ABSTRACT
Plastoglobules (PGs) in chloroplasts are monolayer lipid-protein particles attached to thylakoids. The size and number of PGs per chloroplast respond dynamically to abiotic environmental stresses and developmental transitions. During senescence, the thylakoid membranes and its constituents are dismantled in controlled fashion. Leaf senescence coincides with a dramatic increase in the size of PGs, which is consistent with a functional role of PG in remobilization of thylakoid membrane components. In a recent publication,1 we showed that PG-localized metallopeptidase PGM48 promotes natural senescence. In plants, PGM48 has homologs in mitochondria and the endomembrane system, but PGM48 evolved specifically in photosynthetic organisms. Extensive analysis of Arabidopsis transgenic lines either under- or overexpressing PGM48, showed that PGM48 is a positive regulator of senescence, and we proposed that PG-localized carotenoid cleavage enzyme 4 (CCD4) is a potential substrate of PGM48. Here, we discuss PGM48 function and how it may accelerate natural senescence.
KEYWORDS: Arabidopsis thaliana, chloroplast, metallo-peptidase, plastoglobules, protease, senescence
Abbreviations
- CCD4
Carotenoid Cleavage Enzyme 4
- PGs
Plastoglobules
- Y2H
yeast-2-hybrid
A new subclade of M48 metallo-peptidase unique to photosynthetic organisms
We discovered a M48 peptidase (AT3G27110) in plastoglobules (PGs) isolated from Arabidopsis thaliana leaf chloroplasts2 and we recently assigned it PGM48, for PLASTOGLOBULAR M48.1 According to the peptidase database MEROPS (http://merops.sanger.ac.uk/), the M48 family within the MA (E) clan has 3 clades, M48A, M48B and M48C, each represented by a prototypic enzyme named STE24 or CAAX peptidase, bacterial HTPX and mitochondrial OMA1, respectively. In Arabidopsis there are 3 M48 proteins, one of which (STE24) is located in the ER/nuclear membrane,3 one is PGM48,2 and the third one is a likely mitochondrial homolog of OMA1 in yeast, flies and mammals where it plays a key role in particular in proteolysis of OPTIC ATROPHY 1 (OPA1/MGM1), a key conserved inner membrane dynamin-like GTPase involved in mitochondrial fusion,4,5 but it has not been studied in plants. Based on careful phylogeny we determined that PGM48 forms a separate clade, assigned M48D, that is unique to photosynthetic organisms; we postulate that PGM48 evolved a specific function in the context of plastoglobular metabolism.1 Homology-based modeling supported interaction of PGM48 with the PG monolayer, and pointed out that PGM48 lacks trans-membrane domains, unlike members of the M48A, B and C clades.
PGM48 is an activator of leaf senescence
PGM48 protein and transcript levels strongly increase during natural leaf senescence.1 Extensive analysis of Arabidopsis transgenic lines either under- or overexpressing PGM48, showed that PGM48 is a positive regulator of senescence.1 Loss of PGM48 delayed natural leaf senescence by nearly one week, whereas overexpression of PGM48 visually accelerated senescence by 3–4 d. This was also confirmed by transcript accumulation of the senescence marker genes SAG12 and SAG13. Careful inspection of the transcript levels for most ∼30 PG-localized proteins based on public information assembled in eBAR (http://bar.utoronto.ca/), showed that several PG transcripts are upregulated during leaf senescence. In early stages of leaf senescence in particular the transcripts for CAROTENOID CLEAVAGE ENZYME 4 (CCD4) were upregulated (along with thylakoid protein ZEAXANTHIN EPOXIDASE (ZEP)), whereas during late stage leaf senescence the transcripts for PGM48, the kinases ABC1K3 and 7, PYTHOL ESTERASE 1 AND 2 (PES1 AND 2), PG-localized SENESCENCE ASSOCIATED GENE (PGSAG) and PHEOPHYTINASE (PPH) were strongly upregulated.1
Potential substrates of PGM48 during leaf senescence
The early senescence phenotype of PGM48 overexpression plants and late senescence phenotypes in PGM48 RNAi plants lead us to conclude that PGM48 degrades or inactivates a protein that is either an (indirect) repressor of senescence or activates a protein (indirectly) required for senescence by partial cleavage (Bhuiyan et al., 2016). To find potential substrates or potential regulators/adaptors of PGM48, we performed targeted yeast-2-hybrid (Y2H) experiments, probing interactions between PGM48 and 5 enzymes involved in chlorophyll degradation (including PPH) and 9 PG proteins (including PES1,2, CCD4 and PGSAG). Among those 14 proteins, only PES1, CCD4 and ABC1K3 showed interactions with PGM48.1
For proteins to be substrates or regulators of PGM48, it is reasonable to expect overlapping tissue- and developmental specific transcript accumulation patterns. Fig. 1 showed the transcript accumulation pattern for these 4 proteins across tissues and development (from eBAR). In particular PGM48 and CCD4 have strikingly similar expression patterns across development and tissues types/organs: both accumulate highly in dry seed, the cauline leaf, cotyledons, petals, the oldest leaf on plants of stage 1.12 and senescing leaves. Some quantitative differences can be observed such as that PGM48 expresses higher in sepal than petal, whereas CCD4 expresses higher in petal than sepal in case of older flowers (stage 15 as compared with stage 12). This highly similar, but slightly developmental shifted expression pattern, is compatible with the hypothesis that CCD4 is a direct substrate for PGM48 proteolysis. The expression of PES1 is specific for stamen, older petals, sepals and stamen (likely senescing) and late stage senescence; similar as we observed for the senescing leaf series, PES1 expression peak very sharply, slightly delayed as compared with PGM48; this would be compatible with PGM48 acting as activator of PES1. In contrast, ABC1K3 is far more widely expressed. Finally, none of these PG core proteins are much expressed in roots, mature pollen, carpels or imbibed seeds.
Figure 1.
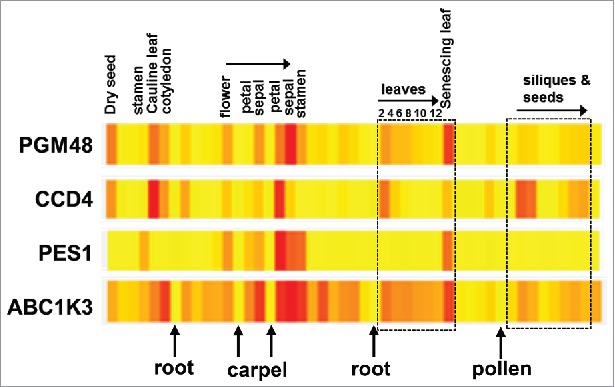
Comparison of the tissue-specific and developmental expression pattern of PGM48, CCD4, ABC1K3 and PES1. Data and individual heat maps are from the BAR Expression Angler. The mRNA expression levels are normalized to the maximum expression levels within each gene. Yellow and Red are respectively the lowest and highest expression levels.
PGM48 substrates and a functional model for PG in senescing leaves
Based on the observations briefly summarized above and others described in more detail in1 and previous studies recently reviewed in,6 we developed a speculative working model for potential functional roles of PGM48 in senescing leaf tissue (Fig. 2). The model concerns senescing chloroplasts in which thylakoids are dismantled in a controlled fashion, resulting in a metabolite flux of lipophilic molecules to the PG. PG core proteins then convert these molecules for further storage or export. Hydrophobic molecules include fatty acids and glycerol derived from thylakoid lipids, chlorophyll and carotenoid breakdown products, and various thylakoid quinones and tocochromanols (e.g. plastoquinone-9, phylloquinone, plastochromanol-8, tocopherols). Each of these metabolites have been shown to accumulate to varying degrees in PGs – extensively reviewed in.6 Here we highlight carotenoid and chlorophyll degradation because of their possible relevance to PGM48 function. Comparison of the metabolite composition of PG isolated from 4 stages of natural senescence in beach leaves showed that the PG content of carotenoids and carotenoid esters was very low in green leaves, but increased in early stages of senescence (while total leaf carotenoid content decreases), and then decreased to low levels in more advanced stages of senescence.7 Thus the PG participates in the controlled removal of carotenoids from the thylakoid membrane during the senescence process; but the size of the flux is unknown. There are multiple lines of evidence that PG-localized CCD4 degrades carotenoids located in PGs: i) in absence of CCD4, higher levels of carotenoids (β-carotene, lutein and violaxanthin) accumulate in PGs and leaf senescence is accelerated,1,8 ii) whereas overexpression of CCD4 results in reduced PG carotenoid content and delayed senescence.8 Furthermore, phytol cleaved from chlorophyll by PPH (found in PGs) is esterified by PG-localized PES1 and accumulates in PGs.6,9
Figure 2.
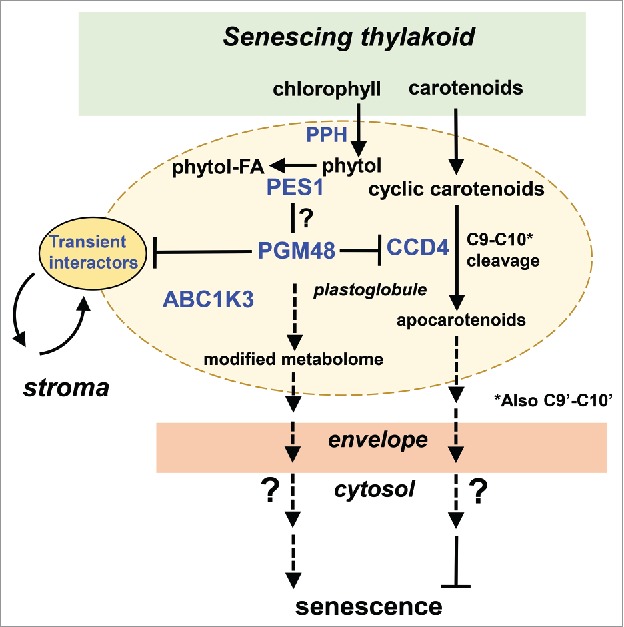
Functional model for PGM48 function in senescing chloroplasts. PES1, CCD4 and ABC1K3 each interacted with PGM48 in Y2H.This model proposes that PGM48 degrades or partially cleaves one of more PG-localized proteins, resulting in modified metabolism and/or metabolite content, thereby accelerating leaf senescence through an unknown retrograde signaling pathway. Possible signals for induction or acceleration of senescence are ABA or JA, whereas carotenoid cleavage products by CCD4 may (indirectly) delay senescence. So far, direct substrates for PGM48 have not been identified but we suggest CCD4 as a strong candidate.
The model in fig. 2 is centered on PGM48 and its interactors ABC1K3, CCD4 and PES1 (as observed from Y2H) and emphasizes 3 main points:
-
1.
The function of ABC1K3 presumably lies in the phosphorylation of one or more PG proteins (or perhaps metabolites), but the ABC1K3 enzymatic activity has yet not been established, and meta-analysis of published phosphoproteomics data in Arabidopsis did not find any solid evidence for phosphorylation of PES1, CCD4 or PGM48.10
-
2.
The model suggests that PGM48 directly degrades CCD4. This speculation is based on i) our observation for strongly reduced CCD4 levels in PG from the PGM48 overexpression line, ii) the development transcriptional dynamics of CCD4 and PGM48 (i.e. CCD4 mRNA accumulations peak immediately before PGM48), and iii) observed physical interactions between PGM48 and CCD4 (even if this was ‘just’ from Y2H). Recombinant PGM48 did not degrade recombinant GST-CCD4 in vitro (data not shown) but this does not exclude in vivo degradation, perhaps because degradation would require a monolayer lipid interface and/or ABC1K3 dependent phosphorylation of PGM48 or CCD4. Arabidopsis CCD4, and its homologs in other plant species, have been extensively investigated showing that CCD4 members cleave a variety of carotenoids, resulting into volatile apocarotenoids that can act as retrograde signals.11-13 For instance, CCD4 is implicated in cleaving of early carotenoid intermediates (i.e., upstream of all-trans-lycopene) that function in a signaling cascade leading to inhibition of chloroplast and leaf development in Arabidopsis.14 However, a recent paper provided strong in vitro evidence that recombinant Arabidopsis CCD4 only cleaves all-trans carotenoids with at least one ionome ring, and not acyclic carotene desaturation intermediates.15 An Arabidopsis genome-wide association study identified CCD4 as a major negative regulator of seed carotenoid content16; ccd4 loss-of-function mutants exhibited increased β-carotene content upon seed desiccation and much higher carotenoid levels than the wild type after dark-induced leaf senescence, resulting in a yellow phenotype. Similarly, lipidomics of isolated PG from senescent Arabidopsis wt and ccd4 null mutants, suggested that CCD4 primarily cleaved β-carotene, lutein and violaxanthin8 consistent with the in vitro analysis.15 Consistent with the reduced accumulation of CCD4 in the PGM48 overexpression line, and the early natural senescence leaf phenotype in the CCD4 null mutant,1 the model proposes that CCD4 directly or indirectly represses senescence through cleavage of carotenoids transiently stored in PG.
-
3.
PES1 was observed as an interactor to PGM48 in the Y2H assays.1 PES1 is preferentially expressed during senescence (both transcript and protein) and has been shown to convert phytol and fatty acids into fatty acid phytol esters and triacylglycerol (TAG) during senescence (Lippold et al., 2012). Fattly acid phytol esters and triacylglycerol levels are reduced in the Arabidopsis pes1 and pes2 mutants and a delayed senescence phenotype was reported in pes1xpes2 double mutant (Lippold et al., 2012). If PES1 (and PES2) was degraded by PGM48, we would expect to see reduced PES levels in PGM48 OE lines and reduced levels in RNAi lines, and consequently delayed and accelerated senescence, respectively. However, our observations are opposite: PGM48 overexpression plants showed more than 2-fold overaccumulation of PES1 protein compared with wild-type, and these lines showed accelerated senescence. An alternative explanation would be that PGM48 activates PES1 through e.g., C-terminal partial cleavage. However, careful inspection of MS/MS data for PES1 did identify the complete predicted C-terminus in wt. The ER-localized M48 homologs in plants and non-photosynthetic eukaryotes cleave isoprenylated C-termini (ending with the residues CAA[X]) of a range of substrates and the C-termini of such substrates enter the M48 active site through a region close to the membrane surface (for references see1). However, based on several previous studies (see references in 1.), it is unlikely that proteins undergo isoprenylation in the chloroplast, and PES1 lacks also a CAAX terminus (instead it ends with ‘THVPSFEP’). It remains therefore to be determined if and how PGM48 and PES1 are functionally related and if they interact to each other in vivo.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Research on chloroplast peptidases in the van Wijk laboratory is supported by a grant from the National Science Foundation (NSF), Division of Molecular and Cellular Biosciences (MCB) #1614629.
References
- 1.Bhuiyan NH, Friso G, Rowland E, Majsec K, van Wijk KJ. The plastoglobule-localized metallopeptidase PGM48 is a positive regulator of senescence in Arabidopsis thaliana. Plant Cell 2016; 28:3020-37; PMID:27895226; https://doi.org/ 10.1105/tpc.16.00745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lundquist P, Poliakov A, Bhuiyan NH, Zybailov B, Sun Q, van Wijk KJ. The functional network of the Arabidopsis thaliana plastoglobule proteome based on quantitative proteomics and genome-wide co-expression analysis. Plant Physiol 2012; 58:1172-92; PMID:22274653; https://doi.org/ 10.1104/pp.111.193144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bracha K, Lavy M, Yalovsky S. The Arabidopsis AtSTE24 Is a CAAX protease with broad substrate specificity. J Biol Chem 2002; 277:29856-64; PMID:12039957; https://doi.org/ 10.1074/jbc.M202916200 [DOI] [PubMed] [Google Scholar]
- 4.Bohovych I, Fernandez MR, Rahn JJ, Stackley KD, Bestman JE, Anandhan A, Franco R, Claypool SM, Lewis RE, Chan SS, et al.. Metalloprotease OMA1 fine-tunes mitochondrial bioenergetic function and respiratory supercomplex stability. Scient Rep 2015; 5:13989; PMID:26365306; https://doi.org/ 10.1038/srep13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korwitz A, Merkwirth C, Richter-Dennerlein R, Troder SE, Sprenger HG, Quiros PM, López-Otín C, Rugarli EI, Langer T. Loss of OMA1 delays neurodegeneration by preventing stress-induced OPA1 processing in mitochondria. J Cell Biol 2016; 212:157-66; PMID:26783299; https://doi.org/ 10.1083/jcb.201507022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Wijk KJ, Kessler F. Plastoglobuli: plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Ann Rev Plant Biol 2017; 68:253-289; PMID:28125283; https://doi.org/ 10.1146/annurev-arplant-043015-111737 [DOI] [PubMed] [Google Scholar]
- 7.Tevini M, Steinmuller D. Composition and function of plastoglobuli II. lipid-composition of leaves and plastoglobuli during beech leaf senescence. Planta 1985; 163:91-6; PMID:24249273; https://doi.org/ 10.1007/BF00395902 [DOI] [PubMed] [Google Scholar]
- 8.Rottet S, Devillers J, Glauser G, Douet V, Besagni C, Kessler F. Identification of plastoglobules as a site of carotenoid cleavage. Front Plant Sci 2016; 7:1855; PMID:28018391; https://doi.org/ 10.3389/fpls.2016.01855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lippold F, vom Dorp K, Abraham M, Hölzl G, Wewer V, Yilmaz JL, Lager I, Montandon C, Besagni C, Kessler F, et al.. Fatty Acid Phytyl ester synthesis in chloroplasts of arabidopsis. Plant Cell 2012; 24:2001-14; PMID:22623494; https://doi.org/ 10.1105/tpc.112.095588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lohscheider JN, Friso G, van Wijk KJ. Phosphorylation of plastoglobular proteins in Arabidopsis thaliana. J Exp Bot submitted 2016; 67(13):3975-84; PMID:26962209; https://doi.org/ 10.1093/jxb/erw091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang FC, Molnar P, Schwab W. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J Exp Botany 2009; 60:3011-22; PMID:19436048; https://doi.org/ 10.1093/jxb/erp137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laloi C, Havaux M. Key players of singlet oxygen-induced cell death in plants. Front Plant Sci 2015; 6:39; PMID:25699067; https://doi.org/ 10.3389/fpls.2015.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Havaux M. Carotenoid oxidation products as stress signals in plants. Plant J 2014; 79:597-606; PMID:24267746; https://doi.org/ 10.1111/tpj.12386 [DOI] [PubMed] [Google Scholar]
- 14.Avendano-Vazquez AO, Cordoba E, Llamas E, San Roman C, Nisar N, De la Torre S, Ramos-Vega M, Gutiérrez-Nava MD, Cazzonelli CI, Pogson BJ, et al.. An uncharacterized apocarotenoid-derived signal generated in zeta-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in arabidopsis. Plant Cell 2014; 26:2524-37; PMID:24907342; https://doi.org/ 10.1105/tpc.114.123349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruno M, Koschmieder J, Wuest F, Schaub P, Fehling-Kaschek M, Timmer J, Beyer P, Al-Babili S. Enzymatic study on AtCCD4 and AtCCD7 and their potential to form acyclic regulatory metabolites. J Exp Botany 2016; 67:5993-6005; PMID:27811075; https://doi.org/ 10.1093/jxb/erw356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Jorge S, Ha SH, Magallanes-Lundback M, Gilliland LU, Zhou A, Lipka AE, Nguyen YN, Angelovici R, Lin H, Cepela J, et al.. CAROTENOID CLEAVAGE DIOXYGENASE4 is a negative regulator of beta-carotene content in arabidopsis seeds. Plant Cell 2013; 25:4812-26; PMID:24368792; https://doi.org/ 10.1105/tpc.113.119677 [DOI] [PMC free article] [PubMed] [Google Scholar]


