
- 1.Kalra D, Lewalle P, Rector A, et al. Semantic interoperability for better health and safer healthcare. Research and deployment roadmap for Europe SemanticHEALTH project report. European Comission. 2009. [Google Scholar]
- 2.Saleem JJ, Flanagan ME, Wilck NR, et al. The next-generation electronic health record: perspectives of key leaders from the US Department of Veterans Affairs. J Am Med Inform Assoc. 2013;20(e1):e175–e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fickenscher KM. President's column: interoperability—the 30% solution: from dialog and rhetoric to reality. J Am Med Inform Assoc. 2013;20(3):593–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisbal J, Berry D. An analysis framework for electronic health record systems. Interoperation and collaboration in shared healthcare. Methods Inf Med. 2011;50(2):180–189. [DOI] [PubMed] [Google Scholar]
- 5. ISO 13606. http://www.iso.org. Accessed January 2015.
- 6. The openEHR Foundation. http://www.openehr.org. Accessed January 2015.
- 7. Health Level Seven International. http://www.hl7.org/. Accessed January 2015.
- 8. The Clinical Element Model. http://clinicalelement.com. Accessed January 2015.
- 9. SemanticHealthNet. http://www.semantichealthnet.eu/. Accessed January 2015.
- 10.Martínez-Costa C, Legaz-García MC, Schulz S, et al. Ontology-based Infrastructure for a Meaningful EHR Representation and Use. In: International Conference on Biomedical & Health Informatics (BHI) . Valencia, 2014. [Google Scholar]
- 11. SemanticHealthNet deliverable 4.2: Ontology/Information models covering the heart failure use case. 2013.
- 12. Clinical Information Modeling Initiative. http://www.opencimi.org/. Accessed January 2015.
- 13. OWL Web Ontology Language. http://www.w3.org/TR/owl-ref/. Accessed January 2015.
- 14.Martínez-Costa C, Menárguez-Tortosa M, Fernández-Breis JT, et al. A model-driven approach for representing clinical archetypes for Semantic Web environments. J Biomed Inform. 2009;42(1):150–164. [DOI] [PubMed] [Google Scholar]
- 15.Iqbal AM. An OWL-DL Ontology for the HL7 Reference Information Model. In Toward Useful Services for Elderly and People with Disabilities. Berlin Heidelberg: Springer; 2011:168–175. [Google Scholar]
- 16.Tao C, Jiang G, Oniki TA, et al. A semantic-web oriented representation of the clinical element model for secondary use of electronic health records data. J Am Med Inform Assoc. 2013;20(3):554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilic O, Dogac A. Achieving Clinical Statement Interoperability Using R-MIM and Archetype-Based Semantic Transformations. IEEE Trans Inf Technol Biomed. 2009;13(4):467–477. [DOI] [PubMed] [Google Scholar]
- 18.Martínez-Costa C, Menárguez-Tortosa M, Fernández-Breis JT. An approach for the semantic interoperability of ISO EN 13606 and OpenEHR archetypes. J Biomed Inform. 2010;43(5):736–746. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Costa C, Menárguez-Tortosa M, Fernández-Breis JT. Clinical data interoperability based on archetype transformation. J Biomed Inform. 2011;44(5):869–880. [DOI] [PubMed] [Google Scholar]
- 20.González C, Blobel BGME, López DM. Ontology-based framework for electronic health records interoperability. Stud Health Technol Inform. 2011;169:694–698. [PubMed] [Google Scholar]
- 21.Sahay RN. An Ontological Framework for Interoperability of Health Level Seven (HL7) Applications. The PPEPR Methodology and System [Thesis]. 2012. http://hdl.handle.net/10379/3034. Accessed January 2015. [Google Scholar]
- 22.Fernández-Breis JT, Maldonado JA, Marcos M, et al. Leveraging electronic healthcare record standards and semantic web technologies for the identification of patient cohorts. J Am Med Inform Assoc. 2013;20:e288–e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangemi A, Presutti V. Ontology design patterns. In: Handbook on Ontologies. Berlin, Heidelberg: Springer; 2009:221–243. [Google Scholar]
- 24.Euzenat J, Shvaiko P. Ontology Matching. Springer; 2007:332p ISBN: 978-3-642-38721-0. [Google Scholar]
- 25.Rea S, Pathak J, Savova G, et al. Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: the SHARPn project. J Biomed Inform. 2012;45(4):763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menárguez-Tortosa M, Fernández-Breis JT. OWL-based reasoning methods for validating archetypes. J Biomed Inform. 2013;46(2):304–317. [DOI] [PubMed] [Google Scholar]
- 27. Manchester OWL Syntax. http://www.w3.org/TR/owl2-manchester-syntax/. Accessed January 2015.
- 28. Ontology Pre-Processing Language version 2. http://oppl2.sourceforge.net/. Accessed January 2015.
- 29.Iannone L, Rector A, Stevens R. Embedding knowledge patterns into OWL. In: Aroyo L, Traverso P, Ciravegna F, et al., eds. The Semantic Web: Research and Applications. Springer; 2009:218–232. [Google Scholar]
- 30.Egaña M, Rector A, Stevens R, et al. Applying Ontology Design Patterns in Bio-ontologies. In: Proceedings of the 16th International Conference on Knowledge Engineering: Practice and Patterns. Berlin, Heidelberg: Springer-Verlag; 2008:7–16. [Google Scholar]