Abstract
The Atacama Desert is the most extreme non-polar biome on Earth, the core region of which is considered to represent the dry limit for life and to be an analogue for Martian soils. This study focused on actinobacteria because they are keystone species in terrestrial ecosystems and are acknowledged as an unrivalled source of bioactive compounds. Metagenomic analyses of hyper-arid and extreme hyper-arid soils in this desert revealed a remarkable degree of actinobacterial ‘dark matter’, evidenced by a detected increase of 34% in families against those that are validly published. Rank-abundance analyses indicated that these soils were high-diversity habitats and that the great majority of designated ‘rare’ genera (up to 60% of all phylotypes) were always rare. These studies have enabled a core actinobacterial microbiome common to both habitats to be defined. The great majority of detected taxa have not been recovered by culture dependent methods, neither, with very few exceptions, has their functional ecology been explored. A microbial seed bank of this magnitude has significance not just for Atacama soil ecosystem resilience but represents an enormous untapped resource for biotechnology discovery programmes in an era where resistance to existing antibiotics is rapidly becoming a major threat to global health.
Introduction
Research on extremophilic and extremotrophic microorganisms has developed rapidly since the mid-1970s as microbiologists recognized that extreme environments were capable of sustaining life. Subsequent delineation of the extremobiosphere has shown it to encompass all of the physico-chemical variables on Earth and that many microorganisms have evolved tolerances to multiple extreme conditions. Our research in this context has shown that members of the phylum Actinobacteria 1 are found throughout the extremobiosphere2, and is based on the rationale of size and diversity of the taxon, its geographic dispersal, and its ecological and biotechnological relevance. We have focused on two extreme environments, the deep-sea3 and more recently hyper-arid desert. Over 22% of the terrestrial environment comprises desert of which a third is classified as hyper-arid (ratio of mean annual rainfall to mean annual evaporation (MAR) less than 0.05) or extreme hyper-arid (ratio < 0.002)4.
The Atacama Desert located in northern Chile is the most ancient and continuously driest non-polar environment on Earth; apart from its aridity it is unique in its range of habitats, its geology and geochemistry, its elevation and topography, and its radiation intensities5. Previous work by our group has enabled cultivable actinobacteria to be recovered from a range of Atacama soils, regoliths and rock surfaces6, 7, including ones purporting to represent “the dry limit of microbial life”8, and from which have been isolated several new species and natural products possessing sought for bioactivities9, 10 and whole genome sequences determined11, 12. However, although there are clear indications from us and other groups that actinobacteria are widespread and frequently dominate prokaryotic communities in the Atacama Desert (for example, 75% and 98% in low13 and high14 elevation soils), this has not been supported by culture-based studies. Consequently the low diversity of actinobacteria observed using selective cultivation techniques6 has provided impetus for the present investigation. As Garza and Dutilh15 have emphasized metagenomics now is enabling comprehensive surveys of microbial communities in diverse environments that culture-based approaches have not achieved and as high throughput molecular techniques become cheaper and more effective they are being adopted as default tools for such investigations16. Roche 454 and Illumina technology platforms have been widely adopted for metagenomic studies, informative comparisons of which have been made on natural microbial communities17, 18. Roche GS-FLX sequencing systems have started to be applied to microbial community analyses in the Atacama Desert and have revealed a dominance of actinobacteria in hyper-arid margin soils13, and bacterial colonization patterns along longitudinal moisture gradients19. As a first step in such an approach specifically to actinobacterial diversity in the Atacama Desert we have carried out community profiling based on 454 pyrosequencing. Our choice of this particular platform in part was based on its high classification efficiency17, and evidence that deep pyrosequencing of 16S tags is well-suited for distinguishing site specific similarities and differences among rare bacteria20.
The principle objectives of this study were to determine how diverse actinobacterial communities are in the Atacama Desert and how big a resource this presents for biotechnology exploitation, and to dissect such diversity in terms of actinobacterial rare and previously undetected dark matter21, 22. To this end, we have applied R H Whittaker’s classic diversity indices23 to actinobacterial diversity of a hyper-arid—extreme hyper-arid landscape at five distinct habitats covering an altitude range of approximately 900–2500 m absl. Based on trial experiments analyses were restricted to generic and family ranks and we have not attempted correlations of local diversity (sensu α-diversity24) with environmental factors. In addition, we hypothesized that this desert landscape would be propitious for exploring the rare biosphere concept, and the notion of actinobacterial signatures defining the habitats. We opine that the results obtained reinforce the view that rather than being devoid of life the Atacama Desert is a microbial treasure trove in the search for the next generation of bioactive natural products.
The purpose of this paper is to report the remarkably high diversity of actinobacteria in the Atacama Desert and highlight its exceptional significance for biodiscovery campaigns. Metagenome sequencing has revealed a strong correlation between global geographic distance and biome type and the biosynthetic diversity residing in soils25, a conclusion that strongly resonates with our decision to explore the unique and poorly studied microbiomes of the Atacama Desert.
Results
Over 90,000 sequence reads were obtained from the 12 sites (Fig. 1) representing 67 actinobacterial families, 16% of which could not be assigned to validly published taxa and therefore, are regarded as putatively novel candidate families (see Table S1). The large majority of identified families belonged to the class Actinobacteria 26 but in addition a few representatives of rare deep lineage actinobacteria belonging to the classes Acidimicrobia 27 and Nitriliruptoria 28 were detected. The total detected diversity at the generic rank numbered 297 only 60% of which were assigned to validly published taxa.
Figure 1.
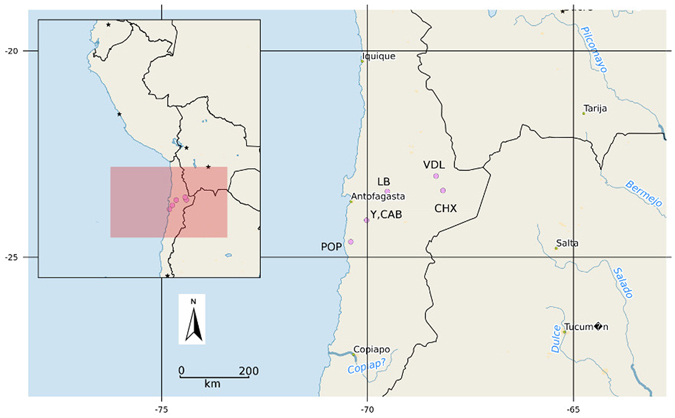
Location of Atacama Desert research in northern Chile. Site codes: Yungay (Y); Cerros Aguas Blancas (CAB); Lomas Bayas (LB); Cerro Paranal (POP); Salar de Atacama (CHX); Cordillera de la Sal (VDL). Map created by RAS from OpenStreetmap cartography (licensed under CC BY-SA http://www.openstreetmap.org/copyright © OpenStreetMap contributors) using QGIS version 2.14 (Open Source Geospatial Foundation http://qgis.org/en/site).
Pyrosequencing data for microbiota diversities in other world deserts show significant variations in the occurrence of actinobacteria. Thus, sandy Asian deserts such as the Gobi, Gurbantünggütt and Taklamaken are reported to have small (2–18%) actinobacterial communities29, 30. In contrast, soils of certain western deserts such as the Atacama and Sonoran are heavily dominated by actinobacterial phylospecies (75–88%13, 19). Culture-dependent values obtained for the sites examined in the present study are 50–70% (hyper-arid) and up to 100% in certain extreme hyper-arid samples (Valle de la Luna6). Some acuity should be exercised in reviewing such measures of dominance, or otherwise, as actinobacterial counts are influenced significantly by the selective procedures used for their isolation6. The taxonomic megadiversity of actinobacteria in the Atacama Desert landscape as revealed for the first time in this study argues strongly for their successful adaptation to extreme resource-depleted conditions.
Rarefaction analysis
Construction of rarefaction curves enabled us to compare genus richness and gauge the extent to which total diversity had been recovered at each of the Atacama sites (Fig. 2). In most cases the major extent of the actinobacterial community diversities had been captured by our sequencing campaign at the 1500-sequence level but in one case (CAB3) the sequence level of 1054 obtained fell well below this value and likely compromised subsequent analyses of this site. Most of the curves closely approached asymptotes; a few others remained curvilinear indicating that further actinobacterial diversity remained to be sequenced. Thus it is important to recognize that taxon counts are strictly comparable only when richness is asymptotic, a situation pertaining to our observations. The efficiencies of sequence capture in a few of the sites examined (Yungay Y6.1, Y6.2, Y6.3; Chaxa Laguna CHX1, CHX2) were not influenced by sampling depth.
Figure 2.
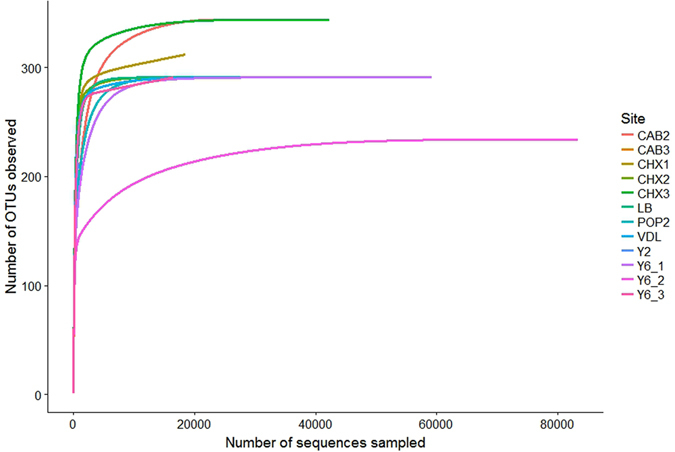
Rarefaction analysis of the extreme hyper-arid and hyper-arid OTUs at 94.5–97% phylogenetic similarity.
Application of Good’s library coverage index indicated that genotype recovery reached 90% to 97% at all sites. Table 1 presents total OTUs (total observed validly published genera, plus unidentified genera) and α-diversity indices. Only a small decrease (ca. 7% average) was found in the observed OTUs recovered from surface soil samples as a function of increased aridity.
Table 1.
Observed OTUs and α-diversity indices.
| Sampling site and depth (cm) | Observed OTUs (Genera) | Chao1 Richness | Shannon Index (H) | Simpson Diversity (1/D) |
|---|---|---|---|---|
| 1. Extreme hyper-arid locations 1 | ||||
| Yungay | ||||
| Y6_1 (2) | 186 | 316 | 4.01 | 19.2 |
| Y6_2 (30) | 87 | 134 | 2.78 | 7.9 |
| Y6_3 (100) | 159 | 222 | 3.87 | 19.6 |
| CAB2 (2) | 175 | 331 | 3.73 | 16.9 |
| CAB3 (2) | 63 | 110 | 3.40 | 20.8 |
| Lomas Bayas | ||||
| LB (1) | 162 | 264 | 3.54 | 10.1 |
| Cordillera de la Sal | ||||
| VDL (2) | 185 | 242 | 4.29 | 27.8 |
| 2. Hyper-arid locations 2 | ||||
| Laguna Chaxa | ||||
| CHX1 (2) | 222 | 306 | 4.48 | 43.5 |
| CHX2 (30) | 208 | 295 | 4.38 | 29.4 |
| CHX33 (2) | 171 | 247 | 3.73 | 14.7 |
| Cerro Paranal | ||||
| POP2 (2) | 157 | 228 | 3.40 | 9.1 |
| Yungay Oasis | ||||
| Y24 (2) | 89 | 137 | 3.21 | 15.4 |
1MAR 0.002, 2MAR 0.009, 3Partly colonized by cyanobacteria, 4Tamarisk grove.
Comparable data for other desert environments are largely lacking but increased salinity was not found to be a major determinant of species richness in saline lakes of the Monegros Desert in NE Spain31. Neither did we observe a major change in overall actinobacterial ecological diversity (H, 1-D) between extreme hyper-arid and hyper-arid locations, a result indicating that they are dominated by relatively few abundant taxa. Chao 1 predicted richness was high at both types of location exceeding the observed OTU numbers by 63% (extreme hyper-arid) and 41% (hyper-arid) and providing further evidence of diversity yet to be sequenced. It should be noted that non-parametric richness estimators such as Chao 1 predict counts of the number of taxa (in this case genera and families) present at a site but cannot be used to compare the genetic diversity between sites32. The reciprocal of the Simpson index (1/D) is sensitive to the degree of dominance in a community33; increases in the index indicate increases in diversity. Values below about 50 have been taken to indicate typical dominance profiles34. With the exception of the CHX1 value which appears to lie near the boundary between dominant and uniform community profiles, all other Atacama Desert communities surveyed here show dominance (see Taxonomic diversity below).
Rank-abundance distributions
For the purpose of defining rank-abundances in Atacama soils we have used a relative cut-off of 0.1%35, 36 below which threshold a rare actinobacterial biosphere can be defined; and an arbitrary threshold of 10% to define abundant taxa36–38. Rank-abundance curves for all of the research sites are shown in Fig. 3.
Figure 3.
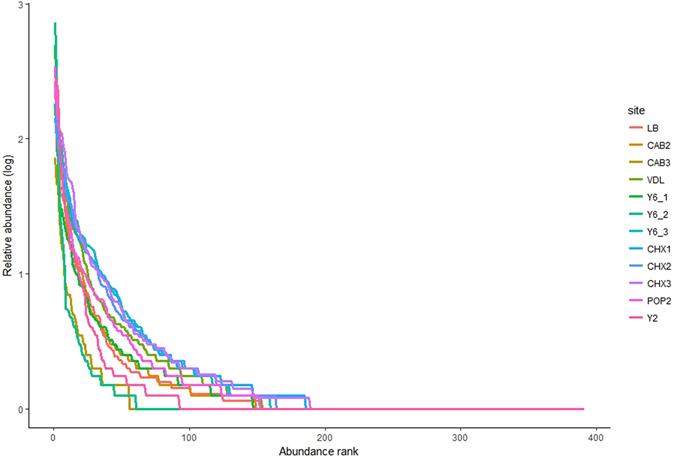
Rank-abundance curves.
Salient points arising from these analyses: (1) the majority of curves are indicative of high-diversity environments, i.e. shallow curve and a long tail represent a rare biosphere; (2) the great majority of rare generic OTUs in the Atacama habitats were always rare (>95%) and were not found as abundant at any of the environments examined; (3) the proportions of abundant and rare actinobacterial taxa were similar at extreme hyper-arid and hyper-arid sites (e.g. 9–17% and 9–15% abundant vs. 46–62% and 48–61% rare genera respectively; (4) on the basis of a small number of comparisons, the composition of the abundant communities in surface, sub-surface and deep samples was similar (e.g., Cerros Aguas Blancas sites at 2, 30 and 100 cm were dominated by Blastococcus, Verrucosispora and unidentified OTUs FJ479147_g, HQ674860_g and HQ910322_g; while Laguna Chaxa sites at 2 and 30 cm were dominated by Arthrobacter, Blastococcus and unidentified OTU HQ910322_g); (5) the taxonomic constancy of abundant OTUs observed as a function of soil depth argues for long term habitat stability and minimal atmospheric aerosol dispersal of Atacama microbiota, a conclusion supported by aerosol optical depth measurements made in our research landscape39; and (6) at genera and family ranks a very much higher number of unassigned OTUs were evident in the rare biosphere confirming that the Atacama landscape represents a vast reservoir of microbial dark matter.
Taxonomic diversity
The profiles of actinobacterial family lineages throughout the landscape locations were similar and dominated by members of the families Acidimicrobiaceae, Geodermatophilaceae, Iamiaceae, Microbacteriaceae, Micrococcaceae, Micromonosporaceae, Nocardiaceae and Nocardioidaceae and two unidentified taxa, FJ479147_f and HQ910322_f. The relative abundance of the top ten families (Fig. 4) shows clearly how community structure was affected by soil depth (Y6 and CHX series) and how at two sites (Y2, CAB3) the communities were dominated by OTUs belonging to the family Micrococcaceae. At this stage we have insufficient data to interpret the shifts in the structures of the latter communities except to note that site Y2 was partially vegetated, and that variations in community structure observed at sites Y6.1, CAB2 and CAB3 reinforce the importance of recognising spatial and temporal factors in habitat sampling. The majority of the most abundant identified families belong to the orders Acidimicrobiales, Geodermatophilales, Micrococcinales and Micromonosporiales 40, 41, actinobacterial taxa notable for their extremophilic and extremotrophic members2. Currently 58 validly published families are recognised in the phylum Actinobacteria (www.bacterio.net at 08/07/2016); consequently the additional 23 taxa recorded as ‘families_f’ constitute a massive element of actinobacterial dark matter in the Atacama landscape.
Figure 4.
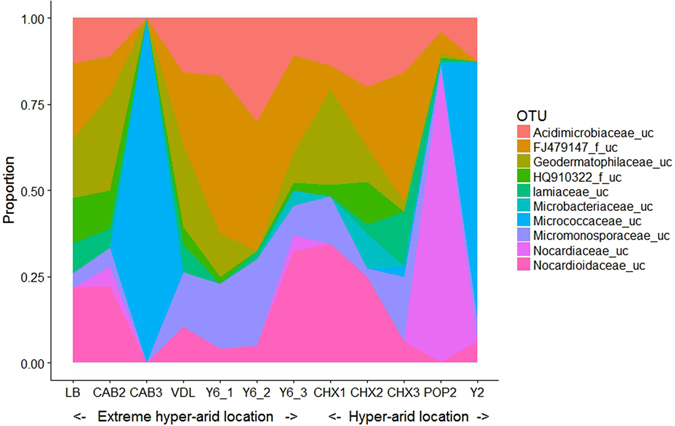
Relative abundance of top 10 most frequently detected actinobacterial families in Atacama Desert locations.
The total number of generic OTUs in each sample is too large to render relative abundance diagrams readily decipherable at the 0.1% cut-off level, consequently data presentation is shown for 2.5% cut-off levels in Fig. 5; the full list of the 297 genera is given in Table S2. The genera dominating the various sites were Arthrobacter, Blastococcus, Geodermatophilus, Gordonia, Microbacterium, Modestobacter and Verrucosispora, and three unidentified putatively novel taxa FJ479147_g, HQ674860_g and HQ910322_g. Although the proportions of these genera varied from site to site, generally they comprised 75% of the community and, predictably, showed high correspondence with family dominance at the same sites. Figure 5 defines the distinctive nature of sites CAB3 and Y2 in which the genus Microbacterium comprised 48% and 58% of these communities respectively. The genera Kocuria, Nocardioides, Sanguibacter and Streptomyces were found to be relatively abundant in both the hyper-arid and extreme hyper-arid core microbiomes (Table 2). The few genera recorded as deep lineage actinobacteria (Aciditerrimonas, Iamia, Ilumatobacter, and Nitriliruptoraceae_uc) were mainly detected at very low read levels at certain sites and can be regarded as constituents of the rare actinobacterial biosphere (see below) in the Atacama landscape. Although Acidimicrobiaceae was found to be among the dominant families, Ilumatobacter was the only member genus detected and in most cases as a constituent of the rare biosphere.
Figure 5.
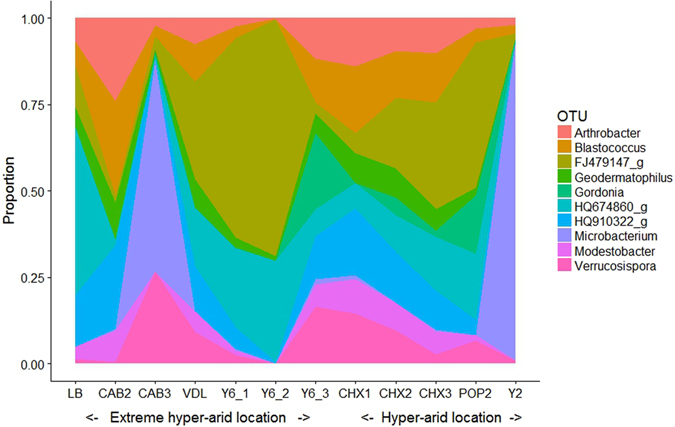
Relative abundance of actinobacterial genera detected in Atacama Desert locations. Relative abundance shown at 2.5% cut off.
Table 2.
Most abundant genera of the extreme hyper-arid and hyper-arid core microbiomes.
| Extreme hyper arid | Hyper arid |
|---|---|
| FJ479147_g | FJ479147_g |
| HQ674860_g | HQ674860_g |
| Blastococcus | Microbacterium |
| HQ910322_g | Kocuria |
| Arthrobacter | Sanguibacter |
| Verrucosispora | Verrucosispora |
| Geodermatophilus | HQ910322_g |
| Modestobacter | Nocardioides |
| Microbacterium | Gordonia |
| Sporichthya | Blastococcus |
| Friedmanniella | Corynebacterium |
| Streptomyces | Arthrobacter |
| Sanguibacter | Streptomyces |
| Kocuria | Aciditerrimonas |
| Nocardioides | FJ478790_g |
The order of genera indicates their comparative dominance within each microbiome.
Habitat specificity and co-occurrence
Venn diagram analyses showed that the proportion of surface community genera shared between sites within extreme hyper-arid (32%) and within hyper-arid (37%) locations was comparable (Fig. 6). However, similar analyses highlighted the effect of vegetation on hyper-arid community composition and of soil depth on extreme hyper-arid shared community composition reducing them to 14% and 22%, respectively.
Figure 6.
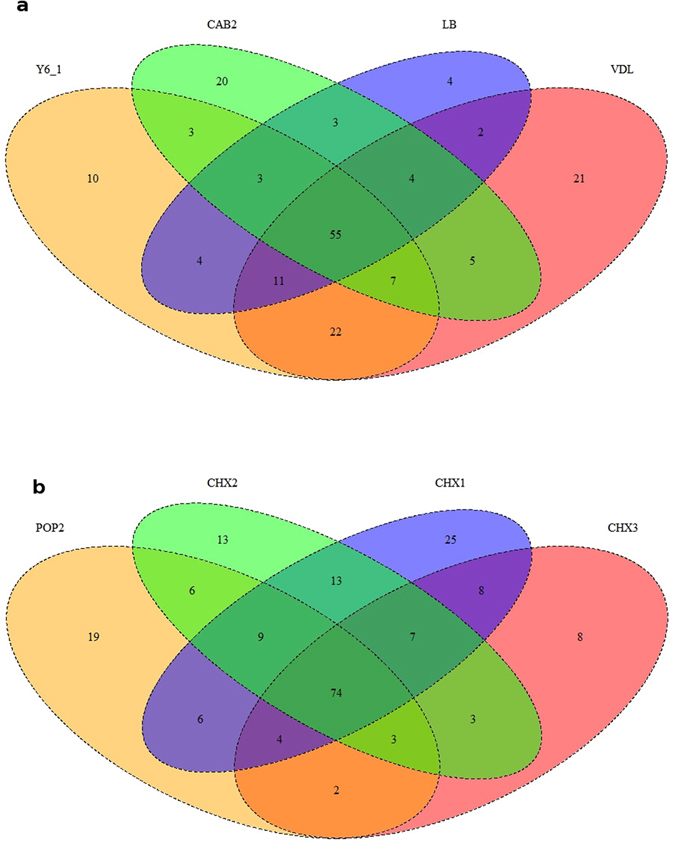
Venn diagrams showing proportions of shared and unique actinobacterial genera at (a) extreme hyper-arid and (b) hyper-arid sites.
Venn diagrams can provide a ‘reasonable first exploration’ of defining the ‘core microbiome’ of a habitat38 and we have adopted this approach to describe organisms shared across the Atacama landscape. Although we have focused solely on the phylum Actinobacteria we opine that it is credible to construct a core microbiome based upon this dominant component of the Atacama microbiota. Figure 6 indicates that 55 and 74 respectively of the OTUs were shared in extreme hyper-arid and hyper-arid surface sites. The most abundant OTUs are almost identical in the two biomes (Table 2) suggesting an actinobacterial signature and postulating that they play a key role in ecosystem function within these Atacama soils. However, the genera Friedmaniella and Sporichthya were abundant in the extreme hyper-arid core microbiome and the genera Aciditerrimonas and Corynebacterium and the putatively novel taxon TJ478790g in the hyper-arid core microbiome (Table 2).
Discussion
The results reported here confirm that actinobacteria constitute a major and frequently dominant component of desert soils. Remarkable, however, is the megadiversity of this phylum throughout hyper-arid and extreme hyper-arid habitats in the world’s driest desert such that our metagenomics data set revealed a 16% greater coverage of actinobacteria at the family rank than that currently recognized. Similar actinobacterial dark matter was evident at the generic level where 40% of the captured diversity was not ascribable to validly published genera. Equally remarkable are the high proportions of unidentified (No-Rank) and very low abundance OTUs, features that are also found in other extreme and poorly researched ecosystems42, 43. Although caution is required in interpreting such metagenomic data44 this study provides a sound base from which to explore an astonishing and unpredicted desert microbiota.
An actionbacterial core microbiome dominated by FJ479147_g, HQ 674860_g, HQ910322_g, Arthrobacter, Blastococcus, Geodermatophilus, Gordonia, Microbacterium and Verrucosispora (a phylogenetic relative of the genus Micromonospora 45) defined the sampling sites. The genera Friedmaniella, Nocardioides, Sanguibacter, Sporichthya and Streptomyces were relatively abundant in the extreme hyper-arid core microbiome and the genera Aciditerramonas and Corynebacterium in the hyper-arid core microbiome. Most of the genera found in these core microbiomes encompassed amycelial actinobacteria classified in the families Geodermatophilaceae (Blastococcus, Geodermatophilus and Modestobacter). Micrococcaceae (Arthrobacter), Microbacteriaceae (Microbacterium), Propionibacteriaceae (Friedmaniella) and Sanguibacteriaceae (Sanguibacter)40. These genera, apart from those classified in the family Geodermatophilaceae, are not usually associated with desert habitats though Arthrobacter strains have been isolated from Yungay soil40 and Friedmaniella antarctica and Sanguibacter antarticus from Antarctic sea sand46, 47.
Exploration of the microbial world has recently been revolutionised by the advent of rapid and inexpensive DNA sequencing technology. In the past assessments of species ‘rarity’ were largely subjective and relied on culture-dependent surveys but now the ability to construct and analyse very large rank-abundance data sets has enabled rare biospheres to be defined with confidence. However, it is important to recognise that truly rare sequences in environmental samples may escape detection due to incomplete sequencing20. The actinobacterial rare biosphere determined within the Atacama landscape is the first to be reported for highly arid environments and accords with the few broader microbial surveys of other deserts (e.g. Sonoran Desert48). A notable feature of the Atacama rare biosphere is the high frequency of taxonomically unassigned OTUs confirming that this landscape contains a vast reservoir of actinobacterial dark matter. Although the functional ecology of the Atacama’s actinobacteria remains to be researched it provides an immense microbial seed bank whose role in soil ecosystem resilience warrants investigation. This hypothesis is given credence by recent reports that members of rare bacterial taxa became dominant in spring sediment microcosm experiments following exposure to environmental stressors49 and are resuscitated in a variety of ecosystems following rewetting of dry soils50.
The results of this study commend several future research imperatives among which we highlight the following: (1) develop innovative isolation/cultivation techniques to enable whole organism physiology of desert microorganisms to be determined; (2) evaluate the viability and metabolic activity of desert microbiomes using, for example, microcosms19, 49, DNA stable isotope probing which to date has not been utilized in desert ecology, and ribosomal tag pyrosequencing51; (3) apply functional genomics to explore ecological traits in desert microorganisms; the genome of Modestobacter caceresii recently isolated from the extreme hyper-arid Yungay region of the Atacama Desert contains 110 genes that are associated with stress responses including those involved in heat and cold shock responses, osmotic stress, and carbon starvation12; and (4) develop the biotechnological potential of this huge reservoir of desert “biosynthetic dark matter”52 access to which has become greatly facilitated by genome and metagenome mining as evidenced by the discovery of new antibacterial/anticancer ansamycins53, synthesized by a novel strain, Streptomyces leeuwenhoekii, isolated from a hyper-arid Atacama Desert soil7.
Methods
Sample collection and research landscape
Soil samples (Table 3) were collected from selected locations in the Atacama Desert landscape (Fig. 1) between 2010 and 2011 (ATB, MG), and additional ones in 2004 (ATB), and 2012 (Professor Luis Cáceres, University of Antofagasta). Samples were collected aseptically using implements sterilised in the field with ethanol and contained in sterile polycarbonate bottles. Sampling was made between 11:00 and 16:00 h during which period temperature and relative humidity at the lower elevation sites ranged between 34°–38 °C and 5–20% respectively, and 30°–33 °C and 3–18% respectively at the higher altitudes. Following transport to the UK all samples were stored at 4 °C.
Table 3.
Sampling locations.
| Research location | Sampling site and code | Collection date | Latitude (°S) | Longitude (°W) | Altitude | Sample description |
|---|---|---|---|---|---|---|
| Yungay | (1) Tamarisk oasis | |||||
| Y2 | 11.11.2010 | 24°04′50.5″ | 69°55′08.3″ | 918 | Degraded tamarisk leaves and surface nitrate soil | |
| (2) Cerros Aguas Blancas | ||||||
| Y6_1 | 13.11.2010 | 24°06′18.4″ | 70°01′15.4″ | 1002 | Extreme hyper-arid site, fine white soil (surface 2 cm) | |
| Y6_2 | 13.11.2010 | 24°06′18.6″ | 70°01′15.6′ | 1002 | Sub-surface (30 cm) | |
| Y6_3 | 13.11.2010 | 24°06′18.6″ | 70°01′15.6′ | 1002 | Sub-surface (100 cm) | |
| CAB2 | 30.10.2011 | 24°05′24.9″ | 69°58′31.9″ | 1060 | Surface (2 cm) | |
| CAB3 | 30.10.2011 | 24°06′40.3″ | 70°02′03.5″ | 1079 | Surface (2 cm) | |
| Lomas Bayas | LB1 | 26.10.2012 | 23°24′27.4″ | 69°31′03.8″ | 1500 | Extreme hyper-arid, surface soil |
| Cerro Paranal | POP2 | 30.10.2011 | 27°75′ | 68°42′ | 1945 | Coarse sandy hyper-arid, surface soil |
| Salar de Atacama | Laguna Chaxa | |||||
| CHX1 | 26.10.2012 | 23°17′33″ | 68°10′99″ | 2219 | Hyper-arid halite (surface 2 cm) | |
| CHX2 | 26.10.2012 | 23°17′33″ | 68°10′99″ | 2219 | Sub-surface (30 cm) | |
| CHX3 | 26.10.2012 | 23°17′36″ | 68°10′83″ | 2222 | Soil colonized by cyanobacteria | |
| Cordillera de la Sal | Valle de la Luna | |||||
| VDL | 05.10.2004 | 23°02′ | 68°20′ | 2450 | Extreme hyper-arid sand | |
The Yungay area is the extreme hyper arid core of the Atacama Desert and is considered to be the closest analogue of Martian soils on Earth54. A large proportion of the area is encrusted with halite and super-rich in nitrates, while certain slopes on the Cerros Aguas Blancas present evidence of water erosion in geological time. Two Yungay sites were sampled: (1) nitrate rich soil from a small tamarisk oasis (Y2); and (2) transect samples from the Cerros Aguas Blancas (CAB series, Y6) WSW of the Cerro Caballo Muerto. The Lomas Bayas region is another extreme hyper-arid environment and a center of copper mining located north east of Antofagasta; samples (LB) were collected at non-mining sites. Sampling was made on the eastern slope of Cerro Paranal adjacent to Route B-710 linking Antofagasta and the coastal village of Paposo. The Salar de Atacama is the largest salt flat in Chile within which the Laguna Chaxa is an area of open water and highly crystalline salt encrusted soils. Samples (CHX) were collected from the halite soils one of which was partially colonized by cyanobacteria. The Valle de la Luna is an extreme hyper-arid area in the Cordillera de la Sal; the sample (VDL) was collected at a sand formation site. Small diversities and low numbers (102–103 cfu g−1 soil) of culturable actinobacteria have been recovered from each of these sampling sites6, 55.
DNA extraction, PCR amplification and pyrosequencing
Total community DNA was extracted from all of the environmental samples using the proprietary UltraClean Soil DNA extraction kit (MoBIO Laboratories, Inc., USA) following the manufacturer’s protocol, and stored at −20 °C. Soil extractions were made in a laminar flow chamber using sterilized equipment in order to avoid contamination. The quality of community DNA preparations was checked by agarose gel electrophoresis and the sizes of the DNA fragments were compared with a molecular size marker (Gene RulerTM, MBI Fermentas, Vilnius, Lithuania).
Actinobacterial specific regions in community DNA preparations were amplified using the primers Com2xf (5′-AAA CTC AAA GGA ATT GAC GG-3′) and Ac1186r (5′-CTT CCT CCG AGT TGA CCC-3′)56. Unique, sample-specific barcodes (10 bp) were attached to the 5′ end of the forward primer. Four replicates were constructed from each of the environmental samples, the latter were assigned individual barcodes.
PCR was made in 25 μl reaction mixtures containing 1 μl DNA template, 1x buffer (10x buffer: 160 mM (NH4)2SO4, 670 mMTris-HCl [pH 8.8 at 25 °C], 0.1% Tween 20), 0.125 mM each of the four dNTPs, 200 µM each of forward and reverse primers, 1.5 µM MgCl2 and 1.25 µM Taq polymerase. Positive and negative controls (Verrucosispora maris DSM 45365T DNA and sterile distilled water, respectively) were incorporated into all PCR runs. The PCR reactions were carried out under the following conditions: initial denaturation at 94 °C for a minute, 30 cycles of 94 °C for a minute, 60 °C for 30 seconds and 72 °C for 30 seconds, and finally 72 °C for 5 minutes. The quality of the amplified products was established using agarose gel electrophoresis. The preparations were kept at −20 °C prior to use. PCR products were purified using the Agencourt AMPure XP PCR Purification System (Beckman Coulter, USA) and MagnaRack (Thermo Fisher Scientific Inc., USA) according to the manufacturers’ protocols. Sample concentrations were quantified using a Qubit® Fluorometer (Invitrogen, CA, USA) and adjusted to give a final concentration of 100 µg/mL; sample quality was checked using an Agilent Bioanalyzer by NewGene Ltd. (Newcastle upon Tyne, UK). Pyrosequencing was made using the Roche GS-FLX+ system (454 Life Sciences, Branford, CT) at the WM Keck Center for Comparative and Functional Genomics, University of Illinois, USA, as described in the manufacturer’s protocol. The library was sequenced at designated regions of a 70 × 75 PicoTiter Plate using flow pattern A with the Roche XL+ sequencing kit (454 Life Sciences) and software version 2.9. Signal processing and base calling were performed using the bundled 454 Data Analysis Software version 2.9 for amplicons.
Bioinformatic analyses
High quality reads were processed using CLCommunity v3.30 software (www.chunlab.com) at ChunLab Inc., Seoul National University, Seoul, Korea). Processing steps involved trimming barcodes, filtering of low quality (<300 bp) and chimera sequences, taxonomic assignment, and statistical calculations using the ChunLab bioinformatics pipeline. The average post-processing sequence length was 283 bp. Taxonomic assignments of the sequence reads were made using the EzTaxon-e database57 (http://www.ezbiocloud.net/eztaxon) and a robust global pairwise sequencing alignment coupled with the BLASTN search tool (CLCommunityTM User Manual, 2015). The similarity ranges used to identify operational taxonomic units (OTUs) were genus: 97-94.5 and family: 94.5-86.5 and taxonomic nomenclature was based on the list of prokaryotic names with standing in nomenclature (http://www.bacterio.net/). Chimera sequences formed during the PCR step were detected and removed from the dataset by UCHIME58. Sequences that could not be related to either validly published genera or families were given an accession number and an underscore suffix (e.g. family FJ479147_f, genus AM991247_g).
Rarefaction and rank-abundance analyses, Shannon diversities and Chao1 richness estimations were calculated using the CLCommunity software. OTU tables were saved in comma delimited (.csv) format prior to further analysis. Identified genera were checked and identified manually from the OTU table and analyzed to determine their composition in each of the environmental samples. Venn diagrams were constructed using the Mothur version 1.37.4 program (http://www.mothur.org/).
Data availability statement
Comma separated value (csv) format data files and R scripts used in this study are available at https://github.com/rasanderson/sci_reports.
Electronic supplementary material
Acknowledgements
The authors are grateful for support from Jongsik Chun, Byung-yong Kim and Eric Kwon (ChunLab), to Roselyn Brown for technical assistance, to Ian Singleton for help with pyrosequencing and Luis Cáceres for provision of soil samples. ATB and JAA thank the Royal Society for International Joint Project Grant JP100654, HI the Malaysian Government for a postgraduate scholarship, and MG the Leverhulme Trust for an Emeritus Fellowship. We thank Martin Embley for constructive comments on the draft paper.
Author Contributions
H.I. conducted the experiments that were conceived by A.T.B., M.G. and H.I.; A.T.B., J.A.A. and M.G. collected environmental samples, R.S. analysed results. The manuscript was written by A.T.B. and reviewed by all authors.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08937-4
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Michael Goodfellow, Email: m.goodfellow@ncl.ac.uk.
Roy Sanderson, Email: roy.sanderson@ncl.ac.uk.
References
- 1.Goodfellow, M.In Bergey’s Manual of Systematic Bacteriology; The Actinobacteria (eds Goodfellow, M. et al.) 33–34 (Springer, 2012).
- 2.Bull, A. T. In Extremophiles Handbook (ed. Koki Horikoshi) 1203–1240 (Springer Japan, 2011).
- 3.Bull AT, Stach JEM, Ward AC, Goodfellow M. Marine actinobacteria: perspectives, challenges, future directions. Antonie van Leeuwenhoek. 2005;87:65–79. doi: 10.1007/s10482-004-6562-8. [DOI] [PubMed] [Google Scholar]
- 4.Houston J. Evaporation in the Atacama Desert: An empirical study of spatio-temporal variations and their causes. J Hydrol. 2006;330:402–412. doi: 10.1016/j.jhydrol.2006.03.036. [DOI] [Google Scholar]
- 5.Bull AT, Asenjo JA, Goodfellow M, Gomez-Silva B. The Atacama Desert: Technical resources and the growing importance of novel microbial diversity. Annu Rev Microbiol. 2016;70:215–234. doi: 10.1146/annurev-micro-102215-095236. [DOI] [PubMed] [Google Scholar]
- 6.Okoro CK, et al. Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek. 2009;95:121–133. doi: 10.1007/s10482-008-9295-2. [DOI] [PubMed] [Google Scholar]
- 7.Busarakam K, et al. Streptomyces leeuwenhoekii sp. nov., the producer of chaxalactins and chaxamycins, forms a distinct branch in Streptomyces gene trees. Antonie van Leeuwenhoek. 2014;105:849–861. doi: 10.1007/s10482-014-0139-y. [DOI] [PubMed] [Google Scholar]
- 8.Navarro-Gonzalez R, et al. Mars-like soils in the Atacama Desert, Chile, and the dry limit of microbial life. Science. 2003;302:1018–1021. doi: 10.1126/science.1089143. [DOI] [PubMed] [Google Scholar]
- 9.Nachtigall J, et al. Atacamycins A-C, 22-membered antitumor macrolactones produced by Streptomyces sp. C38. J Antibiot (Tokyo) 2011;64:775–780. doi: 10.1038/ja.2011.96. [DOI] [PubMed] [Google Scholar]
- 10.Elsayed SS, et al. Chaxapeptin, a lasso peptide from extremotolerant Streptomyces leeuwenhoekii strain C58 from the hyperarid Atacama Desert. J Org Chem. 2015;80:10252–10260. doi: 10.1021/acs.joc.5b01878. [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Escribano JP, et al. The Streptomyces leeuwenhoekii genome: de novo sequencing and assembly in single contigs of the chromosome, circular plasmid pSLE1 and linear plasmid pSLE2. BMC Genomics. 2015;16 doi: 10.1186/s12864-015-1652-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busarakam K, et al. Modestobacter caceresii sp. nov., novel actinobacteria with an insight into their adaptive mechanisms for survival in extreme hyper-arid Atacama Desert soils. Syst Appl Microbiol. 2016;39:243–251. doi: 10.1016/j.syapm.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Neilson JW, et al. Life at the hyperarid margin: novel bacterial diversity in arid soils of the Atacama Desert, Chile. Extremophiles. 2012;16:553–566. doi: 10.1007/s00792-012-0454-z. [DOI] [PubMed] [Google Scholar]
- 14.Lynch, R. C., Darcy, J. L., Kane, N. C., Nemergut, D. R. & Schmidt, S. K. Metagenomic evidence for metabolism of trace atmospheric gases by high-elevation desert Actinobacteria. Front Microbiol5, doi:10.3389/fmicb.2014.00698 (2014). [DOI] [PMC free article] [PubMed]
- 15.Garza DR, Dutilh BE. From cultured to uncultured genome sequences: metagenomics and modeling microbial ecosystems. Cell Mol Life Sci. 2015;72:4287–4308. doi: 10.1007/s00018-015-2004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwager E, Luo C, Huttenhower C, Morgan XC. Sequencing and other tools for studying microbial communities: Genomics and “meta’omic” tools are enabling us to explore the microbiome from three complementary perspectives - taxonomic, functional and ecological. Microbe. 2015;10:419–425. [Google Scholar]
- 17.Claesson, M. J. et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Research38, doi:10.1093/nar/gkq873 (2010). [DOI] [PMC free article] [PubMed]
- 18.Luo C, Tsementzi D, Kyrpides N, Read T, Konstantinidis KT. Direct comparisons of Illumina vs. Roche 454 sequencing technologies on the same microbial community DNA sample. Plos One. 2012;7 doi: 10.1371/journal.pone.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crits-Christoph A, et al. Colonization patterns of soil microbial communities in the Atacama Desert. Microbiome. 2013;1 doi: 10.1186/2049-2618-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowen JL, Morrison HG, Hobbie JE, Sogin ML. Salt marsh sediment diversity: a test of the variability of the rare biosphere among environmental replicates. ISME J. 2012;6:2014–2023. doi: 10.1038/ismej.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skopina MY, Vasileva AA, Pershina EV, Pinevich AV. Diversity at low abundance: The phenomenon of the rare bacterial biosphere. Microbiology. 2016;85:272–282. doi: 10.1134/S0026261716030139. [DOI] [Google Scholar]
- 22.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 23.Whittaker RH. Vegetation of the Siskiyou mountains, Oregon and California. Ecological monographs. 1960;30:279–338. doi: 10.2307/1943563. [DOI] [Google Scholar]
- 24.Whittaker RJ, Willis KJ, Field R. Scale and species richness: towards a general, hierarchical theory of species diversity. J Biogeogr. 2001;28:453–470. doi: 10.1046/j.1365-2699.2001.00563.x. [DOI] [Google Scholar]
- 25.Charlop-Powers Z, et al. Global biogeographic sampling of bacterial secondary metabolism. Elife. 2015;4 doi: 10.7554/eLife.05048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodfellow, M. In Bergey’s Manual of Systematic Bacteriology; The Actinobacteria (eds M. Goodfellow et al.) 34–35 (Springer, 2012).
- 27.Norris, P. R. In Bergey’s Manual of Systematic Bacteriology; The Actinobacteria (eds Goodfellow, M. et al.) 1968 (Springer, 2012).
- 28.Ludwig, W., Euzeby, J. & Whitman, W. in Bergey’s Manual of Systematic Bacteriology; The Actinobacteria (eds Goodfellow, M. et al.) 2001 (Springer, 2012).
- 29.An S, Couteau C, Luo F, Neveu J, DuBow MS. Bacterial diversity of surface sand samples from the Gobi and Taklamaken deserts. Microb Ecol. 2013;66:850–860. doi: 10.1007/s00248-013-0276-2. [DOI] [PubMed] [Google Scholar]
- 30.Li K, Bai Z, Zhang H. Community succession of bacteria and eukaryotes in dune ecosystems of Gurbantünggut Desert, Northwest China. Extremophiles. 2015;19:171–181. doi: 10.1007/s00792-014-0696-z. [DOI] [PubMed] [Google Scholar]
- 31.Casamayor EO, Triado-Margarit X, Castaneda C. Microbial biodiversity in saline shallow lakes of the Monegros Desert, Spain. FEMS Microbiol Ecol. 2013;85:503–518. doi: 10.1111/1574-6941.12139. [DOI] [PubMed] [Google Scholar]
- 32.Stach JE, Bull AT. Estimating and comparing the diversity of marine actinobacteria. Antonie Van Leeuwenhoek. 2005;87:3–9. doi: 10.1007/s10482-004-6524-1. [DOI] [PubMed] [Google Scholar]
- 33.Stach JE, et al. Statistical approaches for estimating actinobacterial diversity in marine sediments. Appl Environ Microbiol. 2003;69:6189–6200. doi: 10.1128/AEM.69.10.6189-6200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, et al. Spatial and resource factors influencing high microbial diversity in soil. Appl Environ Microbiol. 2002;68:326–334. doi: 10.1128/AEM.68.1.326-334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedros-Alio C. The rare bacterial biosphere. Ann Rev Mar Sci. 2012;4:449–466. doi: 10.1146/annurev-marine-120710-100948. [DOI] [PubMed] [Google Scholar]
- 36.Lynch MD, Neufeld JD. Ecology and exploration of the rare biosphere. Nat Rev Microbiol. 2015;13:217–229. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]
- 37.Ugland KI, Gray JS. Lognormal distributions and the concept of community equilibrium. Oikos. 1982;39:171–178. doi: 10.2307/3544482. [DOI] [Google Scholar]
- 38.Shade A, Handelsman J. Beyond the Venn diagram: the hunt for a core microbiome. Environ Microbiol. 2012;14:4–12. doi: 10.1111/j.1462-2920.2011.02585.x. [DOI] [PubMed] [Google Scholar]
- 39.Cordero RR, et al. The Solar Spectrum in the Atacama Desert. Sci Rep. 2016;6 doi: 10.1038/srep22457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodfellow, M. et al. Bergey’s Manual of Systematic Bacteriology; The Actinobacteria, Parts A & B (Springer, 2012).
- 41.Sen A, et al. Phylogeny of the class Actinobacteria revisited in the light of complete genomes. The orders Frankiales and Micrococcales should be split into coherent entities: proposal of Frankiales ord. nov., Geodermatophilales ord. nov., Acidothermales ord. nov and Nakamurellales ord. nov. Int J Syst Evol Micr. 2014;64:3821–3832. doi: 10.1099/ijs.0.063966-0. [DOI] [PubMed] [Google Scholar]
- 42.Yang S, Wen X, Jin H, Wu Q. Pyrosequencing investigation into the bacterial community in permafrost soils along the China-Russia Crude Oil Pipeline (CRCOP) Plos One. 2012;7 doi: 10.1371/journal.pone.0052730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riquelme C, et al. Actinobacterial diversity in volcanic caves and associated geomicrobiological interactions. Front Microbiol. 2014;6:1342–1342. doi: 10.3389/fmicb.2015.01342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hedlund BP, Dodsworth JA, Murugapiran SK, Rinke C, Woyke T. Impact of single-cell genomics and metagenomics on the emerging view of extremophile “microbial dark matter”. Extremophiles. 2014;18:865–875. doi: 10.1007/s00792-014-0664-7. [DOI] [PubMed] [Google Scholar]
- 45.Supong K, et al. Micromonospora sediminicola sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol. 2013;63:570–575. doi: 10.1099/ijs.0.041103-0. [DOI] [PubMed] [Google Scholar]
- 46.Schumann P, Prauser H, Rainey FA, Stackebrandt E, Hirsch P. Friedmanniella antarctica gen. nov., sp. nov., an LL-diaminopimelic acid-containing actinomycete from Antarctic sandstone. Int J Syst Evol Microbiol. 1997;47:278–283. doi: 10.1099/00207713-47-2-278. [DOI] [PubMed] [Google Scholar]
- 47.Hong SG, Lee YK, Yim JH, Chun J, Lee HK. Sanguibacter antarcticus sp. nov., isolated from Antarctic sea sand. Int J Syst Evol Micr. 2008;58:50–52. doi: 10.1099/ijs.0.65031-0. [DOI] [PubMed] [Google Scholar]
- 48.Andrew DR, et al. Abiotic factors shape microbial diversity in Sonoran Desert soils. Appl Environ Microbiol. 2012;78:7527–7537. doi: 10.1128/AEM.01459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coveley S, Elshahed MS, Youssef NH. Response of the rare biosphere to environmental stressors in a highly diverse ecosystem (Zodletone spring, OK, USA) PeerJ. 2015;3 doi: 10.7717/peerj.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aanderud ZT, Jones SE, Fierer N, Lennon JT. Resuscitation of the rare biosphere contributes to pulses of ecosystem activity. Front Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weigold P, Ruecker A, Loesekann-Behrens T, Kappler A, Behrens S. Ribosomal Tag Pyrosequencing of DNA and RNA Reveals “Rare” Taxa with High Protein Synthesis Potential in the Sediment of a Hypersaline Lake in Western Australia. Geomicrobiol J. 2016;33:426–440. doi: 10.1080/01490451.2015.1049304. [DOI] [Google Scholar]
- 52.Katz L, Baltz RH. Natural product discovery: past, present, and future. J Ind Microbiol Biotechnol. 2016;43:155–176. doi: 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- 53.Rateb ME, et al. Chaxamycins A-D, bioactive ansamycins from a hyper-arid desert Streptomyces sp. J Nat Prod. 2011;74:1491–1499. doi: 10.1021/np200320u. [DOI] [PubMed] [Google Scholar]
- 54.Opfell JB, Zebal GP. Ecological patterns of micro-organisms in desert soils. Life sciences and space research. 1966;5:187–203. [PubMed] [Google Scholar]
- 55.Busarakam, K. Novel actinobacterial diversity in arid Atacama Desert soils as a source of new drug leads PhD thesis, Newcastle University (2014).
- 56.Schäfer J, Jackel U, Kämpfer P. Development of a new PCR primer system for selective amplification of Actinobacteria. FEMS Microbiol Lett. 2010;311:103–112. doi: 10.1111/j.1574-6968.2010.02069.x. [DOI] [PubMed] [Google Scholar]
- 57.Kim OS, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Micr. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 58.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Comma separated value (csv) format data files and R scripts used in this study are available at https://github.com/rasanderson/sci_reports.


