Abstract
Methamphetamine addiction is mimicked in rats that self-administer the drug. However, these self-administration (SA) models do not include adverse consequences that are necessary to reach a diagnosis of addiction in humans. Herein, we measured genome-wide transcriptional consequences of methamphetamine SA and footshocks in the rat brain. We trained rats to self-administer methamphetamine for 20 days. Thereafter, lever-presses for methamphetamine were punished by mild footshocks for 5 days. Response-contingent punishment significantly reduced methamphetamine taking in some rats (shock-sensitive, SS) but not in others (shock-resistant, SR). Rats also underwent extinction test at one day and 30 days after the last shock session. Rats were euthanized one day after the second extinction test and the nucleus accumbens (NAc) and dorsal striatum were collected to measure gene expression with microarray analysis. In the NAc, there were changes in the expression of 13 genes in the SRvsControl and 9 genes in the SRvsSS comparison. In the striatum, there were 9 (6 up, 3 down) affected genes in the SRvsSS comparison. Among the upregulated genes was oxytocin in the NAc and CARTpt in the striatum of SR rats. These observations support a regional role of neuropeptides in the brain after a long withdrawal interval when animals show incubation of methamphetamine craving.
Introduction
Methamphetamine (METH) addiction is a biopsychosocial disorder with a very high prevalence throughout the world1. Its abuse is associated with negative impact on the brain and peripheral organs2, 3. These adverse consequences include abnormal blood flow and brain structural pathologies4–6, with consequent neuropsychological deficits7. Although a diversity of drugs can cause the neuropsychiatric syndromes designated as addictions, it is important to identify specific neurobiological processes involved in causing compulsive taking of individual substances. This is relevant to the development of pharmacological agents against addictions because individual substances may impact the brain differentially4. This suggests the importance of identifying and detailing the potential impact of these drugs on distributed systems both within and beyond the so-called reward circuitry8 because changes in these systems might influence or dictate treatment approaches9–11. These statements suggest that addiction may occur as the result of repeated exposure to drugs that lead to neuropathological changes in distributed networks of potentially dissociable reward and non-reward pathways in the mammalian brain4, 8. This conclusion is supported by accumulated evidence from epigenetic, transcriptional, neuroimaging, and neuropsychological studies that have indeed provided support for a role of various brain regions in the development and maintenance of addicted states7, 10, 12–14. This suggestion also envisions the development of preclinical models that include additional criteria because, by definition, addiction diagnoses include more than just drug SA. Improved models may allow for the identification of diverse neuroanatomical structures, epigenetic and transcriptional events, and memory processes that might be involved in the promotion of addiction to specific substances such as methamphetamine.
A better understanding of how individual drugs differentially impact specific brain regions within these distributed networks will require more in-depth knowledge of the specific biochemical and molecular consequences of each addictive drug on different brain regions4. This statement thus hints to the need to decipher molecular changes that may be predictive of resilience to and/or of risk to develop an addictive state9. Elucidation of these molecular signatures has the potential to help in the development of pharmacological interventions to modify the clinical manifestations of substance use disorders. To address these issues, we have begun to use a model that uses footshocks to differentiate rats that continue to self-administer methamphetamine compulsively from those that significantly reduce their intake in the presence of punishment15–17. In the present study, we have used this model to investigate transcriptional changes in the nucleus accumbens (NAc) and dorsal striatum after a month of forced abstinence.
Results
Behaviors
Figure 1a shows the timeline of the behavioral experiments. METH-trained rats increased their drug intake and control rats decreased lever pressing for saline during the SA training phase (Fig. 1b). The repeated measures ANOVA for earned infusions included the between-subject factor reward type (saline, METH) and the within-subject factor of SA day (training days 1–20). The analysis showed a significant effect of training day x reward type [F(19,456) = 25.31, p < 0.0001]. The significant interaction reflects the fact that METH-trained rats continued to increase their drug intake over the training days whereas the saline rats decreased and stabilized their intake to very low values (Fig. 1b).
Figure 1.
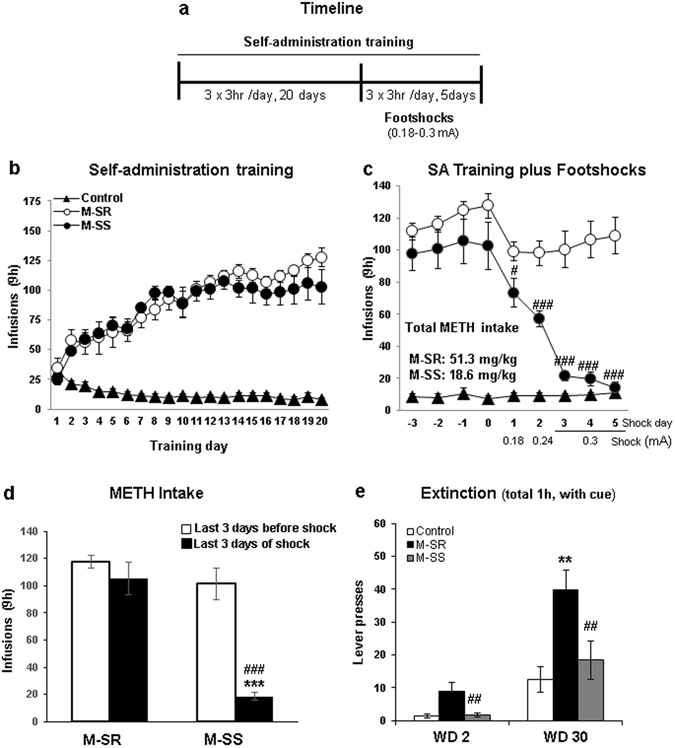
Extended access to methamphetamine and footshock punishment lead to compulsive methamphetamine taking or abstinence in rats. (a) Timeline of behavioral experiments. (b) Rats with long access to METH escalate drug self-administration. The groups were separated post-facto based on their responses to footshocks. (c) Increasing footshock intensity leads to reduction of METH intake in SS but not in SR animals. (d) Mean drug intake on the last 3 days of METH SA compared to last 3 days of shocks. (e) Rats show incubation of METH craving at withdrawal day (WD) 30 in comparison to WD2. SR rats showed higher lever pressing at WD30 in comparison to the SS rats.
During the training plus punishment phase, footshock intensity was increased from 0.18 to 0.30 A over a period of 5 days (Fig. 1c). The increased shock intensity caused reinforced responding to significantly decrease in the shock-sensitive (SS) but not in the shock-resistant (SR) rats (Fig. 1c). The statistical analysis of METH infusions earned included the between-subject factor of groups [(SR) and (SS)] and the within-subject factor of shockday (shockdays 1–5). There was a significant effect of shockday x group [F(4,56) = 16.95, p < 0.0001], with the highest intensity (0.30 mA) showing almost total suppression of METH intake in the SS rats (Fig. 1c). Figure 1d shows the effects of the highest intensity (0.30 mA, 3 days of footshocks) on METH intake in comparison to the last 3 days of training before application of foot-shocks. Rats that were yoked to SR rats to receive non-contingent footshocks (YSR) received significant more footshocks than rats yoked (YSS) to the SS group [(291 ± 39 vs 94 ± 13, respectively), p < 0.001].
We also measured cue-induced reinstatement at withdrawal days 2 and 30 (Fig. 1e) and found that both SR and SS animals increased lever pressing at WD 30 in comparison to WD2. In addition, the SS rats showed less lever pressing than the SR rats at both time points.
Microarray Analyses
We used the Affymetrix array platform that contain a total of 68,842 probes to measure transcriptional changes in the NAc and dorsal striatum of animals euthanized one day after the second extinction test. These probes consist of 24,753 protein coding, 28,724 noncoding, and unassigned pseudogenes. The analysis of raw array data revealed that 9044 genes were expressed in the NAc and 9754 genes were expressed in the dorsal striatum. This was determined by comparing the average signal values of transcripts in the control animals to the average signal value of the bioB gene in the control animals. Gene with average signals above the average signal value of the bioB gene were considered as expressed in the specific brain region. The expressed genes were included in further analyses of differential gene expression between the experimental groups. There were 181 genes differentially expressed in the NAc and 304 genes were differentially expressed in the dorsal striatum.
The results of the microarray analyses in the NAc are shown in Figs 2a and 3a, Table 1, and in supplementary Tables S1 and S1. There were multiple pairwise comparisons that included: shock-resistant versus control (SRvsCT), shock-sensitive versus CT (SSvsCT), and SR versus SS (SRvsSS) groups, yoked SR versus CT (YSRvsCT), yoked SS versus CT (YSRvsCT), and YSRvsYSS (see Figs 2a and 3a and Table 1, and in Supplementary Tables S1 and S1). The differences in gene expression between YSR and YSS are shown in the Supplementary Table S2. In the NAc, there were 13 (6 up- and 7 downregulated) differentially expressed transcripts in the SRvsCT comparison (±1.7-fold change, p < 0.05) (Fig. 2a). The SSvsCT comparison contained only 3 (1 up and 2 down) differentially expressed genes while the SRvsSS comparison had 9 (1 up-and 8 downregulated) differentially expressed transcripts. We used DAVID and literature searches to generate functional annotation and classification analyses for significantly expressed transcripts. The names of these genes and their classification are shown in Table 1. The list includes genes that participate in metabolism and signal transduction as well as several microRNA precursor transcripts. Genes of interest in the SRvsCT comparison include oxytocin that showed 6.19-fold increases in the SR group. OXT was also increased in the SR in comparison to the SS group (5.71-fold) (Table 1). The yoked animals also showed some changes in gene expression in comparison to control (Fig. 3a). Rats yoked to the SR group showed changes in 6 transcripts (3 up and 3 up) in comparison to control. Animals yoked to the SS group showed 9 (4 up and 5 down) differentially expressed genes. The list of genes also included genes involved in metabolism, phosphorylation cascades, and signal transduction (Supplementary Table S1).
Figure 2.
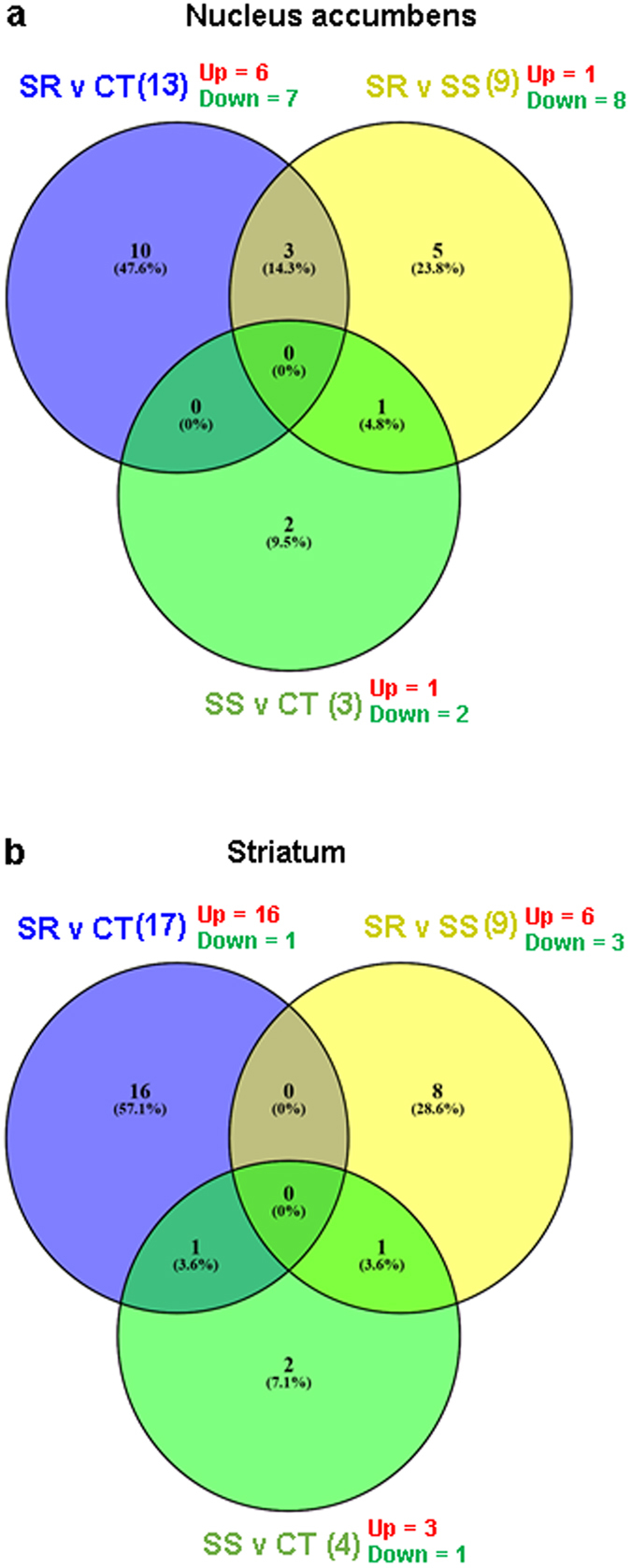
Microarray analysis of gene expression in the NAc and dorsal striatum of rats after one month of withdrawal from methamphetamine SA. Comparison of gene expression in pairwise comparisons between SR, SS, and control rats in NAc (a) in dorsal striatum (b) [n = 6 in each group]. The number of up- and downregulated genes in each comparison is shown in red and green, respectively. The list of genes and their classification are shown in Tables 1 and 2.
Figure 3.
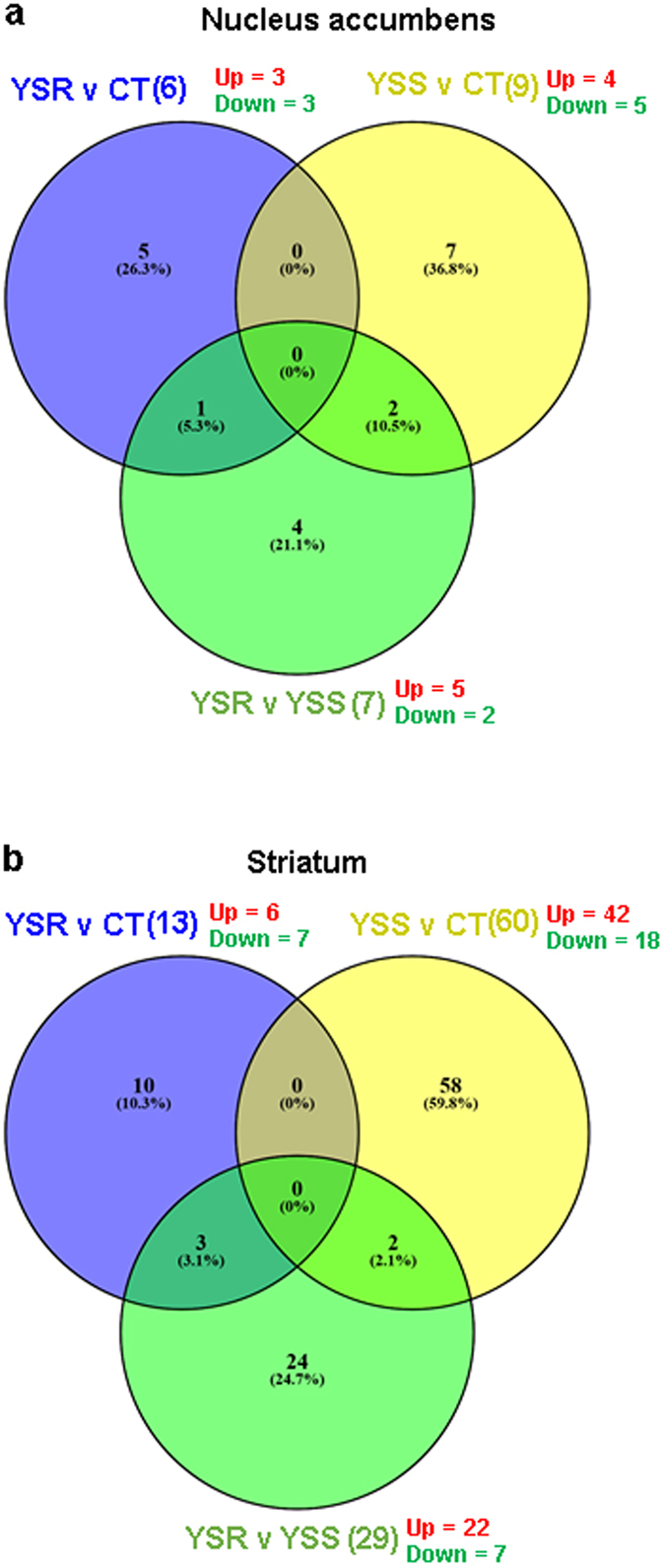
Microarray analysis of gene expression in the NAc and striatum of rats after one month of withdrawal from methamphetamine SA. Comparison of gene expression in pairwise comparisons between YSR (n = 3), YSS (n = 3), and control (n = 6) rats in NAc (a) in dorsal striatum (b). The number of up- and downregulated genes in each comparison is shown in red and green, respectively. The list of genes and their classification are shown in Tables 3 and 4.
Table 1.
Classification of NAc genes with altered expression after METH SA and footshocks.
| GenBank Accession | Symbol | Description | Fold Change SR vs CT | Fold Change SS vs CT | Fold Change SR vs SS |
|---|---|---|---|---|---|
| Metabolism | |||||
| NM_001037355 | Mettl7a | methyltransferase like 7 A | 1.68 | 1.25 | 1.35 |
| MicroRNA | |||||
| NR_030567 | Mir466f-2 | microRNA 466f-2 | −4.71 | −1.03 | −4.55 |
| MTA_TR0500009144.mm | Mir1191 | microRNA 1191 | −1.47 | 1.12 | −1.65 |
| MTA_TR0100005862.mm | Mir297c | microRNA 297c | −2.20 | 1.12 | −2.46 |
| MTA_TR0200016496.mm | Mir3098 | microRNA 3098 | 1.02 | 2.78 | −2.72 |
| MTA_TR1300007696.mm | Mir325 | microRNA 325 | −1.75 | 1.01 | −1.77 |
| NR_030571 | Mir467c | microRNA 467c | −1.71 | 1.23 | −2.10 |
| NR_030645 | Mir467e | microRNA 467e | −1.66 | 1.19 | −1.98 |
| MTA_TR0200000271.mm | Mir669f | microRNA 669 f | −1.86 | −1.13 | −1.64 |
| NR_032296 | Mir770 | microRNA 770 | 1.96 | 1.64 | 1.20 |
| NR_032756 | Mir876 | microRNA 876 | −1.85 | −1.18 | −1.57 |
| NR_032286 | Mir598 | microRNA mir-598 | −1.25 | 1.35 | −1.69 |
| MTA_TR0500000814.mm | Mir362 | microRNA mir-6362 | −2.38 | −1.09 | −2.19 |
| NR_105852 | Mir6481 | microRNA mir-6481 | −1.83 | −1.47 | −1.25 |
| Protein Kinase/Phosphatase | |||||
| NM_031769 | Pdxk | pyridoxal (pyridoxine, vitamin B6) kinase; similar to pyridoxal (pyridoxine, vitamin B6) kinase; pyridoxal kinase-like | 1.73 | 1.63 | 1.07 |
| Proteolysis | |||||
| NM_020091 | Cym | chymosin | −1.38 | −2.39 | 1.74 |
| Signal Transduction | |||||
| NM_001191104 | Tmem223 | transmembrane protein 223 | 2.07 | 1.31 | 1.58 |
| NM_022005 | Fxyd6 | FXYD domain-containing ion transport regulator 6 | 2.10 | 1.35 | 1.56 |
| NM_053859 | Slc17a7 | solute carrier family 17 (vesicular glutamate transporter), member 7 | −1.50 | −3.06 | 2.04 |
| NM_012996 | Oxt | oxytocin/neurophysin 1 prepropeptide | 6.19 | 1.08 | 5.71 |
| Synaptic Plasticity | |||||
| MTA_TR1100003798.mm | Zfp616 | zinc finger protein 616 | −1.81 | −1.56 | −1.16 |
The results of the striatal microarray data are shown in Figs 2b and 3b, Table 2, and in supplementary Tables S3 and S4. Figure 2b shows that there were 17 (16 up and 1 down) differentially expressed transcripts in the SRvsCT comparison, 4 (3 up and 1 down) transcripts in the SSvsCT comparison, and 9 (6 up and 3 down) transcripts in the SRvsSS comparison. Table 2 shows a list of genes that participate cell adhesion, metabolism, signal transduction, and transcription regulation in the dorsal striatum. Figure 3b shows the results of the comparison between the yoked animals and the control group. There were 13 (6 up and 7 down) differentially expressed transcripts in the YSRvsCT comparison, 60 (42 up and 18 down) in the YSSvsCT comparison and 29 (22 up and 7 down) differentially expressed genes in the YSRvs YSS comparison. The genes are listed in supplementary Table S3. The differences in gene expression between YSR and YSS in the dorsal striatum are shown in Supplementary Table S4.
Table 2.
Classification of striatal genes with differential expression after METH SA and footshocks.
| Genbank Accession | Gene Symbol | Description | Fold Change SR vs CT | Fold Change SS vs CT | Fold Change SR vs SS |
|---|---|---|---|---|---|
| Calcium Ion Binding | |||||
| NM_001271195 | Fstl5 | follistatin-like 5 | 1.52 | −1.11 | 1.68 |
| Cell Adhesion | |||||
| NM_001169129 | Pcdh19 | protocadherin 19 | 2.11 | 1.50 | 1.41 |
| XM_001061943.6 | Cdh4 | cadherin 4 | 2.02 | 1.43 | 1.41 |
| DNA Binding | |||||
| NM_153626 | Npas4 | neuronal PAS domain protein 4 | −1.75 | −1.48 | −1.18 |
| Membrane Structure | |||||
| NM_001191104 | Tmem22 | transmembrane protein 223 | 1.98 | 1.30 | 1.52 |
| NM_001109561 | Nrsn2 | neurensin 2 | 1.40 | −1.24 | 1.74 |
| Metabolism | |||||
| NM_001107311 | Enpp6 | ectonucleotide pyrophosphatase/phosphodiesterase 6 | 1.12 | −1.78 | 1.99 |
| NM_001107671 | Plcxd3 | phosphatidylinositol-specific phospholipase C, X domain | 1.69 | 1.22 | 1.39 |
| NM_001191118 | Siah3 | siah E3 ubiquitin protein ligase family member 3 | 1.75 | 1.25 | 1.41 |
| NM_001258237 | Ace3 | angiotensin I converting enzyme (peptidyl-dipeptidase A | −1.14 | 1.54 | −1.75 |
| NM_031655 | Lxn | latexin | 1.05 | 1.71 | −1.63 |
| NM_031776 | Gda | guanine deaminase | 1.81 | 1.33 | 1.36 |
| XM_006247643.3 | LOC102556346 | angiotensin-converting enzyme-like | −1.18 | 1.87 | −2.21 |
| MicroRNA | |||||
| NR_035479 | Mir1954 | microRNA 1954 | −1.14 | −1.88 | 1.66 |
| NR_106093 | Mir7234 | microRNA mir-7234 | −1.42 | 1.34 | −1.91 |
| Protein Binding | |||||
| NM_017110 | Cartpt | CART prepropeptide | 1.33 | −1.59 | 2.11 |
| Signal Transduction | |||||
| XM_008763452.2 | Eps8 | epidermal growth factor receptor pathway substrate 8 | 1.89 | 1.35 | 1.40 |
| NM_001102381 | Nts | neurotensin | 1.82 | 1.13 | 1.61 |
| NM_001108774 | Mrap2 | melanocortin 2 receptor accessory protein 2 | 1.73 | 1.44 | 1.20 |
| NM_001109018 | Rab9b | RAB9B, member RAS oncogene family | 1.66 | 1.06 | 1.57 |
| NM_001287022 | Rgs21 | regulator of G-protein signaling 21 | 1.01 | 1.89 | −1.87 |
| NM_012568 | Glra2 | glycine receptor, alpha 2 | 3.50 | 2.07 | 1.69 |
| NM_017362 | Chrm5 | cholinergic receptor, muscarinic 5 | 3.06 | 1.58 | 1.93 |
| NM_022669 | Scg2 | secretogranin II | 1.81 | 1.25 | 1.45 |
| NM_031823 | Wfs1 | Wolfram syndrome 1 (wolframin) | 2.35 | 1.68 | 1.40 |
| Transcription | |||||
| NM_001107473 | Zim1 | zinc finger, imprinted 1 | 1.78 | 1.14 | 1.57 |
| NM_001191702 | Zfhx4 | zinc finger homeobox 4; Unannotated AceView Transcript; | 1.73 | 1.24 | 1.39 |
| XM_008762737.1 | PEG10 | ENCODES a protein that exhibits poly(A) RNA binding | 1.71 | 1.02 | 1.67 |
Validation of array-identified genes by quantitative PCR
Compulsive METH taking-related genes
To validate the changes in oxytocin observed in the microarray data, we ran quantitative PCR using RNAs from the various groups of rats. Figure 4 presents the effects of METH SA and footshocks on the expression of oxytocin (OXT) in the nucleus accumbens and dorsal striatum. NAc OXT mRNA levels displayed significant changes [F(4,19) = 5.44, p = 0.0043] in expression. Post-hoc analyses demonstrated that the SR group showed substantial increases in OXT mRNA expression in comparison to the CT, SS, and yoked animals (Fig. 4a). YSR and YSS groups showed small decreases that were not significant in comparison to the control group (Fig. 4a). OXT receptor (OXTR) mRNA levels also exhibited significant changes [F(4,19) = 7.19, p = 0.0011] after withdrawal from METH SA, with post-hoc analyses indicating that all groups showed small increases in comparison to the control group (Fig. 4b). In contrast, there were no significant changes in OXT [F(4,24) = 1.643, p = 0.1961] or OXTR [F(4,27) = 1.715, p = 0.1767] mRNA levels in the dorsal striatum (Fig. 4b and d, respectively).
Figure 4.
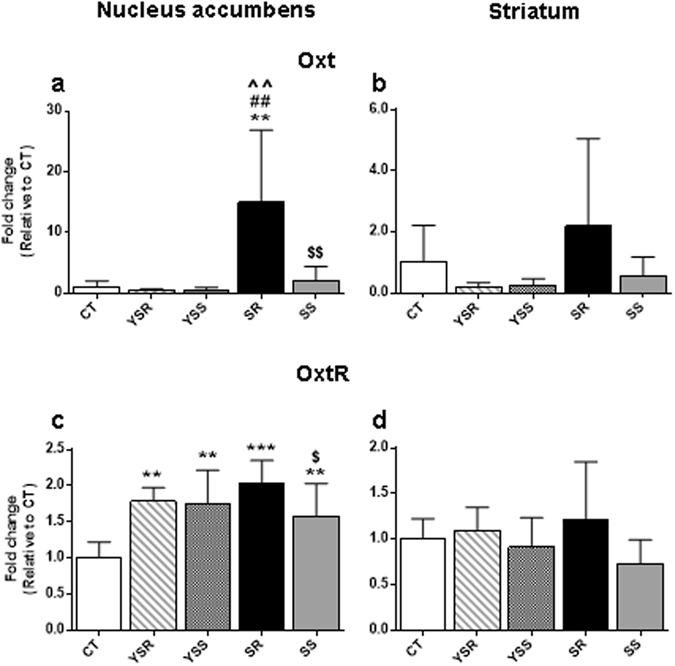
PCR validation of changes in oxytocin mRNA levels in the rat NAc after a month withdrawal from methamphetamine SA and footshocks. We conducted quantitative PCR using individual RNA from the NAc (a,c) and dorsal striatum (b,d) of rats from the various conditions (CT, n = 6–8; SR, n = 6–8; SS, n = 6–9; YSR, n = 3; YSS, n = 3–6). The shock-resistant rats showed significant increases in oxytocin (a) and oxytocin receptor (c) in the NAc but not in the dorsal striatum (b and d, respectively). Values are means ± SEM fold changes relative to the control group. Key to statistics: **p < 0.01, ***p < 0.001, in comparison to the control group; ##p < 0.01, in comparison to YSR rats; ^^p < 0.01, in comparison to the YSS group; $p < 0.05, $$p < 0.01 in comparison to the SR group.
We also validated the expression of striatal CARTpt that showed increased expression between SR and SS in the microarray data (Table 2). Figure 5a showed confirmation of the significant [F(4,28) = 4.220, p = 0.0085] increases in CARTpt in the striatum of SR rats. There were, however, no significant changes in the NAc [F(4,26) = 1.059, p = 0.3966] (Fig. 5b).
Figure 5.
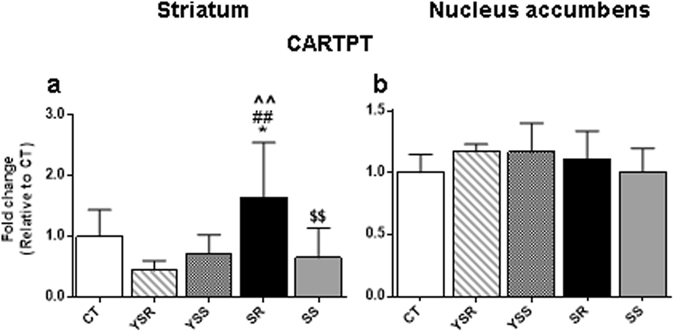
PCR validation of changes in CARTpt mRNA expression in the dorsal striatum after a month withdrawal from methamphetamine SA and footshocks. We conducted quantitative PCR using individual RNA from the dorsal striatum (a) and NAc (b) of rats. The shock-resistant rats showed significant increases in CARTpt in the dorsal striatum (a) but not in the NAc (b). Values are means ± SEM fold changes relative to the control group. Key to statistics: *p < 0.05, in comparison to the control group; ##p < 0.01, in comparison to YSR rats; ^^p < 0.01, in comparison to the YSS group; $$p < 0.01 in comparison to the SR group.
Footshock-responsive genes
We also validated the expression of some genes that showed changes in rats that were yoked to the METH SA to receive similar number of footshocks as the METH SA animals (Fig. 6). Figure 6a and b show that animals that received footshocks experienced no significant changes in FMO2 (flavin-containing monooxygenase) in the NAc but showed significant [F(4,19) = 6.849, p = 0.0014] changes in the dorsal striatum. Post-hoc analyses showed that the YSR group (291 ± 39 footshocks) exhibited significantly higher mRNA levels in the dorsal striatum than the other groups (Fig. 6b). Figure 6c and d show that there were significant changes in the expression of PDK4 (pyruvate dehydrogenase kinase 4) in the NAc [F(4,19) = 6.388, p = 0.0020] and dorsal striatum [F(4,28) = 6.497, p = 0.0008]. In addition to the YSR group showing higher levels than the other groups in the NAc, post-hoc analyses showed that both SR and SS groups [but not the YSS (94 ± 13 footshocks) rats] also showed small increases in comparison to the control animals. The results suggest that METH might have partially attenuated the transcriptional effects of the large number of footshocks that the SR rats had received (compare YSR to SR results in Fig. 6c). In the dorsal striatum (Fig. 6d), YSR rats also showed higher expression than the other groups whereas the SR animals showed lower expression than the control group, suggesting the large amount of METH taken by the SR rats might have significantly suppressed the effects of footshocks alone (YSR) in this brain region. Smaller number of footshocks and lower amount of METH (SS) did not significantly influence the expression of PDK4 in the dorsal striatum (Fig. 6d). These data support the notion that the NAc and dorsal striatum may respond differentially to a diversity of exogenous stimuli.
Figure 6.
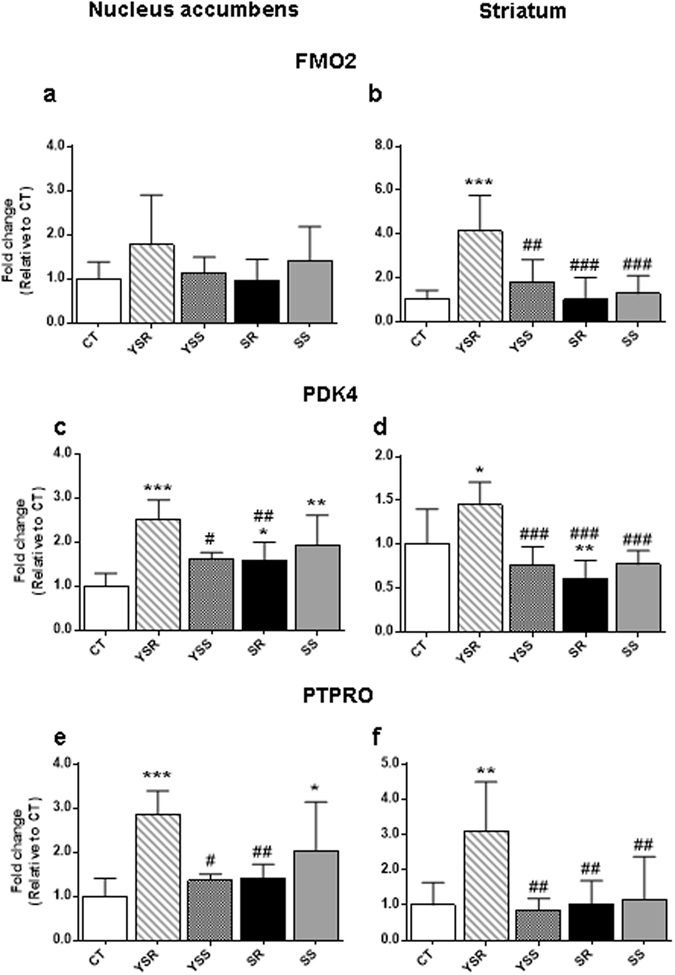
PCR measures of changes in FMO2, PDK4, and PTPRO mRNA levels in the rat NAc and dorsal striatum after a month withdrawal from methamphetamine SA and footshocks. We conducted quantitative PCR using individual RNA from the NAc (a,c,e) and dorsal striatum (b,d,f) of rats from the 5 groups of rats. The YSR that received many non-contingent footshocks (291 ± 39) showed significant increases in FMO2 in the dorsal striatum (b) but not in the NAc (a). YSR rats also experienced increases in PDK4 in both the NAc (c) and dorsal striatum (d). PTPRO mRNA levels were also increased in the NAc (e) and dorsal striatum (f) of YSR rats. Values are means ± SEM fold changes relative to the control group. Key to statistics: *p < 0.05, **p < 0.01, ***p < 0.001, in comparison to the control group; #p < 0.01, ##p < 0.01, ###p < 0.001, in comparison to the YSR rats.
Figure 6e and f show the effects of METH and footshocks on the expression of PTPRO (protein tyrosine phosphatase, receptor type O) in the NAc and dorsal striatum, respectively. PTPRO mRNA expression was significantly [F(4.19) = 4.890, p = 0.0070] affected in the NAc. Post-hoc analyses showed that the YSR and SS groups exhibited higher mRNA expression than controls (Fig. 6e). In addition, SR rats showed lower expression than YSR animals, again suggesting that large amount of METH might have inhibited the effects of footshocks alone on PTPRO mRNA levels in the NAc. In the dorsal striatum, there were also significant [F(4,28) = 3.982, p = 0.0111] increases in the levels of PTPRO mRNA in the YSR group when compared to the other groups (Fig. 6f). Post-hoc tests indicated that PTPRO mRNA levels in the other groups were comparable to those of controls (Fig. 6f).
Discussion
The aim of the present study was to use an unbiased genome-wide approach to identify transcriptional changes in shock-sensitive and -resistant rats that had all undergone METH self-administration training and footshocks. We also compared gene expression in rats that were yoked to the METH SA animals to control for the effects of footshocks during the shock phase of the study. Our aim was to compare gene expression in the NAc and dorsal striatum because these two brain regions are known for their involvement in different aspects and phases of compulsive drug-taking behaviors and during withdrawal from METH SA16, 18–24. We had reasoned that drug-induced alterations of these neuroanatomical systems may negatively impact information processing and, over time, might have led to disturbed responses to environmental cues7, based on their different connections to various brain regions implicated in addiction25. Indeed, we found significant increases in oxytocin mRNA levels in the NAc, but not in the dorsal striatum, 30 days after stopping METH SA. These results are consistent with those of Baracz et al.26 who had reported significant increases in plasma oxytocin levels immediately after and at 15 days of withdrawal from METH self-administration. The observation that animals with reduced METH intake in the presence of footshocks did not show significant increases in oxytocin mRNA levels suggest that these observations may be related specifically to compulsive METH takers that continue to take the drug despite the footshocks (SR rats). The changes in oxytocin mRNA are also not secondary to footshocks alone since the yoked animals did not show any increases in oxytocin in comparison to control rats. We also found significant increases in CARTpt mRNA levels in the dorsal striatum but not in the NAc of compulsive METH takers, suggesting that different neurotransmitter systems may be activated in different brain regions during protracted forced abstinence from METH self-administration.
Potential role of NAc oxytocin in compulsive methamphetamine taking
Oxytocin is a centrally active neuropeptide that is involved in social bonding/attachment27, 28 and in some psychopathological states including stress, affective disorders, and schizophrenia29, 30. A potential role for oxytocin in addiction has also been investigated by several groups of scientists18, 26, 31–34. For example, intracerebroventricular injections of oxytocin were shown to reduce METH-induced increased locomotor activity35 and conditioned place preference (CPP)36 in mice. Carson et al.32 have also reported that peripheral oxytocin administration can dose-dependently reduce the number of self-administered METH infusions by rats. Oxytocin also reduced locomotor activity and drug-induced reinstatement32. Follow-up studies have also documented that infusions of oxytocin in the NAc can reduce METH CPP37 and METH-primed reinstatement31. These studies are consistent with the recent report that oxytocin, given peripherally or intracerebrally, can reduce METH demand and seeking by rats33. When taken together, these observations implicate an important role of this neuropeptide in the effects of this psychostimulant. This conclusion is consistent with the report that METH can cause increased plasma oxytocin levels and decreased oxytocin receptors in the NAc of rodents31. Thus, our findings of increased oxytocin mRNA levels in the NAc may constitute compensatory responses to replace oxytocin that might have been released from the NAc during compulsive METH taking.
Potential role of striatal CARTpt in withdrawal from compulsive methamphetamine taking
We also found significant increases in CARTpt mRNA levels in the striatum, but not in the NAc, after 30 days of withdrawal from METH (see Fig. 5). These results are consistent with recent proposals that the dorsal striatum may be involved in longterm abstinence from METH SA20, 38. CARTpt is a brain/gut neuropeptide initially identified by differential display in the dorsal striatum of rats that were injected with either cocaine or amphetamine39. In the NAc, CARTpt was reported to be localized in D1-containing neurons not in D2 neurons40. In addition to the NAc, CARTpt is widely distributed in the brain41, 42 and appears to play a role in the acute and chronic effects of some drugs of abuse43, 44. For example, intra-NAc administration of CART was shown to attenuate hyperactivity induced by either cocaine or amphetamine45, 46. The effects of CARTpt were reported to be dependent on the modulation of both dopamine (DA) D1 and D2 receptors47. Intra-accumbal CARTpt injections also attenuated cocaine self-administration48. Non-contingent injection of METH caused significant increases in CARTpt mRNA levels in the NAc of rats euthanized at either 2 hours or one month later49. When taken together with our present observations, these results support the notion that CARTpt may play a substantial role in regulating certain aspects of stimulant-induced behaviors40, 44 both acutely and after prolonged abstinence.
Footshock-induced changes in gene expression
In view of the novelty of these observations, it is of interest to briefly discuss the findings that some genes showed significant changes in their expression in animals that had received many non-contingent footshocks (YSR group, see Fig. 6). There is evidence that repeated footshocks can affect DA metabolism in the rodent NAc and dorsal striatum but to different degrees based on the severity of footshocks50–54. These studies had investigated perturbances in dopaminergic systems during short periods of time after application of single or multiple footshocks. In the present study, gene expression was assessed after one month of withdrawal and, yet, we still observed significant changes in FMO2, PDK4 and PTPRO mRNA levels in two DA terminal fields. Although the reasons for these changes remain to be determined, these findings suggest that footshock punishment can have substantial transcriptional effects in these brain regions in a fashion that might affect brain metabolism since FMO2 is a member of a class of enzymes (FMO1–5) that are localized in the membranes of endoplasmic reticulum and are involved in metabolizing drugs, regulating fat and cholesterol metabolism, and breaking down of other dietary components55, 56. Importantly, the presence of FMOs in the brain57 and the increased expression that we observed in the striatum suggests that FMO2 might play a role in the longterm effects of footshocks on the metabolic health of this brain structure. Parenthetically, FMO2 expression is increased by manipulations that increase animal longevity58.
Another important metabolic enzyme affected by repeated footshocks in both the striatum and NAc is PDK4, a member of a class of 4 enzymes (PDK1-PDK4)59, 60 that are involved in the regulation of the mitochondrial pyruvate dehydrogenase (PDH) complex (PDC)61, 62. PDC catalyzes the oxidative decarboxylation of pyruvate to acetyl-coA63 and is inactivated via PDK-dependent phosphorylation59. PDKs are expressed in the brain64 and astrocytes appear to contain higher expression of PDK2 and PDK4 whereas neurons contain higher levels of PDK165. PDKs may be involved in the generation and progression of several neurodegenerative processes that involve inflammation and abnormal mitochondrial functions66. Thus, our findings of increased PDK4 mRNA in the NAc and dorsal striatum suggest that footshocks might have activated mechanisms meant to further downregulate PDC function in glial cells to modify biochemical responses to repeated painful stimuli. This notion is consistent with the report that glial responses to inflammatory stimuli were attenuated in PDK4 knockout mice60.
Protein phosphorylation/dephosphorylation processes are key regulatory processes that serve to activate or inactivate proteins by the actions of protein kinases and protein phosphatases67, 68. Protein tyrosine phosphatases (PTPs) are subdivided into receptor and non-receptor phosphatases and are expressed in the central nervous system69. Thus, it was of interest that the mRNA expression of a protein phosphatase, PTPRO, was significantly increased in rats that received large number of non-contingent footshocks. PTPRO was first identified as a membrane protein that is expressed in the brain70 and is involved in the regulation of axonal guidance and neurodevelopment71–73. The observed increased in PTPRO in the yoked rats suggests that footshocks might have led to activation of mechanisms known to be involved in the regulation of PTPRO-dependent neurodevelopmental processes and, by extension, shock-induced synaptic plasticity in adult rats. In any case, the observations of footshock-induced changes in gene expression are consistent with the idea that adverse consequences can have longterm effects on the brain.
Conclusion
Our present observations support the notion that accumbal oxytocin is a potentially important neuropeptide in the manifestation of METH addiction. Our data also identify a potential role of striatal CARTpt in regulating rat behaviors after protracted withdrawal from compulsive METH taking. When taken together, the present findings point to a potentially significant role of a diversity of neuropeptides in different stages of METH addiction. This conclusion supports the need for further clinical evaluation of these neuropeptides or their analogs in clinical situations.
Methods
Animals and drug treatment
We used male Sprague-Dawley rats (Charles River Labs, Raleigh, NC, USA), weighing 350–400 g in the beginning of the study in all experiments. We housed animals in a humidity and temperature-controlled (22.2 ± 0.2 °C) room with free access to food and water. Our procedures followed the Guide for the Care and Use of Laboratory Animals (ISBN 0–309–05377–3) and were approved by the National Institute of Drug Abuse Animal Care and Use Committee.
Intravenous surgery
We anesthetized rats with ketamine and xylazine (50 and 5 mg/kg, i.p., respectively) and inserted silastic catheters into the jugular veins, as described previously74. We attached the catheters to a modified 22-gauge cannula that was mounted to their skulls with dental cement. We injected buprenorphine (0.1 mg/kg, s.c.) one time after surgery to relieve pain and allowed the rats to recover for 5–7 days before methamphetamine self-administration training. During the recovery, training and punishment phases of the experiment, we flushed the catheters every 24–48 h with gentamicin (Butler Schein; 5 mg/ml) and sterile saline.
Training and punishment phases
We performed the training procedure for methamphetamine self-administration essentially as described74. On the first day of training, we brought rats to the self-administration room and chronically housed them in self-administration chambers. Animals had free access to food and water that were available in water bottles and feeders hanging on the walls of all self-administration chambers. We trained rats to self-administer dl-methamphetamine HCl (NIDA) or saline during three 3-h sessions/day (the sessions were separated by 30 min off intervals) for 20 days under a fixed-ratio-1 with 20-s timeout reinforcement schedule. Presses on the retractable active lever activated the infusion pump. Active lever presses were also accompanied by a 5-s compound tone-light cue. Presses on inactive (stationary) lever had no reinforced consequences. We connected the catheters of rats to a modified cannula (Plastics One, Minneapolis, MN) attached to a liquid swivel (Instech Laboratories, Inc., Plymouth Meeting, PA, USA) using a polyethylene-50 tubing that was protected by a metal spring. Rats self-administered methamphetamine for 5 days a week with weekends off. During the 2 days off, rats remained housed in self-administration chambers but were disconnected from intravenous self-administration lines. Also, the levers were retracted and all other cues were removed. Rats self-administered methamphetamine at a dose of 0.1 mg/kg/infusion over 3.5 s (0.1 ml/infusion). To prevent overdose, we limited the number of infusions per 3-h session to 50. Control rats self-administered saline under the same conditions. We started the self-administration sessions at the onset of the dark cycle and sessions began with the insertion of the active lever and the illumination of a red house light that remained on for the duration of the session. At the end of each 3-h session, the house light was turned off, and the active lever was retracted.
During the training plus punishment phase, rats continued methamphetamine self-administration every day (three 3-h sessions/day separated by 30 min off intervals) under the fixed ratio-1 with 20-s timeout reinforcement schedule that was used during training. For methamphetamine-trained rats, 50% of the reinforced lever-presses also resulted in the concurrent delivery of a 0.5-s footshock through the grid floor. We set the initial footshock at 0.18 mA and increased the shock intensity by 0.06 mA to a final value of 0.3 mA (a total of 5 punishment days). Additionally, some control rats were yoked to the shocked animals so that each time animals in the methamphetamine SA group received a contingent shock, these rats also received a non-contingent shock and a saline infusion. Thus, by the end of the behavioral experiments, there were rats yoked to the corresponding shock-sensitive and -resistant rats, namely yoked SS (YSS) and yoked SR (YSR), respectively.
Withdrawal phase
At the end of the training plus punishment phase, rats were returned to the animal vivarium and individually housed with no access to METH. Intravenous catheters were covered using dust caps and rats had access to home-cage food and water ad libitum. Cue-induced drug craving was then assessed at days 2 and 30 of withdrawal. To test cue-induced craving, rats were brought back to their corresponding SA chambers on the morning of each test. Each test consisted of a single 1-h session during which presses on the drug-associated lever resulted in contingent presentations of the tone and light cues previously paired with METH. However, no METH was available during these tests. Cue-induced drug seeking behavior was assessed using a within-subject design such that all rats tested on day 2 were also tested on day 30 of withdrawal. Animals were euthanized one day after the second cue-induced methamphetamine craving test.
RNA preparation
Thirty days after cessation of methamphetamine self-administration and footshocks, we euthanized the rats by decapitation with guillotine and isolated NAc and dorsal striatal samples from the brains. We extracted total RNA from individual samples using Qiagen RNeasy Mini kit (Qiagen, Valencia, CA, USA). We assessed RNA integrity using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA). We obtained RNA integrity numbers (RIN, see Table S5) prior to RNA amplification and hybridization to the Affymetrix GeneChip™ Rat Transcriptome Array 1.0 (RTA 1.0.) [Thermo Fisher Scientific Inc., Catalog # 902634]. RNA samples showed no degradation with all RINs being greater than 8.4 (see Table S5).
Transcriptomic profiling using Affymetrix microarrays
The RNA was amplified using Affymetrix labelling based on standard Affymetrix protocols (Affymetrix, Santa Clara, CA, USA). The cRNA and single-stranded cDNA size distribution was determined using an Agilent Bioanalyzer to confirm that the cRNA was the correct size range (200–2000 nt) and the single-stranded cDNA was the correct size (approximately 400 nt). Quality control processes were also used for gene expression analysis including but not limited to hybridization controls, labeling controls, internal control genes (housekeeping controls), global array metrics, and algorithm parameters. Hybridization, labelling, scanning, and data extraction were also performed using standard Affymetrix protocols. Differentially expressed genes were chosen if there were ± 1.7-fold changes (p < 0.05) based on pairwise comparisons using Affymetrix software.
Quantitative PCR analysis of mRNA levels
Genes were selected for validation by real-time quantitative polymerase chain reaction (qPCR) essentially as previously described [15]. Briefly, individual total RNA from 6 rats per group was reverse-transcribed into cDNA using Advantage RT for PCR kit (Clontech, Mountain View, CA, USA). Gene-specific PCR primers were generated by the LightCycler probe design software v. 2.0 (Roche Biosystems, Indianapolis, IN, USA) and purchased from the Synthesis and Sequencing Facility of Johns Hopkins University (Baltimore, MD USA). High Capacity cDNA Reverse Transcription Kit (Invitrogen, Waltham, MA, USA) was used with a ViiA 7 instrument (Life Technologies, Waltham, MA, USA). The relative amounts of messenger RNA were normalized to means of clathrin, ornithine decarboxylase antienzyme 1 (OAZ1), and tubulin.
Statistical Analysis
We analyzed the behavioral data with the statistical program SPSS and followed significant effects (p < 0.05) with SPSS post-hoc contrasts with the repeated measures ANOVA. For the training and shock phases, the dependent variables were the number of METH or saline infusions during 20 training days and 5 footshock days. The PCR data were analyzed by one-way ANOVA followed by Fischer’s protected least-significant difference test (PLSD) using StatView (version 4.02, SAS Institute, Cary, NC, USA). The null hypothesis was rejected at p < 0.05.
Availability of data and materials
Gene expression data have been deposited at the NCBI under the accession #GSE95571.
Electronic supplementary material
Acknowledgements
This work was supported by funds of the Intramural Research Program of the DHHS/NIH/NIDA.
Author Contributions
I.N.K., M.C.G., B.L. and M.M. performed the rodent METH SA training. B.L. and D.W. did RNA isolation. D.W. performed the microarray and RT-PCR experiments. S.J. prepared the manuscript. C.B. analyzed the microarray data. J.L.C. supervised the overall project, designed experiments, and wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08898-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Radfar SR, Rawson RA. Current research on methamphetamine: epidemiology, medical and psychiatric effects, treatment, and harm reduction efforts. Addiction & health. 2014;6:146–154. [PMC free article] [PubMed] [Google Scholar]
- 2.Cadet J., Brannock C., Jayanthi S., Krasnova I. Transcriptional and Epigenetic Substrates of Methamphetamine Addiction and Withdrawal: Evidence from a Long-Access Self-Administration Model in the Rat. Molecular neurobiology 1–22 (2014). [DOI] [PMC free article] [PubMed]
- 3.Paratz ED, Cunningham NJ, MacIsaac AI. The Cardiac Complications of Methamphetamines. Heart Lung Circ. 2016;25:325–332. doi: 10.1016/j.hlc.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Cadet JL, Bisagno V, Milroy CM. Neuropathology of substance use disorders. Acta neuropathologica. 2014;127:91–107. doi: 10.1007/s00401-013-1221-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang L, Alicata D, Ernst T, Volkow N. Structural and metabolic brain changes in the striatum associated with methamphetamine abuse. Addiction. 2007;102(Suppl 1):16–32. doi: 10.1111/j.1360-0443.2006.01782.x. [DOI] [PubMed] [Google Scholar]
- 6.Volkow ND, et al. Higher cortical and lower subcortical metabolism in detoxified methamphetamine abusers. Am J Psychiatry. 2001;158:383–389. doi: 10.1176/appi.ajp.158.3.383. [DOI] [PubMed] [Google Scholar]
- 7.Cadet JL, Bisagno V. Neuropsychological Consequences of Chronic Drug Use: Relevance to Treatment Approaches. Front Psychiatry. 2015;6 doi: 10.3389/fpsyt.2015.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koob GF, Volkow ND. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry. 2016;3:760–773. doi: 10.1016/S2215-0366(16)00104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kendler KS, Myers J. Addiction resistance: Definition, validation and association with mastery. Drug Alcohol Depend. 2015;154:236–242. doi: 10.1016/j.drugalcdep.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.London ED, et al. Mood disturbances and regional cerebral metabolic abnormalities in recently abstinent methamphetamine abusers. Arch Gen Psychiatry. 2004;61:73–84. doi: 10.1001/archpsyc.61.1.73. [DOI] [PubMed] [Google Scholar]
- 11.Nathan PE, Conrad M, Skinstad AH. History of the Concept of Addiction. Annu Rev Clin Psychol. 2016;12:29–51. doi: 10.1146/annurev-clinpsy-021815-093546. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein RZ, Volkow ND. Dysfunction of the prefrontal cortex in addiction: neuroimaging findings and clinical implications. Nat Rev Neurosci. 2011;12:652–669. doi: 10.1038/nrn3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014;76 Pt B:259–268. doi: 10.1016/j.neuropharm.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker DM, Cates HM, Heller EA, Nestler EJ. Regulation of chromatin states by drugs of abuse. Curr Opin Neurobiol. 2015;30:112–121. doi: 10.1016/j.conb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cadet, J. L. et al. hydroxymethylation identifies potassium channels in the nucleus accumbens as discriminators of methamphetamine addiction and abstinence. Mol Psychiatry (2016). [DOI] [PMC free article] [PubMed]
- 16.Cadet JL, et al. Increased expression of proenkephalin and prodynorphin mRNAs in the nucleus accumbens of compulsive methamphetamine taking rats. Sci Rep. 2016;6 doi: 10.1038/srep37002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belin-Rauscent, A., Fouyssac, M., Bonci, A., Belin, D. How Preclinical Models Evolved to Resemble the Diagnostic Criteria of Drug Addiction. Biological psychiatry (2015). [DOI] [PMC free article] [PubMed]
- 18.Baracz SJ, Cornish JL. The neurocircuitry involved in oxytocin modulation of methamphetamine addiction. Front Neuroendocrinol. 2016;43:1–18. doi: 10.1016/j.yfrne.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Krasnova IN, et al. CREB phosphorylation regulates striatal transcriptional responses in the self-administration model of methamphetamine addiction in the rat. Neurobiology of disease. 2013;58:132–143. doi: 10.1016/j.nbd.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, et al. Incubation of methamphetamine craving is associated with selective increases in expression of Bdnf and trkb, glutamate receptors, and epigenetic enzymes in cue-activated fos-expressing dorsal striatal neurons. J Neurosci. 2015;35:8232–8244. doi: 10.1523/JNEUROSCI.1022-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubio FJ, et al. Context-induced reinstatement of methamphetamine seeking is associated with unique molecular alterations in Fos-expressing dorsolateral striatum neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2015;35:5625–5639. doi: 10.1523/JNEUROSCI.4997-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheyer AF, et al. AMPA Receptor Plasticity in Accumbens Core Contributes to Incubation of Methamphetamine Craving. Biol Psychiatry. 2016;80:661–670. doi: 10.1016/j.biopsych.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szumlinski, K. K. et al. Methamphetamine Addiction Vulnerability: The Glutamate, the Bad, and the Ugly. Biol Psychiatry (2016). [DOI] [PMC free article] [PubMed]
- 24.Whitfield TW, Jr., et al. kappa Opioid receptors in the nucleus accumbens shell mediate escalation of methamphetamine intake. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2015;35:4296–4305. doi: 10.1523/JNEUROSCI.1978-13.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joel D, Weiner I. The connections of the dopaminergic system with the striatum in rats and primates: an analysis with respect to the functional and compartmental organization of the striatum. Neuroscience. 2000;96:451–474. doi: 10.1016/S0306-4522(99)00575-8. [DOI] [PubMed] [Google Scholar]
- 26.Baracz, S. J. et al. Chronic Methamphetamine Self-Administration Dysregulates Oxytocin Plasma Levels and Oxytocin Receptor Fibre Density in the Nucleus Accumbens Core and Subthalamic Nucleus of the Rat. J Neuroendocrinol28 (2016). [DOI] [PubMed]
- 27.Bernaerts S, et al. Long-term oxytocin administration enhances the experience of attachment. Psychoneuroendocrinology. 2017;78:1–9. doi: 10.1016/j.psyneuen.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Feldman R. The Neurobiology of Human Attachments. Trends Cogn Sci. 2017;21:80–99. doi: 10.1016/j.tics.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 29.McQuaid RJ, et al. Relations between plasma oxytocin and cortisol: The stress buffering role of social support. Neurobiol Stress. 2016;3:52–60. doi: 10.1016/j.ynstr.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintana, D. S., Dieset, I., Elvsashagen, T., Westlye, L. T., Andreassen, O. A. Oxytocin system dysfunction as a common mechanism underlying metabolic syndrome and psychiatric symptoms in schizophrenia and bipolar disorders. Front Neuroendocrinol (2017). [DOI] [PubMed]
- 31.Baracz SJ, Everett NA, McGregor IS, Cornish JL. Oxytocin in the nucleus accumbens core reduces reinstatement of methamphetamine-seeking behaviour in rats. Addict Biol. 2016;21:316–325. doi: 10.1111/adb.12198. [DOI] [PubMed] [Google Scholar]
- 32.Carson DS, Cornish JL, Guastella AJ, Hunt GE, McGregor IS. Oxytocin decreases methamphetamine self-administration, methamphetamine hyperactivity, and relapse to methamphetamine-seeking behaviour in rats. Neuropharmacology. 2010;58:38–43. doi: 10.1016/j.neuropharm.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 33.Cox, B. M. et al. Oxytocin Acts in Nucleus Accumbens to Attenuate Methamphetamine Seeking and Demand. Biol Psychiatry (2016). [DOI] [PMC free article] [PubMed]
- 34.Lee MR, Weerts EM. Oxytocin for the treatment of drug and alcohol use disorders. Behav Pharmacol. 2016;27:640–648. doi: 10.1097/FBP.0000000000000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi J, et al. Inhibition by oxytocin of methamphetamine-induced hyperactivity related to dopamine turnover in the mesolimbic region in mice. Naunyn Schmiedebergs Arch Pharmacol. 2008;376:441–448. doi: 10.1007/s00210-007-0245-8. [DOI] [PubMed] [Google Scholar]
- 36.Qi J, et al. Effects of oxytocin on methamphetamine-induced conditioned place preference and the possible role of glutamatergic neurotransmission in the medial prefrontal cortex of mice in reinstatement. Neuropharmacology. 2009;56:856–865. doi: 10.1016/j.neuropharm.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Baracz SJ, et al. Oxytocin directly administered into the nucleus accumbens core or subthalamic nucleus attenuates methamphetamine-induced conditioned place preference. Behav Brain Res. 2012;228:185–193. doi: 10.1016/j.bbr.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 38.Caprioli D, et al. Role of Dorsomedial Striatum Neuronal Ensembles in Incubation of Methamphetamine Craving after Voluntary Abstinence. J Neurosci. 2017;37:1014–1027. doi: 10.1523/JNEUROSCI.3091-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douglass J, McKinzie AA, Couceyro P. PCR differential display identifies a rat brain mRNA that is transcriptionally regulated by cocaine and amphetamine. The Journal of Neuroscience. 1995;15:2471–2481. doi: 10.1523/JNEUROSCI.15-03-02471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubert GW, Kuhar MJ. Colocalization of CART with substance P but not enkephalin in the rat nucleus accumbens. Brain Res. 2005;1050:8–14. doi: 10.1016/j.brainres.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 41.Koylu EO, Couceyro PR, Lambert PD, Kuhar MJ. Cocaine- and amphetamine-regulated transcript peptide immunohistochemical localization in the rat brain. J Comp Neurol. 1998;391:115–132. doi: 10.1002/(SICI)1096-9861(19980202)391:1<115::AID-CNE10>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 42.Subhedar NK, Nakhate KT, Upadhya MA, Kokare DM. CART in the brain of vertebrates: circuits, functions and evolution. Peptides. 2014;54:108–130. doi: 10.1016/j.peptides.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Kuhar MJ, Jaworski JN, Hubert GW, Philpot KB, Dominguez G. Cocaine- and amphetamine-regulated transcript peptides play a role in drug abuse and are potential therapeutic targets. AAPS J. 2005;7:E259–265. doi: 10.1208/aapsj070125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogge G, Jones D, Hubert GW, Lin Y, Kuhar MJ. CART peptides: regulators of body weight, reward and other functions. Nat Rev Neurosci. 2008;9:747–758. doi: 10.1038/nrn2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaworski JN, Kozel MA, Philpot KB, Kuhar MJ. Intra-accumbal injection of CART (cocaine-amphetamine regulated transcript) peptide reduces cocaine-induced locomotor activity. J Pharmacol Exp Ther. 2003;307:1038–1044. doi: 10.1124/jpet.103.052332. [DOI] [PubMed] [Google Scholar]
- 46.Kim JH, Creekmore E, Vezina P. Microinjection of CART peptide 55–102 into the nucleus accumbens blocks amphetamine-induced locomotion. Neuropeptides. 2003;37:369–373. doi: 10.1016/j.npep.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Moffett MC, Song J, Kuhar MJ. CART peptide inhibits locomotor activity induced by simultaneous stimulation of D1 and D2 receptors, but not by stimulation of individual dopamine receptors. Synapse. 2011;65:1–7. doi: 10.1002/syn.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaworski JN, Hansen ST, Kuhar MJ, Mark GP. Injection of CART (cocaine- and amphetamine-regulated transcript) peptide into the nucleus accumbens reduces cocaine self-administration in rats. Behav Brain Res. 2008;191:266–271. doi: 10.1016/j.bbr.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cadet JL, et al. Enhanced upregulation of CRH mRNA expression in the nucleus accumbens of male rats after a second injection of methamphetamine given thirty days later. PloS one. 2014;9 doi: 10.1371/journal.pone.0084665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chrapusta SJ, Wyatt RJ, Masserano JM. Effects of single and repeated footshock on dopamine release and metabolism in the brains of Fischer rats. J Neurochem. 1997;68:2024–2031. doi: 10.1046/j.1471-4159.1997.68052024.x. [DOI] [PubMed] [Google Scholar]
- 51.Deutch AY, Tam SY, Roth RH. Footshock and conditioned stress increase 3,4-dihydroxyphenylacetic acid (DOPAC) in the ventral tegmental area but not substantia nigra. Brain Res. 1985;333:143–146. doi: 10.1016/0006-8993(85)90134-9. [DOI] [PubMed] [Google Scholar]
- 52.Dunn AJ. Stress-related activation of cerebral dopaminergic systems. Ann N Y Acad Sci. 1988;537:188–205. doi: 10.1111/j.1749-6632.1988.tb42106.x. [DOI] [PubMed] [Google Scholar]
- 53.Kalivas PW, Duffy P. Similar effects of daily cocaine and stress on mesocorticolimbic dopamine neurotransmission in the rat. Biol Psychiatry. 1989;25:913–928. doi: 10.1016/0006-3223(89)90271-0. [DOI] [PubMed] [Google Scholar]
- 54.Kramarcy NR, Delanoy RL, Dunn AJ. Footshock treatment activates catecholamine synthesis in slices of mouse brain regions. Brain Res. 1984;290:311–319. doi: 10.1016/0006-8993(84)90949-1. [DOI] [PubMed] [Google Scholar]
- 55.Cruciani G, et al. Flavin monooxygenase metabolism: why medicinal chemists should matter. J Med Chem. 2014;57:6183–6196. doi: 10.1021/jm5007098. [DOI] [PubMed] [Google Scholar]
- 56.Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther. 2005;106:357–387. doi: 10.1016/j.pharmthera.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Cashman JR. Quantitative analysis of FMO gene mRNA levels in human tissues. Drug Metab Dispos. 2006;34:19–26. doi: 10.1124/dmd.105.006171. [DOI] [PubMed] [Google Scholar]
- 58.Leiser SF, et al. Cell nonautonomous activation of flavin-containing monooxygenase promotes longevity and health span. Science. 2015;350:1375–1378. doi: 10.1126/science.aac9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jha MK, Jeon S, Suk K. Pyruvate Dehydrogenase Kinases in the Nervous System: Their Principal Functions in Neuronal-glial Metabolic Interaction and Neuro-metabolic Disorders. Curr Neuropharmacol. 2012;10:393–403. doi: 10.2174/157015912804499528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jha MK, et al. Pyruvate dehydrogenase kinase 2 and 4 gene deficiency attenuates nociceptive behaviors in a mouse model of acute inflammatory pain. J Neurosci Res. 2016;94:837–849. doi: 10.1002/jnr.23727. [DOI] [PubMed] [Google Scholar]
- 61.Patel MS, Nemeria NS, Furey W, Jordan F. The pyruvate dehydrogenase complexes: structure-based function and regulation. J Biol Chem. 2014;289:16615–16623. doi: 10.1074/jbc.R114.563148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smolle M, Lindsay JG. Molecular architecture of the pyruvate dehydrogenase complex: bridging the gap. Biochem Soc Trans. 2006;34:815–818. doi: 10.1042/BST0340815. [DOI] [PubMed] [Google Scholar]
- 63.Harris RA, Bowker-Kinley MM, Huang B, Wu P. Regulation of the activity of the pyruvate dehydrogenase complex. Adv Enzyme Regul. 2002;42:249–259. doi: 10.1016/S0065-2571(01)00061-9. [DOI] [PubMed] [Google Scholar]
- 64.Nakai N, et al. The abundance of mRNAs for pyruvate dehydrogenase kinase isoenzymes in brain regions of young and aged rats. Life Sci. 2000;68:497–503. doi: 10.1016/S0024-3205(00)00947-4. [DOI] [PubMed] [Google Scholar]
- 65.Halim ND, et al. Phosphorylation status of pyruvate dehydrogenase distinguishes metabolic phenotypes of cultured rat brain astrocytes and neurons. Glia. 2010;58:1168–1176. doi: 10.1002/glia.20996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jha MK, Lee IK, Suk K. Metabolic reprogramming by the pyruvate dehydrogenase kinase-lactic acid axis: Linking metabolism and diverse neuropathophysiologies. Neurosci Biobehav Rev. 2016;68:1–19. doi: 10.1016/j.neubiorev.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 67.Hunter T. The genesis of tyrosine phosphorylation. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tonks NK. Protein tyrosine phosphatases–from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013;280:346–378. doi: 10.1111/febs.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stoker AW. RPTPs in axons, synapses and neurology. Semin Cell Dev Biol. 2015;37:90–97. doi: 10.1016/j.semcdb.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 70.Thomas PE, et al. A renal glomerular epithelial cell (podocyte) membrane protein-tyrosine phosphatase. Identification, molecular cloning, and characterization in rabbit. J Biol Chem. 1994;269:19953–19962. [PubMed] [Google Scholar]
- 71.Beltran PJ, Bixby JL, Masters BA. Expression of PTPRO during mouse development suggests involvement in axonogenesis and differentiation of NT-3 and NGF-dependent neurons. J Comp Neurol. 2003;456:384–395. doi: 10.1002/cne.10532. [DOI] [PubMed] [Google Scholar]
- 72.Kurusu M, Zinn K. Receptor tyrosine phosphatases regulate birth order-dependent axonal fasciculation and midline repulsion during development of the Drosophila mushroom body. Mol Cell Neurosci. 2008;38:53–65. doi: 10.1016/j.mcn.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao WH, et al. Protein tyrosine phosphatase receptor type O (Ptpro) regulates cerebellar formation during zebrafish development through modulating Fgf signaling. Cell Mol Life Sci. 2013;70:2367–2381. doi: 10.1007/s00018-013-1259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krasnova IN, et al. Incubation of methamphetamine and palatable food craving after punishment-induced abstinence. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2014;39:2008–2016. doi: 10.1038/npp.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data have been deposited at the NCBI under the accession #GSE95571.


