Abstract
NK3 homeobox 1 (Nkx3.1), a transcription factor expressed in the prostate epithelium, is crucial for maintaining prostate cell fate and suppressing tumor initiation. Nkx3.1 is ubiquitously expressed in luminal cells of hormonally intact prostate but, upon androgen deprivation, exclusively labels a type of luminal stem cells named castration-resistant Nkx3.1-expressing cells (CARNs). During prostate cancer initiation, Nkx3.1 expression is frequently lost in both humans and mouse models. Therefore, investigating how Nkx3.1 expression is regulated in vivo is important for understanding the mechanisms of prostate stem cell specification and cancer initiation. Here, using a transgenic mouse line with destabilized GFP, we identified an 11-kb genomic region 3′ of the Nkx3.1 transcription start site to be responsible for alterations in Nkx3.1 expression patterns under various physiological conditions. We found that androgen cell-autonomously activates Nkx3.1 expression through androgen receptor (AR) binding to the 11-kb region in both normal luminal cells and CARNs and discovered new androgen response elements in the Nkx3.1 3′ UTR. In contrast, we found that, in Pten−/− prostate tumors, loss of Nkx3.1 expression is mediated at the transcriptional level through the 11-kb region despite functional AR in the nucleus. Importantly, the GFP reporter specifically labeled CARNs in the regressed prostate only in the presence of cell-autonomous AR, supporting a facultative model for CARN specification.
Keywords: androgen, phosphatase and tensin homolog (PTEN), prostate cancer, stem cells, transcription, Nkx3.1
Introduction
The homeodomain-containing protein Nkx3.1 is the earliest known transcription factor expressed in prostate development (1, 2) and orchestrates a transcriptional regulatory network important for prostate cell fate specification (3). The mature prostate gland contains stromal layers and an epithelium that is composed of secretory luminal cells, basal cells, and rare neuroendocrine cells. Nkx3.1 is primarily expressed in luminal epithelial cells of the adult prostate, with low levels present in a small subset of basal cells (4, 5). It plays a tumor-suppressing role, as both homozygous and heterozygous Nkx3.1 mutant mice display increases in epithelial hyperplasia and defects in prostate branching and protein secretion as they age (6–9). In humans, Nkx3.1 down-regulation is considered one of the earliest events in prostate cancer initiation (10). Mechanistically, Nkx3.1 has been shown to play a critical role in protecting prostate cells from DNA damage (11–14).
Despite its functional significance, how Nkx3.1 expression is regulated in normal and tumorigenic prostate in vivo remains elusive. Nkx3.1 mRNA is detected in prostatic buds in E17.5 mouse embryos (2), and studies using urogenital sinus explant culture have demonstrated the involvement of the Fgf10 and Wnt signaling pathways in activating Nkx3.1 expression during prostate organogenesis (15–18). In postnatal and adult prostate, androgen receptor (AR)3 signaling has been shown to maintain Nkx3.1 expression. In particular, androgen deprivation via castration in mice induced prostate regression accompanied by apoptosis in the majority of luminal cells and loss of Nkx3.1 expression in the ones that survived (1, 2, 5), although the relative contribution of cell-autonomous luminal AR versus non-cell-autonomous stromal AR in this process has yet to be determined. Notably, a small subset of the surviving luminal cells retained Nkx3.1 expression in the regressed prostate. Those cells, named castration-resistant Nkx3.1-expressing cells (CARNs), were shown to behave as luminal stem cells that contribute to prostate regeneration upon androgen readministration and could also serve as a cell of origin for prostate cancer (5). How CARNs are specified is unclear; the retention of their Nkx3.1 expression could be due to an intrinsically different cellular program from other luminal cells or, alternatively, stochastically determined by the local microenvironment. Another important question concerning Nkx3.1 expression arises from studies of prostate cancer. Under prostate tumor–initiating conditions, such as the loss of Pten, luminal Nkx3.1 expression is abolished in both human samples and mouse models (19–22). How this is accomplished remains unclear. The decrease of Nkx3.1 mRNA in Pten−/− tumors suggests that the regulation may occur at the transcriptional level (20, 23), whereas the loss of Nkx3.1 protein, but not mRNA, in Pten−/+ tumors suggests the existence of translational or posttranslational regulation (19).
We reason that studying Nkx3.1 transcriptional regulation by analysis of the Nkx3.1 gene locus should shed light on the above questions. A pioneer study using transgenic LacZ reporter mice discovered that a 32-kb fragment containing 20 kb upstream and 12 kb downstream of the Nkx3.1 transcription start site (TSS) could drive the endogenous Nkx3.1 expression pattern in most organs during embryogenesis (4). Within this fragment, the downstream-most 5-kb region acted as a urogenital enhancer that partially restored prostatic Nkx3.1 expression (4). Based on this finding, we hypothesize that change of Nkx3.1 expression in adult prostate in vivo is regulated transcriptionally through its 3′ local genomic region. Our data below support this hypothesis by testing it in the contexts of both prostate regression–regeneration and Pten loss–induced cancer initiation. They also support a facultative model for CARN specification.
Results
An 11-kb Nkx3.1 region drives normal gene expression in adult prostate
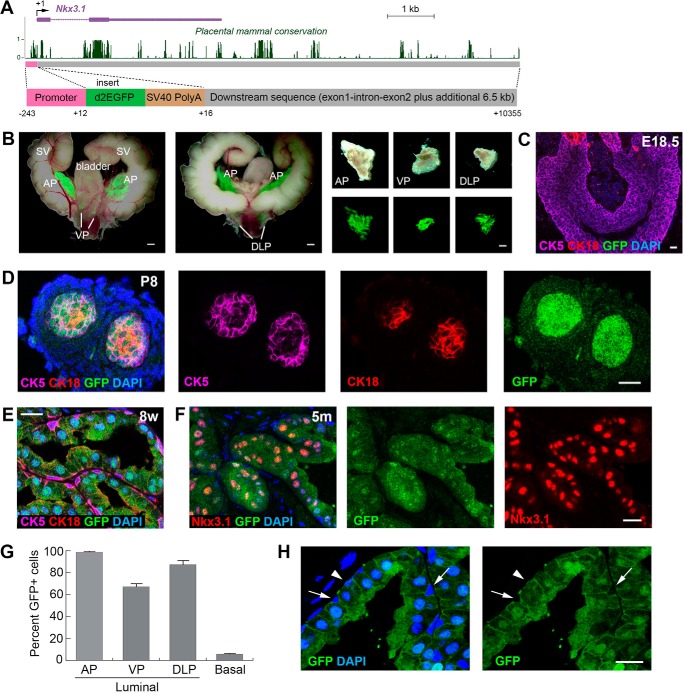
To test the hypothesis that change of prostatic Nkx3.1 expression is mediated transcriptionally through its 3′ genomic sequence in vivo, we took a transgenic reporter mouse approach by constructing an ∼11-kb DNA fragment containing the 240-bp Nkx3.1 proximal promoter, the 4-kb Nkx3.1 gene sequence, and its adjacent 3′ 6.5-kb intergenic region (Fig. 1A). In addition, for visualizing loss of Nkx3.1 expression under androgen deprivation and tumor initiation conditions, we inserted the d2EGFP sequence encoding a destabilized GFP (24) with an SV40 poly(A) sequence right after the start codon (Fig. 1A). With a half-life of 2 h, the d2EGFP should dynamically report the Nkx3.1 transcriptional activities in vivo. The final construct (named Nkx3.1(11)-d2EGFP) was used for pronuclear injection, and five independent transgenic mouse lines were obtained. A robust GFP signal was observed specifically in the adult prostate; four of the five lines showed strong to moderate GFP expression in various prostate lobes, whereas one line showed weak expression (Fig. 1B and supplemental Fig. S1 and Table S1). For subsequent analyses, we focused on transgenic line 18 (Ntg18) because it displayed the strongest expression in all lobes of the prostate (Fig. 1B).
Figure 1.
Nkx3.1(11)-d2EGFP recapitulates Nkx3. 1 expression in adult prostate. A, the DNA construct used for generating Nkx3.1(11)-d2EGFP transgenic mice, showing the relative position of its sequences in the mouse Nkx3.1 gene locus. Conservation of DNA in multiple placental mammal species is shown as in the UCSC genome browser. B, white-field and GFP overlay images of dissected urogenital tissues, showing strong prostate-specific GFP expression in the Ntg18 line. C, immunofluorescence (IF) images showing no GFP reporter expression in E18.5 prostate bud. D, IF showing GFP expression in all prostate epithelial cells at P8. E, IF showing GFP expression primarily in luminal cells at 8 weeks (8w). F, IF showing co-localization of reporter GFP and Nkx3.1 signals in luminal cells at 5 months. G, quantitation of the percentages of GFP+ cells in luminal cells of different lobes and in basal cells. H, direct visualization of the GFP signal, showing that most basal cells were GFP-negative (arrows), whereas a small proportion was GFP+ (arrowheads) in adult prostate. SV, seminal vesicle. Scale bars = 1 mm (B) and 20 μm (C–F and H). Error bars correspond to 1 S.D.
We next examined reporter gene expression during embryogenesis and prostate development. No GFP was observed in somites of E10.5 embryos in any of the lines, consistent with a previous report showing that key elements for somite Nkx3.1 expression lie in the 5′ region of the gene locus (4). We also did not detect GFP in E18.5 prostate buds, where cells mostly expressed the basal marker CK5 (Fig. 1C). GFP began to be expressed in the prostate around neonatal stage P4. By neonatal stage P8, GFP was expressed throughout the prostate epithelium, which contained basal (CK5+), luminal (CK18+) and intermediate cells (CK5+CK18+) and co-localized with Nkx3.1 expression (Fig. 1D and supplemental Fig. S2A). Throughout the adult stage, GFP expression was persistently present in the luminal layer and co-localized with Nkx3.1 expression (Fig. 1, E and F). The proportions of luminal cells that were GFP+ in the anterior, ventral, and dorsal-lateral prostate lobes (AP, VP, and DLP, respectively) were 98.2% (n = 2019 of 2055), 67.2% (n = 839 of 1248), and 87.1% (n = 1283 of 1473), respectively (Fig. 1G and supplemental Fig. S2, B–D). Notably, ∼5.8% (n = 95 of 1638) of basal cells were also GFP+ (Fig. 1, G and H), in agreement with the percentage of Nkx3.1+ basal cells reported previously (4, 5). Overall, Nkx3.1(11)-d2EGFP recapitulates the endogenous Nkx3.1 expression pattern in adult prostate homeostasis.
Androgen activates Nkx3.1 transcription through cell-autonomous AR in both luminal cells and CARNs
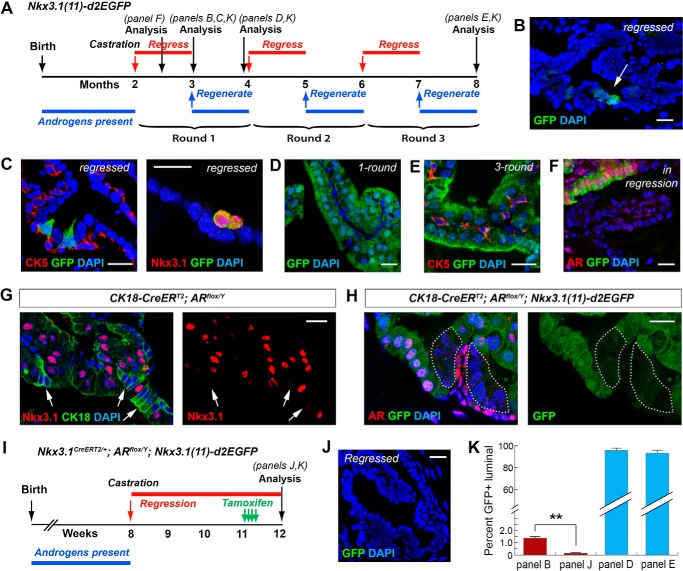
To test whether androgen regulates Nkx3.1 expression through the 11-kb region, we castrated Nkx3.1(11)-d2EGFP mice at 8 weeks of age and performed three rounds of androgen-mediated serial prostate regression–regeneration (Fig. 2A). We found that the GFP signal was dramatically decreased in the regressed prostate (Fig. 2, B and C), consistent with loss of Nkx3.1 expression in most luminal cells upon androgen deprivation. Importantly, cells that retained GFP expression were strictly luminal and expressed Nkx3.1 (Fig. 2C), and they comprised 1.4% (n = 29 of 2053) of all luminal cells (Fig. 2K), suggesting that they correspond to the previously identified CARNs (5). Upon androgen readministration, the GFP signal was again detected in most luminal cells (95.6%, n = 1883 of 1970) of the regenerated prostate, and the same results were observed for both one-round and three-round regeneration experiments (Fig. 2, D, E, and K). These data demonstrate that the Nkx3.1 11-kb region is fully responsive to androgen regulation.
Figure 2.
AR-dependent Nkx3. 1 expression in luminal cells and CARNs revealed by Nkx3.1(11)-d2EGFP during prostate regression–regeneration. A, timeline of the serial prostate regression–regeneration experiment for Nkx3.1(11)-d2EGFP mice. B, direct visualization showing relatively few GFP+ cells (arrow) in the regressed prostate. C, IF images showing that the rare GFP+ cells in the regressed prostate were luminal (CK5−, left) and Nkx3.1+ (right). D, direct visualization showing that most luminal cells recovered GFP expression after one-round prostate regeneration. E, IF image showing that most luminal cells were GFP+ after three-round regression–regeneration. F, IF image showing co-localization of GFP and nuclear AR in prostate that is undergoing regression. G, IF image showing loss of Nkx3.1 expression in AR-null luminal cells (revealed by enhanced CK18 expression, arrows). H, the GFP reporter signal was strongly reduced in AR-null luminal cells in the AP of tamoxifen-induced CK18-CreERT2; ARflox/Y; Nkx3.1(11)-d2EGFP mice. I, timeline of the experiment for AR ablation in CARNs in Nkx3.1CreERT2/+; ARflox/Y; Nkx3.1(11)-d2EGFP mice. J, almost no GFP signal was observed by direct visualization of the prostate in I. K, quantitation of the proportions of GFP+ cells among total luminal cells for the experiments in B, D, E, and J. **, p < 0.01 by Student's t test. Scale bars = 20 μm. Error bars correspond to 1 S.D.
Androgen-regulated luminal Nkx3.1 expression could be mediated cell-autonomously through luminal AR and/or through paracrine signals induced by stromal AR. To distinguish these two possibilities, we analyzed prostate in the midst of regression. We found that, 2 weeks after castration of Nkx3.1(11)-d2EGFP mice (Fig. 2A), the shrinking prostate showed a mosaic GFP expression pattern, with the GFP+ regions overlapping with luminal nuclear AR signals (Fig. 2F). To further confirm the cell-autonomous role of androgen, we deleted AR specifically in luminal cells using the CK18-CreERT2 driver (25) by tamoxifen induction of 8-week-old CK18-CreERT2; ARflox/Y mice. Two weeks after induction, AR-null luminal cells could be identified as clusters with condensed nuclei and enhanced membrane CK18 expression, as we reported previously (26), and we found that Nkx3.1 expression was diminished in those cells (Fig. 2G). Direct staining of AR and Nkx3.1 in adjacent sections also confirmed this result (supplemental Fig. S3A). Importantly, we found that the GFP signal was significantly reduced in AR-null luminal cells of tamoxifen-induced CK18-CreERT2; ARflox/Y; Nkx3.1(11)-d2EGFP mice (Fig. 2H and supplemental Fig. S3B), indicating that luminal cell-autonomous AR directly activates Nkx3.1 transcription through the 11-kb region. To determine whether such a mechanism also applies to CARNs, we ablated AR specifically in CARNs using the Nkx3.1CreERT2/+ driver (5) in the fully regressed prostate and analyzed GFP reporter activity. In Nkx3.1CreERT2/+; ARflox/Y; Nkx3.1(11)-d2EGFP mice that first underwent castration and then tamoxifen induction (Fig. 2I), the percentage of GFP+ luminal cells in the regressed prostate was dramatically reduced by almost 10-fold to 0.16% (n = 4 of 2450) (Fig. 2, J and K), and no Nkx3.1+ cells were found by immunofluorescence staining. Therefore, Nkx3.1 expression in both luminal cells and CARNs are transcriptionally regulated by cell-autonomous AR through the 11-kb region.
AR preferentially binds to the Nkx3.1 3′ gene locus in vivo and in vitro
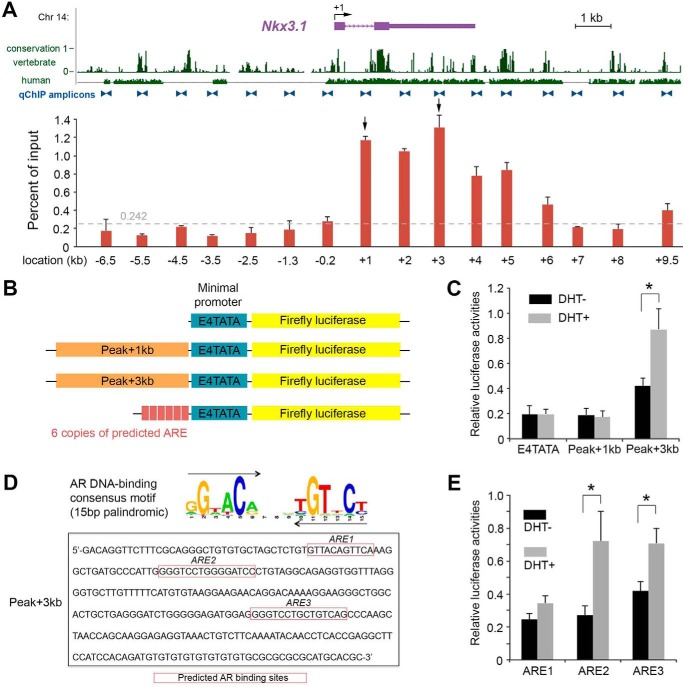
To confirm that AR interacts with the Nkx3.1 3′ gene locus, we next performed quantitative ChIP (qChIP) analyses across a 17-kb Nkx3.1 region that encompasses both 7-kb upstream and 10-kb downstream sequences to its TSS (Fig. 3A). We chose to analyze this region because it was shown previously to be capable of driving prostatic Nkx3.1 expression (4). Consistent with our reporter mouse data, we found that AR had low affinity for the region 5′ to the Nkx3.1 TSS but, instead, occupied multiple loci in the 3′ 11-kb region (Fig. 3A). In particular, our statistical analysis (see “Experimental procedures”) highlighted two binding sites with the highest affinity in the 3′ UTR (“Peak+3kb”) and the intron (“Peak+1kb”), respectively (Fig. 3A). To test whether these two peaks could serve as transcriptional enhancers, we performed a luciferase reporter assay by transfection of synthetic constructs containing an E4TATA minimal promoter in combination with the peak regions into prostate LnCaP cells (Fig. 3B), which express AR and are androgen-responsive (27). Upon dihydrotestosterone (DHT) treatment, luciferase activity for the construct containing the ∼290-bp Peak+3kb sequence was significantly enhanced, whereas no difference was observed for the construct containing the Peak+1kb sequence (Fig. 3C). To test any potential spatial effect of the binding site arrangement, we also generated constructs in which the Peak+1kb and Peak+3kb sequences were positioned downstream of the luciferase sequence and obtained similar results (supplemental Fig. S4). We next used the FIMO software (28) to search for potential AR-responsive elements (AREs) within the Peak+3kb fragment based on the AR DNA-binding consensus motif obtained from TRANSFAC (29). Three new candidate AREs were identified, with ARE1 being a composite site containing two 6-bp half-sites with one overlapping nucleotide and ARE2 and ARE3 both being optimal palindromic sequences 15 bp in length (Fig. 3D). In a series of luciferase assays using synthetic reporters containing six copies of the individual predicted AREs (Fig. 3B), we found that both ARE2 and ARE3, but not ARE1, significantly enhanced luciferase activities upon DHT treatment (Fig. 3E). Cumulatively, these molecular analyses support that AR directly binds to the 11-kb region, particularly the Nkx3.1 3′ UTR, to activate Nkx3.1 transcription.
Figure 3.
AR binds to the Nkx3. 1 11-kb 3′ region to enhance its gene expression. A, distribution of AR detected by qChIP analysis over the 17-kb mouse Nkx3.1 locus in prostate chromatin. The relative enrichments are shown as the percentage of the DNA input. Locations of primer pairs for qChIP amplicons are marked by blue triangles. Conservations of DNA in vertebrate species and in human are shown as in the UCSC genome browser. Specific AR enrichments above the background noise (0.242, dashed gray line, see “Experimental procedures” for a description) were concentrated in the 11-kb region 3′ next to the TSS, with the highest two peaks at locations +3kb and +1kb, respectively (arrows). B, constructs for transient transfection and luciferase reporter assays. C, luciferase reporter assay comparing the construct with the minimal promoter with those containing Peak+1kb and Peak+3kb sequences, showing that the Peak+3kb region is androgen-responsive. D, sequences of three candidate AREs in the Peak+3kb region. E, luciferase assay comparing constructs containing six copies of each predicted ARE, showing that ARE2 and ARE3 are androgen-responsive. Error bars correspond to 1 S.D. *, p < 0.05 by Student's t test.
Pten promotes Nkx3.1 expression in vivo through transcriptional regulation of the 11-kb region
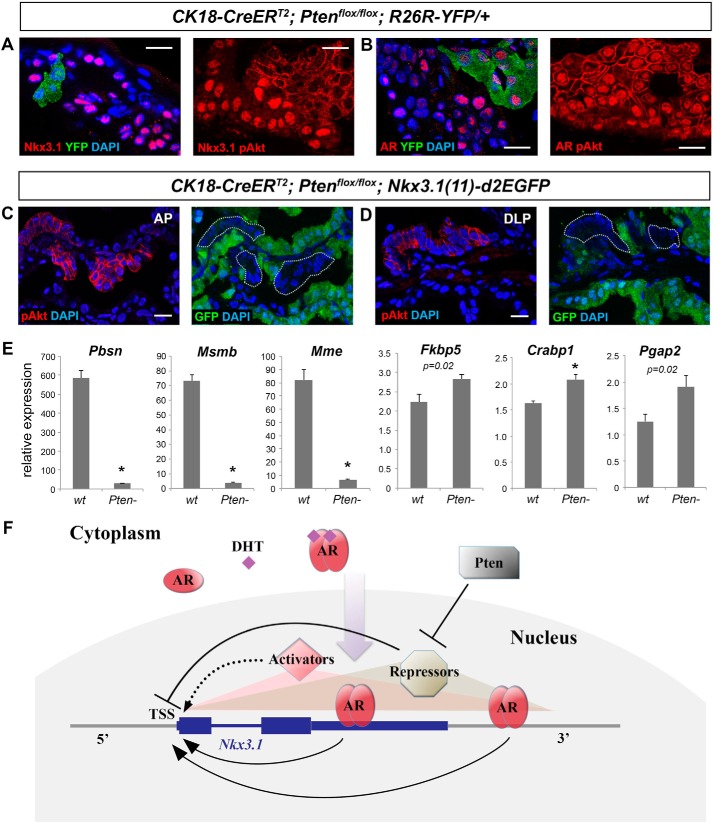
Finally, we sought to address the question how loss of the tumor suppressor Pten leads to down-regulation of Nkx3.1 expression in prostate tumors (19–21). We generated lineage-marked Pten-null prostate cell clones that were identifiable as YFP+ or phospho-Akt+ by tamoxifen-induction of CK18-CreERT2; Ptenflox/flox; R26R-YFP/+ mice and confirmed that Nkx3.1 immunostaining was abolished in those cells (Fig. 4A). Notably, this phenomenon is not due to AR loss because strong nuclear AR staining was still present in the Pten-null cells (Fig. 4B). Many potential causes could result in this phenotype, including decrease of Nkx3.1 transcription, Nkx3.1 mRNA degradation/modification, and Nkx3.1 protein degradation. To determine whether Pten regulation of Nkx3.1 expression occurs at the transcriptional level, we analyzed GFP reporter expression in tamoxifen-induced CK18-CreERT2; Ptenflox/flox; Nkx3.1(11)-d2EGFP mice. Pten-null clones were identified by positivity of phospho-Akt staining 2 weeks after induction (Fig. 4, C and D). Strikingly, the GFP signal was abolished in those cells but not in neighboring wild-type cells (Fig. 4, C and D). Triple staining of AR, phospho-Akt, and GFP also confirmed the above findings (supplemental Fig. S5). To determine whether the nuclear AR is functional in Pten-null cells, we isolated wild-type and Pten-null luminal cells by flow-sorting of YFP+ cells from tamoxifen-induced CK18-CreERT2; R26R-YFP/+ and CK18-CreERT2; Ptenflox/flox; R26R-YFP/+ mice, respectively, and performed qRT-PCR for several well-established prostate AR target genes: Crabp1, Fkbp5, Mme, Msmb, Pbsn, and Pgap2 (23, 30). Consistent with previous reports (23), we found that expression of Pbsn, Msmb, and Mme was significantly reduced in Pten-null cells (Fig. 4E). However, expression of Fkbp5, Crabp1, and Pgap2 was unchanged or modestly up-regulated (Fig. 4E), indicating that AR is transcriptionally active in these cells and that additional transcriptional repressors are involved in suppressing a subset of AR target genes, including Nkx3.1.
Figure 4.
Transcriptional regulation of Nkx3. 1 by Pten and the overall model. A, IF images showing loss of Nkx3.1 expression in Pten-null luminal cells marked by either YFP positivity (left panel) or phospho-Akt expression (right panel) of tamoxifen-induced CK18-CreERT2; Ptenflox/flox; R26R-YFP/+ mice. Phospho-Akt staining (red) can be distinguished from Nkx3.1 or AR staining (red) by its membrane localization. B, IF images showing nuclear AR expression in Pten-null luminal cells. C and D, adjacent sections of phospho-Akt staining and GFP direct visualization in AP (C) and DLP (D) of tamoxifen-induced CK18-CreERT2; Ptenflox/flox; Nkx3.1(11)-d2EGFP mice, showing that the GFP signal is abolished in Pten-null cells. Scale bars = 20 μm. E, qRT-PCR analyses comparing the expression levels of selected AR target genes in wild-type and Pten-null luminal cells by Welch t test. *, p < 0.01. Error bars correspond to 1 S.D. F, model of Nkx3.1 transcriptional regulation in adult prostate epithelial cells. Androgen cell-autonomously maintains Nkx3.1 expression in adult prostate epithelial cells. Upon DHT binding, AR proteins in the cytoplasm dimerize and translocate into the nucleus. They preferentially bind to the 11-kb Nkx3.1 3′ region to the TSS, particularly the 3′ UTR, to activate Nkx3.1 transcription. Other transcription activators also bind to this region (shaded red triangle) and play a minor role in activating Nkx3.1 transcription (dashed arrow). Transcriptional repressors that are normally inhibited by Pten can also bind to this region (shaded yellow triangle) and strongly inhibit Nkx3.1 transcription, even in the presence of nuclear AR.
Discussion
In this study, we show that the 11-kb region 3′ to the Nkx3.1 TSS is responsible for Nkx3.1 expression alterations in adult prostate under various physiological conditions (Fig. 4F). The 11-kb transgene was able to recapitulate the normal Nkx3.1 expression pattern in adult prostate and induce the same levels of expression as the previously characterized 32-kb and 17-kb transgenes (4), suggesting that the 5′ sequence of the Nkx3.1 locus is dispensable for this purpose. Notably, all three fragments induced Nkx3.1 expression in most but not all luminal cells of the DLP and VP, suggesting that a distant cis-regulatory elements exist beyond the 32-kb region to strengthen Nkx3.1 expression in those lobes. In contrast, the 11-kb fragment induced much stronger expression in all lobes than the previously identified 5-kb urogenital enhancer at its distal 3′ location (4), suggesting that the 3′ region adjacent to the Nkx3.1 TSS is also important. Indeed, our qChIP experiments identified multiple AR binding sites in vivo in the 11-kb region but not in the 5′ region. In particular, the Peak+3kb sequence in the 3′ UTR was discovered as a new important androgen response region. Using prostate cancer LnCaP cells, previous studies have identified potential AREs in various locations of the Nkx3.1 locus, including the intron (31), the 3′ UTR (32, 33), and a site 5′ to the Nkx3.1 TSS (34). However, whether those sites are functional in vivo are unknown. Using reporter mice, our prostate regression–regeneration experiments and AR conditional knock-out experiments showed that androgen regulation of Nkx3.1 expression in the adult prostate is primarily mediated by cell-autonomous AR through the 11-kb region. We note that the reporter GFP signal was not completely abolished in some AR-null luminal cell clones, indicating that other transcription factors can also bind to the 11-kb region to enhance Nkx3.1 expression in the absence of AR (Fig. 4F).
An interesting finding from our study is that the reporter mice labeled rare GFP+ luminal cells in the regressed prostate. The frequency of those cells and their positivity for Nkx3.1 suggest that they are the previously identified luminal stem cell CARNs. CARNs are AR+ (5), and we recently showed that they require AR for proper daughter cell differentiation (26). However, how CARNs are specified remains unclear. Is their exclusive Nkx3.1 expression in the regressed prostate a result of AR expression or some other intrinsic properties that are different from the rest of the luminal cells? Our conditional knock-out and reporter mouse experiments indicate that Nkx3.1 expression in CARNs is also mediated by cell-autonomous AR through the 11-kb region. We hypothesize that, after castration, residual levels of androgen secreted from the adrenal gland may induce AR nuclear translocation in a few localized niches, thereby specifying CARNs in the regressed prostate and sensitizing them to AR levels. Such a model would argue that CARNs are facultative stem cells dependent on microenvironmental cues (local androgen) rather than a pre-existing fixed population. This model can also explain the previously reported phenomena that CARNs tend to be clustered and that some initially YFP+ lineage-marked CARNs lost Nkx3.1 expression over time (5). Our Nkx3.1(11)-d2EGFP mice should provide a useful and convenient tool for isolation of bona fide CARNs from the regressed prostate at any particular time and studying their molecular and functional properties. Notably, CARNs are not the only source for prostate luminal cell regeneration because many regressed luminal cells contribute to this process (22, 35), and Bmi1+ cells were recently shown to be a distinct type of luminal progenitor cells competent for regeneration (36). Future work should shed light on how this new type of progenitor cell is specified and its relationship with CARNs.
Nkx3.1 down-regulation has been reported in a variety of mouse prostate cancer models (20, 21, 37, 38) and is well documented in human prostate cancer initiation (10). In human prostate cancer samples, decreases in Nkx3.1 and Pten expression levels were reported to be significantly correlated (21, 39). Our data showing that loss of Pten abolished the GFP reporter signal provide definitive evidence that Pten promotes Nkx3.1 expression at the transcriptional level. The presence of nuclear AR in Pten-null clones as well as the down-regulation of a subset but not all of the AR target genes indicate that AR protein is still functional but that unknown transcriptional repressors activated by the PI3K pathway are involved and exert their negative effects through the 11-kb region (Fig. 4F). It was proposed that Egr1, c-Jun, and Ezh2 are up-regulated upon Pten deletion and interact with AR to suppress its transcriptional output (23). Further analyses of the 11-kb sequence and molecular analyses of Pten-null luminal cells should provide potential candidates to test. It will also be interesting to determine whether similar transcriptional regulatory mechanisms exist for Nkx3.1 down-regulation under other tumor-initiating conditions.
Experimental procedures
Generation of Nkx3.1(11)-d2EGFP mice
The Nkx3.1(11)-d2EGFP construct is comprised of the 10.5-kb mouse genomic sequence (UCSC genome browser, mm9 Chr 14: 69808515-69819104) at the Nkx3.1 locus (−234/+10356) containing a 240-bp proximal promoter, exon 1, intron, exon 2, and ∼6.5-kb 3′ region as well as the d2EGFP (Clontech) and SV40 poly(A) sequences inserted right after the Nkx3.1 start codon. The construct (11671-bp total length) was synthesized by GenScript and cloned into a pUC57 vector. MluI and XhoI restriction enzyme sites were engineered at both ends of the synthesized sequence for plasmid linearization. The transgenic mouse lines were generated by pronuclear injection of fertilized eggs by Cyagen Biosciences.
Mouse genotyping and operations
The CK18-CreERT2 transgenic line (25), Nkx3.1CreERT2/+ targeted allele (5), ARflox allele (40), Ptenflox allele (41), and R26R-YFP line (42) were described previously. Animals were maintained in C57BL/6N background. Genotyping was performed by PCR using tail genomic DNA; primer sequences are listed in supplemental Table S2. The Nkx3.1(11)-d2EGFP transgene was genotyped by a set of primers amplifying the junction between the proximal promoter and the d2EGFP sequence. The following primers generated a 319-bp PCR product: 5′-AAGGGCTCTGGAGCCTAATC-3′ (forward) and 5′-GAACTTCAGGGTCAGCTTGC-3′ (reverse).
For tamoxifen induction, mice were administered 9 mg/40 g of body weight tamoxifen (Sigma) suspended in corn oil by oral gavage once daily for 4 consecutive days. Castration of adult male mice was performed using standard techniques. For prostate regeneration, testosterone (Sigma) was dissolved at 25 mg/ml in 100% ethanol and diluted in PEG-400 to a final concentration of 7.5 mg/ml. Testosterone was administered for 4 weeks at a rate of 1.875 μg/h, delivered by subcutaneous implantation of mini-osmotic pumps (Alzet). All animal experiments received approval from the Institutional Animal Care and Use Committee at UCSC.
GFP visualization, immunofluorescence staining, and cell quantitation
Prostate tissue dissection and direct GFP visualization were performed under a Nikon SMZ-1000 stereomicroscope with fluorescence and charge-coupled device digital camera. Tissues were fixed in 4% paraformaldehyde for subsequent cryo-embedding in OCT compound (Sakura). Immunofluorescence staining was performed as described previously (26). Slides were mounted with VectaShield mounting medium with DAPI (Vector Laboratories). Images were obtained using a Leica TCS SP5 spectral confocal microscope in the Microscopy Shared Facility of UCSC. All primary antibodies and dilutions used are listed in supplemental Table S3. Nkx3.1, AR, and pAkt primary antibodies were all raised in rabbits, but their expression can be distinguished in co-staining because pAkt signal is on the membrane, whereas AR and Nkx3.1 signals are nuclear. For AR and Nkx3.1 co-staining, adjacent cryosections of 3-μm thickness were used.
Cell numbers were counted manually using confocal ×40 and ×63 photomicrographs across tissue sections. Three animals were counted for each group. Basal cells were identified based on lack of CK18 staining, positivity for CK5 staining, and/or the shape of the cells (oval or triangular) and their positions at the basement of the epithelium. Luminal cells were determined based on positive CK18 staining and/or the shape of the cells (columnar) and their positions at the apical side of the epithelium.
Flow cytometry
To sort luminal cells, lineage-marked prostate tissues were dissected and minced into small clumps, followed by enzymatic dissociation with 0.2% collagenase I (Invitrogen) in DMEM with 5% FBS for 3 h at 37 °C. Tissues were digested with 0.25% trypsin-EDTA (StemCell Technologies) for 1 h at 4 °C, passed through 21- to 26-gauge syringes, and filtered through a 40-μm cell strainer to obtain single-cell suspensions. Dissociated prostate cells were suspended in Hanks' balanced salt solution modified/2% FBS. The Rho-associated protein kinase (ROCK) inhibitor Y-27632 (StemCell Technologies) was added at 10 μm throughout the whole process to inhibit luminal cell death, and dead cells were excluded by propidium iodide staining. Luminal cells were sorted based on YFP positivity on a BD FACS Aria II instrument in the flow cytometry shared facility of UCSC.
Quantitative real-time PCR analysis
Total RNA from FACS-purified luminal cells was isolated using the RNeasy micro kit (Qiagen) and reverse-transcribed into cDNA using SuperScript IV VILO Master Mix (Invitrogen). Quantitative real-time PCR was carried out using Power SYBR Green PCR Master Mix (Life Technology) in the ViiA 7 real-time PCR instrument. cDNA samples were diluted 1:100 for all analyses, which were performed in quadruplicate. Expression values were obtained using the ΔΔCT method and normalized to β-actin expression; average values are shown as the mean ± S.D. Welch two-sample t test was performed. The primer sequences for qRT-PCR are provided in supplemental Table S4.
Quantitative ChIP and statistics
The Magna ChIPTM HiSens chromatin immunoprecipitation kit (Millipore) was used according to the instructions of the manufacturer. Formaldehyde cross-linked chromatin was obtained from adult mouse prostate and sheared by sonication (average size, 600 bp). Aliquots of chromatin from 2 mg of prostate tissue were incubated with 2 μg of anti-AR antibody (Sigma, catalog no. A9853) bound to 40 μl of protein A/G-coated magnetic beads from the kit. The immunoprecipitates were washed three times by SCW buffer of the kit with protease inhibitor mixture and resuspended in ChIP elution buffer. After 1 h of incubation at 55 °C, the cross-links were reversed by 3-h incubation at 65 °C. Genomic DNA was resuspended into 150 μl of water.
The amounts of each specific DNA fragment in immunoprecipitates were determined by quantitative PCR reactions using a standard curve generated for each primer set with 0.04%, 0.2%, and 1% input DNA samples. The primer sequences are provided in supplemental Table S5. The PCR products were between 80 and 120 bp. Reactions were carried out using the ViiATM 7 real-time PCR system and 2× SYBR mixture (ABI). Cycle threshold values were transformed into DNA copy numbers using a standard curve. The copy number of each specific DNA fragment was compared with the value before immunoprecipitation (input DNA). The control antibody (rabbit normal IgG, Millipore, catalog no. CS200581) was included for each set of the quantitative PCR experiments. The enrichment obtained with IgG was subtracted from the corresponding value obtained with anti-AR antibody. To determine a threshold value that distinguishes real binding signals from background noise, statistical analysis was conducted using R Software (version 2.13.1). Analysis of variance was performed for the AR signals obtained from all 16 qChIP amplicon sites (seven amplicons 5′ to and nine amplicons 3′ to the TSS), and Fisher's least significant difference test was performed to separate the 16 sites into the following six groups with a descending order in the signal intensity: +3 kb (3′ UTR) > +1 kb (intron), +2 kb (3′ UTR) > +5 kb, +4 kb > +6 kb, +10 kb > −0.2 kb > the other eight sites (background noise). Student's t test for the last group showed the 99% confidence interval of the background signal value to be 0.107–0.242, indicating that all regions having signals higher than 0.242 should be considered specific AR-binding regions.
Plasmid construction, transfection, and luciferase reporter assays
The sequences of the Peak+1kb and Peak+3kb regions and six copies of the predicted AREs with a minimal E4TATA promoter were synthesized by GenScript and then subcloned into the firefly luciferase reporter vector in the pGL3-Basic plasmid (Promega). Transient transfections were conducted by Lipofectamine 2000 (Invitrogen) in LnCaP cells (ATCC, CRL-1740). The cells were cultured in RPMI 1640 medium with 10% FBS in 24-well plates for 36 h before transfection to ensure complete cell attachment and growth to ∼70% confluence. For transfection, 1.6 μg of firefly luciferase reporter plasmid and 40 ng of Renilla-TK (Promega) internal control plasmid were mixed with 2.5 μl of Lipofectamine 2000 in 100 μl of Opti-MEM (Life Technologies). The mixture was then incubated with the cells for 5 h before being replaced with cell culture medium with or without 10−6 m DHT. The Dual-Luciferase reporter assays (Promega) were performed after 48 h of DHT stimulation according to the instructions of the manufacturer and were repeated twice in triplicates. Statistical evaluation of luciferase activities was performed using two-sided Student's t test.
Prediction of AREs in the Peak+3kb region
To search for candidate AREs in the Peak+3kb region, Find Individual Motif Occurrence (FIMO), a motif-based sequence analysis software of MEME Suite 4.11.3 (http://meme-suite.org/index.html)4 (43) was run with a 15-bp AR consensus motif (V$AR_01 - AR) from TRANSFAC, a manually curated database of eukaryotic transcription factor binding sites. The 15-bp optimal AR binding motif is a palindromic sequence containing two 6-bp AR binding half-sites separated by 3-bp random nucleotides with a head-to-head arrangement. Prediction was also performed by FIMO using the AR binding half-site sequence. Filter criteria with the match p < 10−4 was used to identify the three potential AREs.
Author contributions
Q. X. conducted the experiments. Q. X. and Z. A. W. conceived the project, analyzed the data, and wrote the manuscript.
Supplementary Material
Acknowledgment
We thank UCSC for a special research grant enabling generation of the Nkx3.1(11)-d2EGFP mice.
This work was supported by National Institutes of Health Grant GM116872 (to Z. A. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S5 and Tables S1–S5.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- AR
- androgen receptor
- CARN
- castration-resistant Nkx3.1-expressing cell
- TSS
- transcription start site
- E17.5
- embryonic day 17.5
- P4
- postnatal day 4
- AP
- anterior prostate
- VP
- ventral prostate
- DLP
- dorsal-lateral prostate
- qChIP
- quantitative ChIP
- DHT
- dihydrotestosterone
- ARE
- androgen receptor–responsive element
- qRT-PCR
- quantitative real-time PCR.
References
- 1. Bieberich C. J., Fujita K., He W. W., and Jay G. (1996) Prostate-specific and androgen-dependent expression of a novel homeobox gene. J. Biol. Chem. 271, 31779–31782 [DOI] [PubMed] [Google Scholar]
- 2. Sciavolino P. J., Abrams E. W., Yang L., Austenberg L. P., Shen M. M., and Abate-Shen C. (1997) Tissue-specific expression of murine Nkx3.1 in the male urogenital system. Dev. Dyn. 209, 127–138 [DOI] [PubMed] [Google Scholar]
- 3. Dutta A., Le Magnen C., Mitrofanova A., Ouyang X., Califano A., and Abate-Shen C. (2016) Identification of an NKX3.1-G9a-UTY transcriptional regulatory network that controls prostate differentiation. Science 352, 1576–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen H., Mutton L. N., Prins G. S., and Bieberich C. J. (2005) Distinct regulatory elements mediate the dynamic expression pattern of Nkx3.1. Dev. Dyn. 234, 961–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang X., Kruithof-de Julio M., Economides K. D., Walker D., Yu H., Halili M. V., Hu Y. P., Price S. M., Abate-Shen C., and Shen M. M. (2009) A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 461, 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhatia-Gaur R., Donjacour A. A., Sciavolino P. J., Kim M., Desai N., Young P., Norton C. R., Gridley T., Cardiff R. D., Cunha G. R., Abate-Shen C., and Shen M. M. (1999) Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 13, 966–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tanaka M., Komuro I., Inagaki H., Jenkins N. A., Copeland N. G., and Izumo S. (2000) Nkx3.1, a murine homolog of Drosophila bagpipe, regulates epithelial ductal branching and proliferation of the prostate and palatine glands. Dev. Dyn. 219, 248–260 [DOI] [PubMed] [Google Scholar]
- 8. Schneider A., Brand T., Zweigerdt R., and Arnold H. (2000) Targeted disruption of the Nkx3.1 gene in mice results in morphogenetic defects of minor salivary glands: parallels to glandular duct morphogenesis in prostate. Mech. Dev. 95, 163–174 [DOI] [PubMed] [Google Scholar]
- 9. Abate-Shen C., Banach-Petrosky W. A., Sun X., Economides K. D., Desai N., Gregg J. P., Borowsky A. D., Cardiff R. D., and Shen M. M. (2003) Nkx3.1; Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res. 63, 3886–3890 [PubMed] [Google Scholar]
- 10. Shen M. M., and Abate-Shen C. (2010) Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 24, 1967–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bowen C., and Gelmann E. P. (2010) NKX3.1 activates cellular response to DNA damage. Cancer Res. 70, 3089–3097 [DOI] [PubMed] [Google Scholar]
- 12. Bowen C., Ju J. H., Lee J. H., Paull T. T., and Gelmann E. P. (2013) Functional activation of ATM by the prostate cancer suppressor NKX3.1. Cell Rep. 4, 516–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bowen C., Zheng T., and Gelmann E. P. (2015) NKX3.1 Suppresses TMPRSS2-ERG gene rearrangement and mediates repair of androgen receptor-induced DNA damage. Cancer Res. 75, 2686–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang H., Zheng T., Chua C. W., Shen M., and Gelmann E. P. (2016) Nkx3.1 controls the DNA repair response in the mouse prostate. Prostate 76, 402–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang L., Pu Y., Alam S., Birch L., and Prins G. S. (2005) The role of Fgf10 signaling in branching morphogenesis and gene expression of the rat prostate gland: lobe-specific suppression by neonatal estrogens. Dev. Biol. 278, 396–414 [DOI] [PubMed] [Google Scholar]
- 16. Simons B. W., Hurley P. J., Huang Z., Ross A. E., Miller R., Marchionni L., Berman D. M., and Schaeffer E. M. (2012) Wnt signaling though β-catenin is required for prostate lineage specification. Dev. Biol. 371, 246–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Francis J. C., Thomsen M. K., Taketo M. M., and Swain A. (2013) β-catenin is required for prostate development and cooperates with Pten loss to drive invasive carcinoma. PLoS Gen. 9, e1003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Julio M. K., Shibata M., Desai N., Reynon M., Halili M. V., Hu Y. P., Price S. M., Abate-Shen C., and Shen M. M. (2013) Canonical Wnt signaling regulates Nkx3.1 expression and luminal epithelial differentiation during prostate organogenesis. Dev. Dyn. 242, 1160–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim M. J., Cardiff R. D., Desai N., Banach-Petrosky W. A., Parsons R., Shen M. M., and Abate-Shen C. (2002) Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 2884–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang S., Gao J., Lei Q., Rozengurt N., Pritchard C., Jiao J., Thomas G. V., Li G., Roy-Burman P., Nelson P. S., Liu X., and Wu H. (2003) Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 4, 209–221 [DOI] [PubMed] [Google Scholar]
- 21. Lei Q., Jiao J., Xin L., Chang C. J., Wang S., Gao J., Gleave M. E., Witte O. N., Liu X., and Wu H. (2006) NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell 9, 367–378 [DOI] [PubMed] [Google Scholar]
- 22. Wang Z. A., Mitrofanova A., Bergren S. K., Abate-Shen C., Cardiff R. D., Califano A., and Shen M. M. (2013) Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 15, 274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mulholland D. J., Tran L. M., Li Y., Cai H., Morim A., Wang S., Plaisier S., Garraway I. P., Huang J., Graeber T. G., and Wu H. (2011) Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 19, 792–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X., Zhao X., Fang Y., Jiang X., Duong T., Fan C., Huang C. C., and Kain S. R. (1998) Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 273, 34970–34975 [DOI] [PubMed] [Google Scholar]
- 25. Van Keymeulen A., Mascre G., Youseff K. K., Harel I., Michaux C., De Geest N., Szpalski C., Achouri Y., Bloch W., Hassan B. A., and Blanpain C. (2009) Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. J. Cell Biol. 187, 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xie Q., Liu Y., Cai T., Horton C., Stefanson J., and Wang Z. A. (2017) Dissecting cell-type-specific roles of androgen receptor in prostate homeostasis and regeneration through lineage tracing. Nat. Commun. 8, 14284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horoszewicz J. S., Leong S. S., Kawinski E., Karr J. P., Rosenthal H., Chu T. M., Mirand E. A., and Murphy G. P. (1983) LNCaP model of human prostatic carcinoma. Cancer Res. 43, 1809–1818 [PubMed] [Google Scholar]
- 28. Grant C. E., Bailey T. L., and Noble W. S. (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wingender E., Dietze P., Karas H., and Knüppel R. (1996) TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 24, 238–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang X. D., Wang B. E., Soriano R., Zha J., Zhang Z., Modrusan Z., Cunha G. R., and Gao W. Q. (2007) Expression profiling of the mouse prostate after castration and hormone replacement: implication of H-cadherin in prostate tumorigenesis. Differentiation 75, 219–234 [DOI] [PubMed] [Google Scholar]
- 31. Kojima C., Zhang Y., and Zimmer W. E. (2010) Intronic DNA elements regulate androgen-dependent expression of the murine Nkx3.1 gene. Gene Expr. 15, 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thomas M. A., Hodgson M. C., Loermans S. D., Hooper J., Endersby R., and Bentel J. M. (2006) Transcriptional regulation of the homeobox gene NKX3.1 by all-trans retinoic acid in prostate cancer cells. J. Cell. Biochem. 99, 1409–1419 [DOI] [PubMed] [Google Scholar]
- 33. Thomas M. A., Preece D. M., and Bentel J. M. (2010) Androgen regulation of the prostatic tumour suppressor NKX3.1 is mediated by its 3′ untranslated region. Biochem. J. 425, 575–583 [DOI] [PubMed] [Google Scholar]
- 34. Yoon H. G., and Wong J. (2006) The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol. Endocrinol. 20, 1048–1060 [DOI] [PubMed] [Google Scholar]
- 35. Choi N., Zhang B., Zhang L., Ittmann M., and Xin L. (2012) Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell 21, 253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoo Y. A., Roh M., Naseem A. F., Lysy B., Desouki M. M., Unno K., and Abdulkadir S. A. (2016) Bmi1 marks distinct castration-resistant luminal progenitor cells competent for prostate regeneration and tumour initiation. Nat. Commun. 7, 12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bethel C. R., and Bieberich C. J. (2007) Loss of Nkx3.1 expression in the transgenic adenocarcinoma of mouse prostate model. Prostate 67, 1740–1750 [DOI] [PubMed] [Google Scholar]
- 38. Ellwood-Yen K., Graeber T. G., Wongvipat J., Iruela-Arispe M. L., Zhang J., Matusik R., Thomas G. V., and Sawyers C. L. (2003) Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4, 223–238 [DOI] [PubMed] [Google Scholar]
- 39. Nodouzi V., Nowroozi M., Hashemi M., Javadi G., and Mahdian R. (2015) Concurrent down-regulation of PTEN and NKX3.1 expression in Iranian patients with prostate cancer. Int. Braz. J. Urol. 41, 898–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Gendt K., Swinnen J. V., Saunders P. T., Schoonjans L., Dewerchin M., Devos A., Tan K., Atanassova N., Claessens F., Lécureuil C., Heyns W., Carmeliet P., Guillou F., Sharpe R. M., and Verhoeven G. (2004) A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc. Natl. Acad. Sci. U.S.A. 101, 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lesche R., Groszer M., Gao J., Wang Y., Messing A., Sun H., Liu X., and Wu H. (2002) Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis 32, 148–149 [DOI] [PubMed] [Google Scholar]
- 42. Srinivas S., Watanabe T., Lin C. S., William C. M., Tanabe Y., Jessell T. M., and Costantini F. (2001) Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., and Noble W. S. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.