Abstract
OBJECTIVE
We tested the associations between genetic background and selected environmental exposures with respect to islet autoantibodies and type 1 diabetes.
RESEARCH DESIGN AND METHODS
Infants with HLA-DR high-risk genotypes were prospectively followed for diabetes-related autoantibodies. Single nucleotide polymorphisms (SNPs) came from the Illumina ImmunoChip and environmental exposure data were by parental report. Children were followed to age 6 years.
RESULTS
Insulin autoantibodies occurred earlier than GAD antibody (GADA) and then declined, while GADA incidence rose and remained constant (significant in HLA-DR4 but not in the DR3/3 children). The presence of SNPs rs2476601 (PTPN22) and rs2292239 (ERBB3) demonstrated increased risk of both autoantibodies to insulin (IAA) only and GADA only. SNP rs689 (INS) was protective of IAA only, but not of GADA only. The rs3757247 (BACH2) SNP demonstrated increased risk of GADA only. Male sex, father or sibling as the diabetic proband, introduction of probiotics under 28 days of age, and weight at age 12 months were associated with IAA only, but only father as the diabetic proband and weight at age 12 months were associated with GADA only. Mother as the diabetic proband was not a significant risk factor.
CONCLUSIONS
These results show clear differences in the initiation of autoimmunity according to genetic factors and environmental exposures that give rise to IAA or GADA as the first appearing indication of autoimmunity.
Introduction
We previously published that diabetes-related persistent confirmed islet autoantibodies first appear singly, with autoantibodies to insulin (IAA) appearing at an earlier age than GAD autoantibodies (GADA) (1). The order of appearance was shown to be related to HLA-DR-DQ genotypes. The observed difference in the incidence of GADA and IAA as first-appearing autoantibodies was quite dramatic: it was consistent with the generally held view that IAA appears first (2–4). Yet the appearance of IAA first would identify only a fraction of the very young who developed diabetes-related autoimmunity, as the incidence dropped precipitously through the first 6 years of life, despite the observation that autoantibodies may be detected as early as 3 months of age. In addition, the appearance of a second or third autoantibody after GADA as the first was slower than after the appearance of IAA as the first. These distinct phenotypes may point to different genetic risk factors and prenatal or perinatal exposures and different etiological pathways for islet autoimmunity (IA) and progression to type 1 diabetes compared with the appearance of autoantibodies occurring later in life. This article explores those associations in the TEDDY (The Environmental Determinants of Diabetes in the Young) population, followed up to 6 years of age. The factors considered are taken from the body of previously published results from the entire TEDDY study cohort (5–7). We test the hypothesis that the genetic and environmental etiology differs between either IAA or GADA appearing as the first autoantibody (IAA only or GADA only).
Research Design and Methods
Participants
TEDDY is a prospective cohort study funded by the National Institutes of Health with the primary goal to identify environmental causes of type 1 diabetes. It includes six clinical research centers: three in the U.S. (Colorado, Georgia/Florida, and Washington) and three in Europe (Finland, Germany, and Sweden). Detailed study design and methods have previously been published (8,9). Written informed consents were obtained for all study participants from a parent or primary caretaker, separately, for genetic screening and participation in the prospective follow-up beginning at birth. The high-risk genotypes for participants screened from the general population (GP) were as follows: DR3-DQ2.5/4-DQ8, DR4/4, DR4/8, and DR3/3. An additional six genotypes were included for first-degree relatives (FDRs) to a subject with type 1 diabetes: DR4/4, DR4/1, DR4/13, DR4/4, DR4/9, and DR3/9. The study was approved by local institutional review or ethics boards at each site (University of Washington, Seattle; University of Colorado; Medical College of Georgia, Augusta; University of South Florida, Tampa; University of Turku, Finland; Technische Universität, Munich, Germany; Lund University, Malmö, Sweden) and is monitored by an External Evaluation Committee formed by the National Institutes of Health.
Non-HLA Genotyping
As previously described (1), when the child was 9–12 months of age (n = 7,463), the HLA-DR-DQ genotypes were confirmed by reverse blot hybridization at the central HLA Reference Laboratory at Roche Molecular Systems, Oakland, CA (10), along with the INS-23Hph1 (rs689), CTLA4 T17A (rs231775), and PTPN22 R620W (rs2476601) single nucleotide polymorphism (SNP) primer pairs. Quality control included the removal of those with a low call rate (>5% SNPs missing), those with a Hardy-Weinberg equilibrium P value <10−6, those who were monomorphic, or those with an insertion-deletion. Further SNP analysis was performed by the Center for Public Health Genomics at the University of Virginia using the Illumina ImmunoChip, which is a custom array for genotyping of SNPs selected from regions of the human genome firmly associated with autoimmune diseases (11). The final selection of SNPs, including 186,000 SNPs in 186 regions for 12 autoimmune diseases, was decided by the ImmunoChip Consortium. As part of the development of the chip quality control, 634 biallelic SNPs were assayed in duplicate at those SNPs, resulting in a genotype discordance rate of 0.0012% (12). TEDDY also examined whether any non-HLA SNPs previously shown to be associated with type 1 diabetes in a genome-wide association study (GWAS) and meta-analysis conferred risk for IA (6). Six additional SNPs were identified and have been included in this report: rs2816316 in RGS1, rs10517086 in a region on chromosome 4p15.2 without any known genes, rs2292239 in ERBB3, rs3184504 in SH2B3, rs4948088 in COBL, and rs12708716 in CLEC16A. One of the GWAS SNPs, rs11755527 (BACH2), not available on the ImmunoChip, was replaced with SNP rs3757247 (BACH2), which was in high linkage disequilibrium (R2 = 0.94). For each SNP above, the minor allele frequency was determined in the study population, and based on previous findings, a dominant model with risk of outcomes was chosen (6).
Islet Autoantibodies
IAA, GADA, or antibodies to insulinoma antigen-2 (IA-2A) were measured in two laboratories by radiobinding assays. In the U.S., all sera were assayed at the Barbara Davis Center for Childhood Diabetes at the University of Colorado Denver; in Europe, all sera were assayed at the University of Bristol in the U.K. Both laboratories show high sensitivity and specificity as well as concordance (13). All positive islet autoantibodies and 5% of negative samples were retested in the other reference laboratory and deemed confirmed if concordant. The laboratories were 93%, 98%, and 92% concordant for GADA, IA-2A, and IAA, respectively. Persistent IA was defined as confirmed positive IAA, GADA (specifically, autoantibodies to GAD65), or IA-2A on at least two consecutive visits.
Autoantibody findings attributed to maternal IgG transmission were excluded in defining the child’s autoantibody status (1). While important to distinguish true child autoantibodies from maternal autoantibodies, this approach is limited in that the child’s positivity during the first 18 months of life could be masked by maternal autoantibodies.
Environmental Exposures and Phenotypes
Our previous publication showed that introduction to probiotics before the age 28 days was associated with lower risk of islet autoantibodies (5). Probiotic exposure was defined as timing of first introduction to probiotics from either dietary supplementation or infant formula that was monitored from birth using questionnaire and diaries. Weight after the first year of life was associated with increased risk of IA. Standardized weight z scores were derived using the Centers for Disease Control and Prevention standardized growth charts and SAS programs as previously described (7). Early infectious-related conditions are also of interest in this study and were recorded at the 3-month clinic visit. The common reported infectious conditions were rash, diarrhea, and respiratory tract–related illnesses. Upper-respiratory infection for the infant was defined as having a cold or runny nose in addition to having either an ear infection or eye discharge. A lower-respiratory infection was defined as having either pneumonia, difficulty breathing, or respiratory problems.
Statistical Methods
The age-specific incidence of IA was described as a rate of per 1,000/person-years. CIs (95% CI) were calculated using the χ2 relationship to the Poisson distribution (14). Differences in the cumulative incidence of type 1 diabetes between groups were examined in Kaplan-Meier analyses. Multivariate proportional hazards (PH) models evaluated sex, family history, HLA, SNPs, country of residence, and environmental exposures for their independent association with risk of all-cause IA (first appearance of any autoantibody), cause-specific IA (IAA only, GADA only, or more than one of IAA, GADA, or IA-2A [termed “multiple IA”] at seroconversion), and type 1 diabetes. All predictors were categorical and considered time invariant except for weight at 12 months, which was continuous (z score) and treated as time varying up to 12 months and fixed thereafter. Children negative for IA were right censored after the date of the last negative sample or the 72-month blood draw—whichever came first. In the cause-specific PH models, children who seroconverted for a competing IA event that was not of interest were censored after the date of seroconversion. Children negative for type 1 diabetes were censored after the last visit or 72-month visit. The strength of the associations was described by hazard ratios (HRs) and 95% CIs. P values <0.05 were considered significant. For further testing of whether risk factors for GADA only and IAA only differed, a multiple logistic regression examined whether the factors significantly influenced the IAA only–to–GADA only ratio. All factors included in the PH models were also included in the logistic model in addition to age of seroconversion. Environmental exposures showing a significant independent association with outcome were tested for effect modification by other factors by inclusion of interaction terms in the models. All analyses were preplanned, and multivariate analysis was performed where possible. Therefore, no correction for multiple comparisons was necessary.
HLA-DR genotypes were generally used as abbreviations for the full HLA- DR-DQ genotypes used in the study, and SNPs were described by the presence of the minor allele. SAS 9.4 (SAS Institute, Cary, NC) was used for the statistical analyses and GraphPad PRISM 5.03 (GraphPad Software, San Diego, CA) for graphs.
Results
Included in this analysis were 8,503 of the 8,676 children enrolled in TEDDY (118 who did not have the TEDDY high-risk HLA genotypes and 55 with either no sample results or indeterminate autoantibody status were excluded), all of whom had been followed to age 6 years, had developed IA before age 6 years, or had been lost to the study. As of 30 April 2016, 589 children (6.9%) had persistent confirmed autoantibodies during 36,964 person-years of follow-up to 6 years of age (15.9/1,000 person-years). Of the 589, 42.8% (252) had IAA only as the first appearing IA, 38.4% (226) had GADA only, 13.8% (81) had GADA and IAA, 1.5% (9) had IA-2A only, and 3.5% (21) had other combinations at initial presentation (Table 1).
Table 1.
Characteristics of HLA-eligible TEDDY children followed to age 6 years
| IA negative (N = 7,914) | Any IA (N = 589) | IAA only (N = 252) | GADA only (N = 226) | IA-2A only (N = 9) | Multiple IA (N = 102) | Type 1 diabetes (N = 172) | |
|---|---|---|---|---|---|---|---|
| Sex | |||||||
| Female | 3,928 (49.6) | 264 (44.8) | 108 (42.9) | 105 (46.5) | 2 (22.2) | 49 (48.0) | 85 (49.4) |
| Male | 3,986 (50.4) | 325 (55.2) | 144 (57.1) | 121 (53.5) | 7 (77.8) | 53 (52.0) | 87 (50.6) |
| Family history | |||||||
| GP | 7,112 (89.9) | 465 (79.0) | 196 (77.8) | 183 (81.0) | 7 (77.8) | 79 (77.5) | 118 (68.6) |
| FDR mother | 307 (3.9) | 31 (5.3) | 10 (4.0) | 12 (5.3) | 1 (11.1) | 8 (7.8) | 12 (7.0) |
| FDR father | 384 (4.9) | 65 (11.0) | 29 (11.5) | 25 (11.1) | 0 (0) | 11 (10.8) | 31 (18.0) |
| FDR sibling | 111 (1.4) | 28 (4.8) | 17 (6.8) | 6 (2.7) | 1 (11.1) | 4 (3.9) | 11 (6.4) |
| HLA | |||||||
| DR3/3 | 1,706 (21.6) | 76 (12.9) | 17 (6.8) | 54 (23.9) | 1 (11.1) | 4 (3.9) | 18 (10.5) |
| DR3/X | 22 (0.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0.6) |
| DR3/4 | 3,019 (38.2) | 298 (50.6) | 125 (49.6) | 111 (49.1) | 3 (33.3) | 59 (57.8) | 97 (56.4) |
| DR4/X | 1,609 (20.3) | 112 (19.0) | 66 (26.2) | 27 (12.0) | 2 (22.2) | 17 (16.7) | 34 (19.8) |
| DR4/4 | 1,558 (19.7) | 103 (17.5) | 44 (17.5) | 34 (15.0) | 3 (33.3) | 22 (21.6) | 22 (12.8) |
| Country | |||||||
| U.S. | 3,428 (43.3) | 203 (34.5) | 73 (29.0) | 98 (43.4) | 2 (22.2) | 30 (29.4) | 49 (28.5) |
| Finland | 1,658 (21.0) | 144 (24.5) | 78 (31.0) | 40 (17.7) | 2 (22.2) | 24 (23.5) | 54 (31.4) |
| Germany | 525 (6.6) | 48 (8.2) | 20 (7.9) | 10 (4.4) | 1 (11.1) | 17 (16.7) | 25 (14.5) |
| Sweden | 2,303 (29.1) | 194 (32.9) | 81 (32.1) | 78 (34.5) | 4 (44.4) | 31 (30.4) | 44 (25.6) |
| Started probiotics | |||||||
| No or age >365 days | 6,290 (78.5) | 453 (76.9) | 191 (75.8) | 182 (80.5) | 7 (77.8) | 73 (71.6) | 124 (72.1) |
| Yes, age 28–365 days | 1,077 (13.6) | 100 (17.0) | 46 (18.3) | 33 (14.6) | 1 (1.1) | 20 (19.6) | 33 (19.2) |
| Yes, age <28 days | 547 (6.9) | 36 (6.1) | 15 (6.0) | 11 (4.9) | 1 (11.1) | 9 (8.8) | 15 (8.7) |
| Maternal IA | |||||||
| No | 7,702 (97.3) | 568 (96.4) | 243 (96.4) | 220 (97.4) | 8 (88.9) | 97 (95.1) | 162 (94.2) |
| Yes | 212 (2.7) | 21 (3.6) | 9 (3.6) | 6 (2.7) | 1 (11.1) | 5 (4.9) | 10 (5.8) |
| Weight at 12 months, mean z score (SD) | −0.13 (1.02) | 0.00 (1.05) | −0.06 (1.05) | 0.11 (1.09) | 0.03 (1.08) | −0.11 (0.96) | −0.06 (1.05) |
| Child conditions before first clinical visit (age 3 months) | |||||||
| Upper resp. (yes) | 1,857 (23.5) | 144 (24.5) | 68 (27.0) | 47 (20.8) | 3 (33.3) | 26 (26.0) | 50 (29.2) |
| Lower resp. (yes) | 970 (12.3) | 66 (11.2) | 27 (10.7) | 31 (13.9) | 2 (22.2) | 6 (5.9) | 16 (9.4) |
| Diarrhea (yes) | 740 (9.4) | 52 (8.9) | 26 (10.4) | 16 (7.1) | 0 (0) | 10 (9.8) | 14 (8.1) |
| Rash (yes) | 1,804 (22.8) | 126 (21.4) | 55 (21.8) | 42 (18.7) | 2 (22.2) | 27 (26.5) | 27 (15.7) |
| rs689 (INS) | |||||||
| No | 3,721 (53.7) | 381 (65.6) | 182 (73.4) | 120 (53.6) | 8 (88.9) | 71 (71.0) | 116 (72.1) |
| Yes | 3,206 (46.3) | 200 (34.4) | 66 (26.6) | 104 (46.4) | 1 (11.1) | 29 (29.0) | 45 (28.0) |
| rs231775 (CTLA4) | |||||||
| No | 2,176 (31.4) | 174 (30.0) | 80 (32.3) | 56 (25.0) | 3 (33.3) | 35 (35.0) | 46 (28.6) |
| Yes | 4,751 (68.6) | 407 (70.1) | 168 (67.7) | 168 (75.0) | 6 (66.7) | 65 (65.0) | 115 (71.4) |
| rs2476601 (PTPN22) | |||||||
| No | 5,550 (80.1) | 401 (69.1) | 168 (67.7) | 159 (71.3) | 5 (55.6) | 69 (69.0) | 103 (63.9) |
| Yes | 1,376 (19.9) | 179 (30.9) | 80 (32.3) | 64 (28.7) | 4 (44.4) | 31 (31.0) | 58 (26.0) |
| rs2816316 (RGS1) | |||||||
| No | 4,271 (66.9) | 360 (66.5) | 160 (68.7) | 140 (67.3) | 5 (62.5) | 55 (59.8) | 107 (69.9) |
| Yes | 2,110 (33.1) | 181 (33.5) | 73 (31.3) | 68 (32.7) | 3 (37.5) | 37 (40.2) | 46 (30.1) |
| rs10517086 | |||||||
| No | 3,281 (51.4) | 259 (47.9) | 111 (47.6) | 106 (51.0) | 3 (37.5) | 39 (42.4) | 58 (37.9) |
| Yes | 3,100 (48.6) | 282 (52.1) | 122 (52.4) | 102 (49.0) | 5 (62.5) | 53 (57.6) | 95 (62.1) |
| rs4948088 (COBL) | |||||||
| No | 5,801 (90.9) | 508 (93.9) | 221 (94.9) | 192 (92.3) | 8 (100) | 87 (94.6) | 144 (94.1) |
| Yes | 580 (9.1) | 33 (6.1) | 12 (5.1) | 16 (7.7) | 0 (0) | 5 (5.4) | 9 (5.9) |
| rs2292239 (ERBB3) | |||||||
| No | 2,964 (46.5) | 200 (37.0) | 87 (37.3) | 71 (34.1) | 5 (62.5) | 37 (40.2) | 50 (32.7) |
| Yes | 3,417 (53.6) | 341 (63.0) | 146 (62.7) | 137 (65.9) | 3 (37.5) | 55 (59.8) | 103 (67.3) |
| rs3184504 (SH2B3) | |||||||
| No | 2,022 (31.7) | 129 (23.8) | 62 (26.6) | 41 (19.7) | 1 (12.5) | 25 (27.2) | 40 (26.1) |
| Yes | 4,359 (68.3) | 412 (76.2) | 171 (73.4) | 167 (80.3) | 7 (87.5) | 67 (72.8) | 113 (73.9) |
| rs12708716 (CLEC16A) | |||||||
| No | 2,788 (43.8) | 263 (48.6) | 105 (45.1) | 104 (50.0) | 4 (50.0) | 50 (54.4) | 74 (48.4) |
| Yes | 3,577 (56.2) | 278 (51.4) | 128 (54.9) | 104 (50.0) | 4 (50.0) | 42 (45.7) | 79 (51.6) |
Data are N (%) unless otherwise indicated. Lower resp., lower-respiratory infection; Upper resp., upper-respiratory infection.
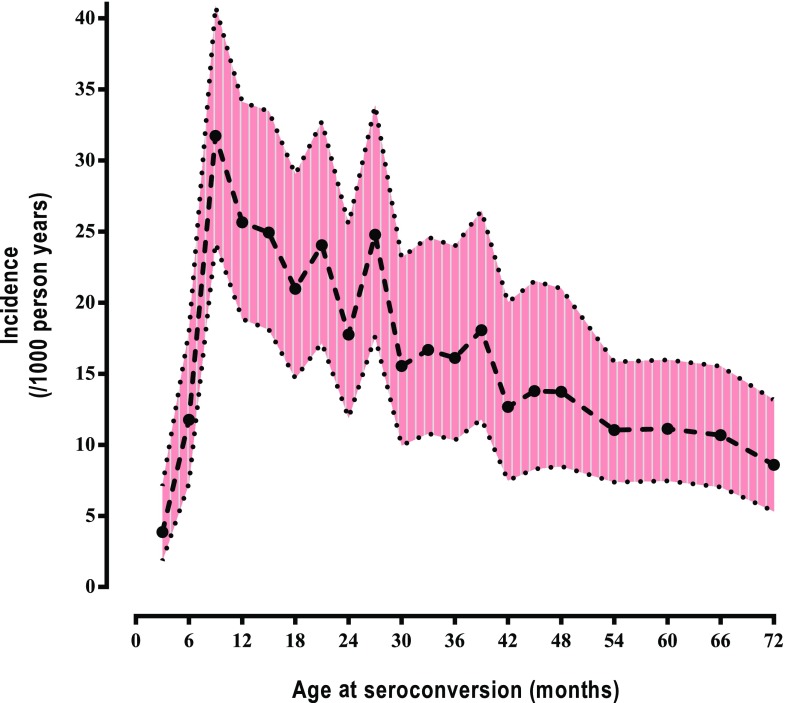
The incidence of islet autoantibodies rose sharply until 9 months of age and declined slowly thereafter (Fig. 1). The median age of seroconversion was 27.0 months (interquartile range [IQR] 14.9–43.2).
Figure 1.
Incidence of antibodies among 0- to 6-year-old children in TEDDY by age of seroconversion (incidence and 95% piecewise confidence bands). Autoantibodies appeared in 589 of 8,503 children.
By age 6 years, 172 children (2.0%) were diagnosed with type 1 diabetes. Median age of diagnosis was 38.5 months (IQR 24.0–56.1). Prior to diagnosis, 156 of 172 of the children had developed IA, with median age of seroconversion of 14.9 months (IQR 10.1–22.2). Of these 156, 49.4% (77) had IAA only as the first appearing IA, 19.2% (30) had GADA only, 23.1% (36) had GADA and IAA, 0.6% (1) had IA-2A only, and 7.7% (12) had other combinations. Also, 82.1% (128 of 156) of children with IA and type 1 diabetes had seroconverted before the median age of seroconversion (27 months).
Factors previously shown to be associated with IA (5–7) were reassessed in the population, limited to those under age 6 years and examined in relation to type 1 diabetes, in a multivariate analysis that adjusted for country of birth (Table 2). Factors associated with a significantly higher risk of IA included male sex (HR 1.32 [95% CI 1.11, 1.56], P = 0.002), having a father or sibling with type 1 diabetes (HR 2.34 [95% CI 1.77, 3.09], P < 0.0001, and HR 3.38 [95% CI 2.28, 5.01], P < 0.0001, respectively) but not a mother with type 1 diabetes (HR 1.24 [95% CI 0.84, 1.84], P = 0.27) compared with the GP, HLA DR3/4 but not 3/3,X (HR 1.44 [95% CI 1.19, 1.73], P = 0.0001, and HR 0.66 [95% CI 0.50, 0.87], P = 0.003, respectively) compared with DR 4/4,X, and weight at 12 months (HR 1.21 [95% CI 1.11, 1.32], P < 0.0001). Probiotic usage at age <28 days of age was associated with significantly lower IA risk (HR 0.66 [95% CI 0.46, 0.96], P = 0.03). None of the categories of infectious conditions occurring before 3 months of age were significantly associated with IA. SNP associations with IA were consistent with previous reports with the exception of the SNPs CTLA4, RGS1, and rs10517086 (P = 0.51, 0.70, and 0.43, respectively).
Table 2.
HRs from a multivariate proportional hazards model of TEDDY, published risk factors for IA and type 1 diabetes by age 6 years, and child infectious-related conditions before first clinical visit
| Factor | Comparison | Any IA |
Type 1 diabetes |
||
|---|---|---|---|---|---|
| HR | P | HR | P | ||
| Sex | Male vs. female | 1.32 | 0.002 | 1.07 | 0.69 |
| Family history | FDR mother vs. GP | 1.24 | 0.27 | 1.70 | 0.11 |
| FDR father vs. GP | 2.34 | <0.0001 | 3.71 | <0.001 | |
| FDR sibling vs. GP | 3.38 | <0.0001 | 5.32 | <0.0001 | |
| HLA | DR3/4 vs. DR4/4,X | 1.44 | 0.0001 | 1.76 | 0.002 |
| DR3/3,X vs. DR4/4,X | 0.66 | 0.003 | 0.63 | 0.11 | |
| Country | Finland vs. U.S. | 1.23 | 0.09 | 1.78 | 0.01 |
| Germany vs. U.S. | 1.27 | 0.19 | 1.97 | 0.02 | |
| Sweden vs. U.S. | 1.08 | 0.48 | 0.99 | 0.96 | |
| Probiotics at age 28 days | Yes vs. no or ≥28 days | 0.66 | 0.03 | 0.86 | 0.61 |
| Weight at age 12 months | z score | 1.21 | <0.0001 | 1.17 | 0.06 |
| Child conditions before first clinical visit (age 3 months) | Upper resp. (yes vs. no) | 1.13 | 0.24 | 1.49 | 0.03 |
| Lower resp. (yes vs. no) | 0.93 | 0.62 | 0.81 | 0.47 | |
| Diarrhea (yes vs. no) | 1.14 | 0.40 | 1.10 | 0.73 | |
| Rash (yes vs. no) | 0.95 | 0.64 | 0.64 | 0.06 | |
| rs689 (INS) | Yes vs. no | 0.67 | <0.001 | 0.55 | 0.0006 |
| rs231775 (CTLA4) | Yes vs. no | 1.07 | 0.51 | 1.05 | 0.78 |
| rs2476601 (PTPN22) | Yes vs. no | 1.73 | <0.0001 | 1.91 | 0.0002 |
| rs2816316 (RGS1) | Yes vs. no | 1.04 | 0.70 | 0.92 | 0.64 |
| rs10517086 | Yes vs. no | 1.07 | 0.43 | 1.53 | 0.01 |
| rs4948088 (COBL) | Yes vs. no | 0.67 | 0.03 | 0.75 | 0.41 |
| rs2292239 (ERBB3) | Yes vs. no | 1.45 | <0.0001 | 1.68 | 0.003 |
| rs3184504 (SH2B3) | Yes vs. no | 1.40 | 0.001 | 1.18 | 0.39 |
| rs12708716 (CLEC16A) | Yes vs. no | 0.82 | 0.02 | 0.87 | 0.38 |
Data set in boldface indicate statistical significance. Lower resp., lower-respiratory infection; Upper resp., upper-respiratory infection.
The significance of the associations with IA in the first 6 years of life did not always extend to type 1 diabetes over the same age period. Sex (HR 1.07 [95% CI 0.77, 1.47], P = 0.69), HLA-DR3/3,X vs. DR4/4,X (HR 0.63 [95% CI 0.35, 1.12], P = 0.11), early exposure to probiotics (HR 0.86 [95% CI 0.49, 1.53], P = 0.61), and SNPs SH2B3 (HR 1.18 [95% CI 0.82, 1.69], P = 0.39), COBL (HR 0.75 [95% CI 0.38, 1.49], P = 0.41), and CLEC16A (HR 0.87 [95% CI 0.63, 1.20], P = 0.38) were not associated with type 1 diabetes.
Early upper-respiratory infections (HR 1.49 [95% CI 1.04, 2.14], P = 0.03) and SNP rs10517086 (HR 1.53 [95% CI 1.10, 2.13], P = 0.01) were associated with increased risk of type 1 diabetes, although no significant association with IA was found. To understand why the significance of the risk factors for IA and type 1 diabetes differed, we next examined how each of the factors related to the type of first-appearing IA.
IAA-Only Versus GADA-Only Autoimmunity
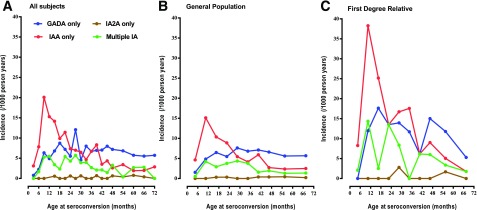
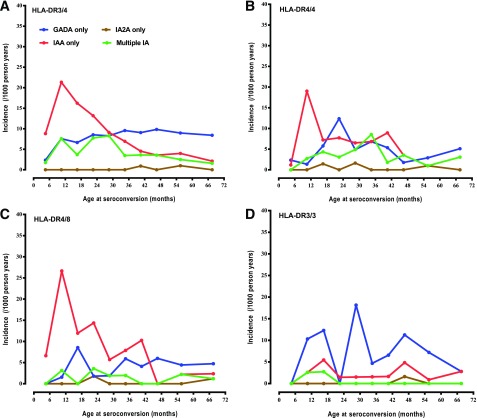
The incidence of IAA-only as first-appearing IA was greatest within the first year of life and declined over the following 5 years. In contrast, the incidence of first-appearing GADA only rose until the second year of life and remained relatively constant through 6 years of age in both FDRs and children from the GP (Fig. 2). The incidence of simultaneous IAA and GADA as first-appearing IA was much less discernable and seemed to occur before 3 years of age. The delay in the incidence of GADA only, relative to IAA only, was significant in all HLA-DR4 (P < 0.001) but not DR3/3 (P = 0.72) children, as previously described (1) (Fig. 3).
Figure 2.
Incidence of IAA only, GADA only, and IA-2A only as first-appearing IA and multiple IA at seroconversion in all participants (A), the general population (B), and FDR of a proband with type 1 diabetes (C). A: IAA only appeared in 252 of 8,503 children, GADA only in 226 of 8,503, and IA-2A only in 9 of 8,503 and multiple IA in 102 of 8,503. B: IAA only appeared in 196 of 7,577 children, GADA only in 183 of 7,577, and IA-2A only in 7 of 7,577 and multiple IA in 79 of 7,577. C: IAA only appeared in 56 of 926 children, GADA only in 43 of 926, and IA-2A only in 2 of 926 and multiple IA in 23 of 926.
Figure 3.
Incidence of IAA only, GADA only, and IA-2A only as first-appearing IA and multiple IA at seroconversion by HLA genotype DR3/4 (A), DR4/4 (B), DR4/8 (C), and DR3/3 (D). A: IAA only appeared in 125 of 3,317 children, GADA only in 111 of 3,317, IA-2A only in 3 of 3,317, and multiple IA in 59 of 3,317. B: IAA only appeared in 44 of 1,661 children, GADA only in 34 of 1,661, IA-2A only in 3 of 1,661, and multiple IA in 22 of 1,661. C: IAA only appeared in 53 of 1,470 children, GADA only in 24 of 1,470, IA-2A only in 2 of 1,470, and multiple IA in 9 of 1,470. D: IAA only appeared in 17 of 1,782 children, GADA only in 54 of 1,782, IA-2A only in 1 of 1,782, and multiple IA in 4 of 1,782.
The ratio of children developing IAA only compared with GADA only was strongly influenced by HLA-DR genotypes (P < 0.001), INS (P < 0.0001), and, to a lesser degree, CTLA4 (P = 0.03). HLA-DR4 was associated predominately with IAA only and HLA-DR3 with GADA only (Table 3) (Fig. 3). The SNPs PTPN22 (HR 1.77 [95% CI 1.34, 2.33], P < 0.0001, and HR 1.63 [95% CI 1.20, 2.22], P = 0.002, respectively) and ERBB3 (HR 1.42 [95% CI 1.08, 1.85], P = 0.01, and HR 1.60 [95% CI 1.20, 2.15], P = 0.002) were associated with increased risk of both IAA only and GADA only. Also, the INS was protective of IAA only (HR 0.47 [95% CI 0.35, 0.63], P < 0.0001) but not of GADA only (HR 1.06 [95% CI 0.80, 1.40], P = 0.68). The BACH2 SNP was discovered to be significantly associated with increased risk of GADA only (HR 1.49 [95% CI 1.08, 2.06], P = 0.01) but tended to lower the risk of IAA only (HR 0.78 [95% CI 0.60, 1.02], P = 0.07) and, thus, significantly modified the IAA only–to–GADA only ratio (P = 0.002). Male sex (HR 1.36 [95% CI 1.05, 1.77], P = 0.02), having a father or sibling as the FDR with diabetes (HR 2.33 [95% CI 1.53, 3.55], P < 0.0001, and HR 5.32 [95% CI 3.20, 8.83], P < 0.0001, respectively), introduction of probiotics at age <28 days (HR 0.54 [95% CI 0.31, 0.95], P = 0.03), and increased weight at 12 months of age (HR 1.16 [95% CI 1.02, 1.33], P = 0.02) were all significantly associated with the incidence of IAA only. However, only having a father as the FDR with diabetes (HR 2.49 [95% CI 1.59, 3.89], P < 0.001) and weight at 12 months of age (HR 1.29 [95% CI 1.13, 1.48], P = 0.0003) were significantly associated with GADA only. Having a mother as the FDR with diabetes was not a significant risk factor for either.
Table 3.
Cause-specific HRs from a multivariate proportional hazards model for first-appearing IAA only and GADA only and multiple IA at seroconversion by age 6 years
| Factor | Comparison | IAA only |
GADA only |
Multiple IA |
|||
|---|---|---|---|---|---|---|---|
| HR | P | HR | P | HR | P | ||
| Sex | Male vs. female | 1.36 | 0.02 | 1.25 | 0.11 | 1.15 | 0.51 |
| Family history | FDR mother vs. GP | 0.87 | 0.66 | 1.56 | 0.15 | 1.42 | 0.32 |
| FDR father vs. GP | 2.33 | <0.0001 | 2.49 | <0.0001 | 2.16 | 0.02 | |
| FDR sibling vs. GP | 5.32 | <0.0001 | 1.60 | 0.33 | 2.73 | 0.05 | |
| HLA | DR3/4 vs. DR4/4,X | 1.18* | 0.23 | 1.92* | 0.0001 | 1.52 | 0.06 |
| DR3/3,X vs. DR4/4,X | 0.27* | <0.0001 | 1.60* | 0.02 | 0.23 | 0.005 | |
| Country | Finland vs. U.S. | 1.93* | 0.0003 | 0.73* | 0.14 | 1.31 | 0.37 |
| Germany vs. U.S. | 1.70* | 0.05 | 0.53* | 0.09 | 2.59 | 0.008 | |
| Sweden vs. U.S. | 1.32 | 0.12 | 0.91 | 0.57 | 1.03 | 0.91 | |
| Probiotics at age 28 days | Yes vs. no or ≥28 days | 0.54 | 0.03 | 0.64 | 0.21 | 0.89 | 0.75 |
| Weight at age 12 months | z score | 1.16 | 0.02 | 1.29 | 0.0003 | 1.13 | 0.26 |
| Child conditions before first clinical visit (age 3 months) | Upper resp. (yes vs. no) | 1.23 | 0.18 | 0.94 | 0.70 | 1.25 | 0.36 |
| Lower resp. (yes vs. no) | 0.97 | 0.87 | 1.10 | 0.64 | 0.44 | 0.07 | |
| Diarrhea (yes vs. no) | 1.36 | 0.16 | 0.94 | 0.82 | 1.20 | 0.60 | |
| Rash (yes vs. no) | 1.01 | 0.94 | 0.83 | 0.31 | 1.10 | 0.71 | |
| rs689 (INS) | Yes vs. no | 0.47* | <0.0001 | 1.06* | 0.68 | 0.56 | 0.01 |
| rs231775 (CTLA4) | Yes vs. no | 0.88* | 0.38 | 1.33* | 0.07 | 1.00 | 0.99 |
| rs2476601 (PTPN22) | Yes vs. no | 1.77 | <0.0001 | 1.63 | 0.002 | 1.70 | 0.02 |
| rs2816316 (RGS1) | Yes vs. no | 0.97 | 0.83 | 0.99 | 0.92 | 1.40 | 0.12 |
| rs10517086 | Yes vs. no | 1.05 | 0.70 | 0.98 | 0.88 | 1.31 | 0.21 |
| rs4948088 (COBL) | Yes vs. no | 0.56 | 0.05 | 0.86 | 0.56 | 0.61 | 0.29 |
| rs2292239 (ERBB3) | Yes vs. no | 1.42 | 0.01 | 1.60 | 0.002 | 1.30 | 0.23 |
| rs3184504 (SH2B3) | Yes vs. no | 1.25 | 0.15 | 1.76 | 0.001 | 1.12 | 0.64 |
| rs12708716 (CLEC16A) | Yes vs. no | 0.96 | 0.78 | 0.76 | 0.06 | 0.67 | 0.06 |
| rs3757247 (BACH2) | Yes vs. no | 0.78* | 0.07 | 1.49* | 0.01 | 1.10 | 0.68 |
Data set in boldface indicate statistical significance. Lower resp., lower-respiratory infection; Upper resp., upper-respiratory infection.
*IAA only–to–GADA only ratio significantly changed (P < 0.05) by factor as determined in multivariate logistic regression.
Type 1 Diabetes
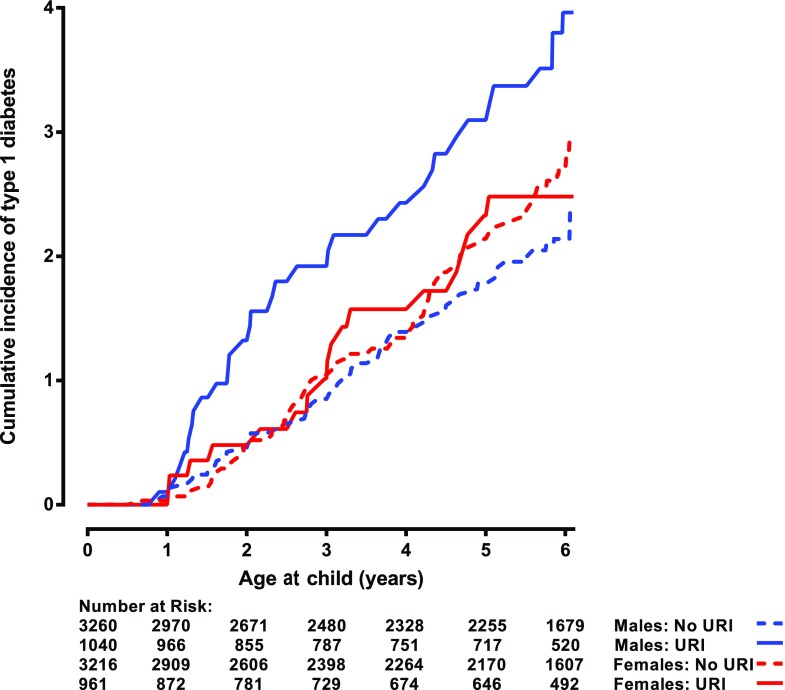
Finally, we looked for effects of interactions between genetic and nongenetic factors on risk of type 1 diabetes and similar interactions on IAA only and GADA only. The association between upper-respiratory infection by 3 months of age and the development of type 1 diabetes by 6 years of age (Table 2) was significantly modified by the child’s sex (test of interaction P = 0.02) (Fig. 4). Among males, an upper-respiratory infection was significantly associated with increased incidence of type 1 diabetes (HR 2.24 [95% CI 1.38, 3.63], P = 0.001); however, among females no association was observed (HR 0.88 [95% CI 0.49, 1.57], P = 0.67). No significant interaction was observed with IAA only, GADA only, or multiple IA. Nevertheless, it was noted that male sex (HR 1.65 [95% CI 1.18, 2.33], P = 0.004) and upper-respiratory infections (HR 1.45 [95% CI 1.01, 2.09], P = 0.05) were both significantly associated with IAA only among younger children (<3 years), an age when the effect of interaction between these two factors on risk of type 1 diabetes was primarily seen (Fig. 4). Weight at 12 months interacted with INS for risk of type 1 diabetes (P = 0.03), as it increased risk of type 1 diabetes (HR 1.48 [95% CI 1.10, 2.00], P = 0.01) among children with INS, but no association was seen among children who did not have INS (HR 1.06 [95% CI 0.87, 1.28], P = 0.59). The same interaction was also associated with IAA only among the children younger than 2 years old (P < 0.001). Similarly, weight at 12 months was associated with higher IAA only (HR 1.72 [95% CI 1.23, 2.40], P = 0.001) among children with INS, but no association was observed among children who did not have INS (HR 0.90 [95% CI 0.74, 1.09], P = 0.26).
Figure 4.
Cumulative incidence of type 1 diabetes among 0- to 6-year-old children in TEDDY by males reporting upper-respiratory infection (URI) before 3 months of age, males not reporting upper-respiratory infection before 3 months of age, females reporting upper-respiratory infection before 3 months of age, and females not reporting upper-respiratory infection before 3 months of age.
Conclusions
The strong association between HLA and seroconversion to a specific islet autoantibody found in TEDDY children up to 6 years of age argues that the association between HLA and type 1 diabetes at the time of clinical diagnosis is secondary to a primary association between HLA and an autoimmune response to either IAA only or GADA only.
The differences seen between HLA-DR genotypes and between the effects of INS and CTLA4 gene polymorphisms suggest specific interactions or immunogenic pathways, e.g., the total absence of DR3/3 and a higher proportion of DR3/4 among those who seroconverted at 3 or 6 months compared with those who seroconverted at 9 months of age or after. Although genes in the HLA region remain the most important genetic risk factors for type 1 diabetes, other non-HLA genetic factors also contribute to seroconversion to IAA, GADA, or both.
TEDDY has carefully collected possible exposures that may be important to IA including prenatal and perinatal events, the introduction of first foods, breastfeeding practices, and early childhood illness; changes in the gut microbiome; and gene expression in blood cells, as well as vaccinations and immunizations. The advantage of delineating different subgroups from the larger TEDDY cohort is that the subgroups may be more homogeneous with respect to gene-environment interactions, which may facilitate the discovery of triggers from a relatively age-restricted set of possibilities. The present TEDDY data suggest for the first time that triggers of IAA only may differ from those of GADA only as a first-appearing autoantibody. It cannot be excluded that the mechanisms by which a trigger is inducing either one of the two islet autoantibodies is associated with HLA-DR or DQ haplotypes or both. The findings are consistent in both FDRs and the general population. Further studies are needed to analyze the second-, third-, and fourth-appearing islet autoantibody and whether it is associated with progression to clinical onset of type 1 diabetes.
The contribution that the SNPs make to the appearance of a first-appearing islet autoantibody, the appearance of a second autoantibody, and the progression to clinical onset of type 1 diabetes differs depending on the end point. The type 1 diabetes genome consortium made an effort to identify non-HLA genetic polymorphisms that would affect the risk for type 1 diabetes. No subjects were studied prior to the clinical diagnosis of type 1 diabetes. The current study makes it possible to investigate this relationship separately. The relative incidence of IAA alone (peaking early and declining) compared with GAD alone (peaking later and plateauing) accounts for some of the associations described. Age at autoantibody development is an important predictor of type 1 diabetes, and the limited follow-up focuses the results of this article on events occurring early in life. Hence, the associations or lack thereof may be different than reported by others owing to the young age and limited follow-up.
We would hypothesize that the relationship seen between autoantibody appearance (and, thus, HLA and autoantibody incidence) would also be reflected in the occurrence and timing of cytokines and other markers of inflammation (15). The association of autoantibodies and cytokines reflecting the innate or adaptive immunity would be apparent in an analysis of the difference in the timing of the first appearance of autoantibodies and would partially explain the lack of association seen in individuals at type 1 diabetes diagnosis (16), reflecting the power of a longitudinal analysis over a cross-sectional analysis. However, the role of the β-cell, autoantibody secretion, and autoantigen presentation leading to enhanced T-cell response, all of which contribute to cytokine secretion, is complex and still to be elucidated (rev. in 17).
Conclusion
These results expand our previous findings (1) that showed clear differences in the initiation of autoimmunity by HLA to include non-HLA genetic factors and environmental exposures occurring early in life. While the associations could be modified by extended follow-up, the strength of these results is that they provide evidence of immunological changes relatively soon after environmental exposures, suggesting a possible causative effect. In particular, they show that the environmental effects may vary according to genotype and that there are significant gene-environment interactions that modify IA risk. A caveat to these findings is that TEDDY includes only high-risk genotypes. The lack of finding a significant association with IA or type 1 diabetes risk with the mother of a TEDDY child as the FDR with diabetes and the associated finding of increased type 1 diabetes (but not IA) risk associated with upper-respiratory infections occurring before 3 months of age requires further analysis. Preliminary analysis (Fig. 4) suggests an interaction between sex and upper-respiratory infections occurring before age 3 months. The systems biology approach of TEDDY is to combine these results with observations of the changes in the microbiome (for dietary and dietary supplement factors), gene expression, proteomics, and metabolomics to establish a plausible pathway by which autoimmunity arises.
Supplementary Material
Article Information
Acknowledgments. The authors give a special acknowledgment to the TEDDY families for their continued participation in this wonderful study.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. All authors attest to meeting the International Committee of Medical Journal Editors uniform requirements for authorship by making substantial contributions to the conception and design of the manuscript; acquisition, analysis, and interpretation of data; drafting or revising the article for intellectual content; and giving final approval of the published version. J.P.K. designed the study, proposed the analysis, interpreted the findings, and wrote the manuscript. K.F.L. performed the analysis and drafted and revised the manuscript. Å.L., W.A.H., M.J.R., J.-X.S., J.T., A.-G.Z., and B.A. designed the study and reviewed and edited the manuscript. J.P.K. and K.F.L. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Funding. This study was funded by grants U01-DK-63829, U01-DK-63861, U01-DK-63821, U01-DK-63865, U01-DK-63863, U01-DK-63836, U01-DK-63790, UC4-DK-63829, UC4-DK-63861, UC4-DK-63821, UC4-DK-63865, UC4-DK-63863, UC4-DK-63836, and UC4-DK-95300 and contract no. HHSN267200700014C from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Allergy and Infectious Diseases, National Institute of Child Health and Human Development, National Institute of Environmental Health Sciences, JDRF, and Centers for Disease Control and Prevention. This work was also supported in part by National Institutes of Health/National Center for Advancing Translational Sciences Clinical and Translational Science Awards to the University of Florida (UL1 TR000064) and the University of Colorado (UL1 TR001082).
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc17-0238/-/DC1.
A complete list of the TEDDY Study Group can be found in the Supplementary Data online.
References
- 1.Krischer JP, Lynch KF, Schatz DA, et al.; TEDDY Study Group . The 6 year incidence of diabetes-associated autoantibodies in genetically at-risk children: the TEDDY study. Diabetologia 2015;58:980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ziegler AG, Bonifacio E; BABYDIAB-BABYDIET Study Group . Age-related islet autoantibody incidence in offspring of patients with type 1 diabetes. Diabetologia 2012;55:1937–1943 [DOI] [PubMed] [Google Scholar]
- 3.Ilonen J, Hammais A, Laine AP, et al. . Patterns of β-cell autoantibody appearance and genetic associations during the first years of life. Diabetes 2013;62:3636–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ziegler AG, Rewers M, Simell O, et al. . Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 2013;309:2473–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uusitalo U, Liu X, Yang J, et al.; TEDDY Study Group . Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr 2016;170:20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Törn C, Hadley D, Lee HS, et al.; TEDDY Study Group . Role of type 1 diabetes-associated SNPs on risk of autoantibody positivity in the TEDDY study. Diabetes 2015;64:1818–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elding Larsson H, Vehik K, Haller MJ, et al.; TEDDY Study Group . Growth and risk for islet autoimmunity and progression to type 1 diabetes in early childhood: The Environmental Determinants of Diabetes in the Young Study. Diabetes 2016;65:1988–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.TEDDY Study Group The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatr Diabetes 2007;8:286–298 [DOI] [PubMed] [Google Scholar]
- 9.TEDDY Study Group The Environmental Determinants of Diabetes in the Young (TEDDY) study. Ann N Y Acad Sci 2008;1150:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagopian WA, Erlich H, Lernmark A, et al.; TEDDY Study Group . The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatr Diabetes 2011;12:733–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet 2013;14:661–673 [DOI] [PubMed] [Google Scholar]
- 12.Trynka G, Hunt KA, Bockett NA, et al.; Spanish Consortium on the Genetics of Coeliac Disease (CEGEC); PreventCD Study Group; Wellcome Trust Case Control Consortium (WTCCC) . Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet 2011;43:1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonifacio E, Yu L, Williams AK, et al. . Harmonization of glutamic acid decarboxylase and islet antigen-2 autoantibody assays for National Institute of Diabetes and Digestive and Kidney Diseases consortia. J Clin Endocrinol Metab 2010;95:3360–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulm K. A simple method to calculate the confidence interval of a standardized mortality ratio (SMR). Am J Epidemiol 1990;131:373–375 [DOI] [PubMed] [Google Scholar]
- 15.Bakay M, Pandey R, Hakonarson H. Genes involved in type 1 diabetes: an update. Genes (Basel) 2013;4:499–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanifi-Moghaddam P, Schloot NC, Kappler S, Seissler J, Kolb H. An association of autoantibody status and serum cytokine levels in type 1 diabetes. Diabetes 2003;52:1137–1142 [DOI] [PubMed] [Google Scholar]
- 17.Hampe CS. B cell in autoimmune diseases. Scientifica (Cairo) 2012;2012:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.