Abstract
Vibrio cholerae is a natural inhabitant of aquatic environments and converts to a pathogen upon infection by a filamentous phage, CTXΦ, that transmits the cholera toxin-encoding genes. This toxigenic conversion of V. cholerae has evident implication in both genome plasticity and epidemic risk, but the early stages of the infection have not been thoroughly studied. CTXΦ transit across the bacterial periplasm requires binding between the minor coat protein named pIII and a bacterial inner-membrane receptor, TolA, which is part of the conserved Tol-Pal molecular motor. To gain insight into the TolA–pIII complex, we developed a bacterial two-hybrid approach, named Oxi-BTH, suited for studying the interactions between disulfide bond-folded proteins in the bacterial cytoplasm of an Escherichia coli reporter strain. We found that two of the four disulfide bonds of pIII are required for its interaction with TolA. By combining Oxi-BTH assays, NMR, and genetic studies, we also demonstrate that two intermolecular salt bridges between TolA and pIII provide the driving forces of the complex interaction. Moreover, we show that TolA residue Arg-325 involved in one of the two salt bridges is critical for proper functioning of the Tol-Pal system. Our results imply that to prevent host evasion, CTXΦ uses an infection strategy that targets a highly conserved protein of Gram-negative bacteria essential for the fitness of V. cholerae in its natural environment.
Keywords: bacterial pathogenesis, bacteriophage, molecular motor, protein complex, protein/protein interaction, Vibrio cholerae
Introduction
Phage/bacterium interaction is one of the driven forces for gene acquisition and bacterial host adaptation to their environment, and it has been frequently associated with increased virulence of the bacterial host. A striking example of this parasitism-dependent adaptation is Vibrio cholerae, a bacterial natural inhabitant of estuaries, and the causative agent of epidemic disease cholera. Although there are more than 200 O-antigen serogroups described, only two have been reported to cause the pandemic disease cholera, the O1 and O139 serotypes, due to the production of two essential virulence factors, the toxin co-regulated pilus (TCP)3 and the cholera toxin (CT). Interestingly, the genes ctxAB encoding the enterotoxin CT are not carried by the core genome of the bacterium but can be acquired after infection by a lysogenic bacteriophage known as CTXΦ (1). Once infected, the bacterium produces CT and assembles new phage particles (carrying the ctxAB genes) that will be secreted in the environment, and it may convert non-pathogenic V. cholerae cells to pathogenicity.
Most of the knowledge on CTXΦ infection have been extrapolated from the canonical model of Escherichia coli F-pilus-specific coliphages Ff (including f1Φ, fdΦ, and M13Φ). CTXΦ and FfΦ both belong to the genus Inovirus, which are filamentous particles containing a circular single-stranded DNA genome. The genome of inoviruses includes about 10 genes and is generally organized in a conserved modular structure in which functionally related genes are grouped together (2, 3). FfΦ and CTXΦ binding and uptake into the host cell rely primarily on the minor coat protein pIII located at the distal tip of the phage and present at three to five copies. Although there is no sequence conservation between pIIIFf and pIIICTX, both proteins are composed of three distinct domains separated by two low-complexity regions that serve as linkers. Although the N-terminal (N1) and the central domains (N2) are exposed at the capsid surface, the C-terminal domain (N3) anchors the pIII protein to the phage particle through hydrophobic interactions (4–6).
Filamentous phage infection of the bacterial host is seen as a sequential two-step process. First, phage recruitment occurs upon specific binding between the phage capsid pIII-N2 domain and a primary receptor exposed at the surface of the cell host, the conjugative F pilus for E. coli (3, 7) and the TCP for V. cholerae (1, 5). In E. coli, ATP-dependent retraction of the F pilus brings the phage in contact with the cell envelope, promoting its transport across the outer membrane (OM) by an unknown mechanism. Then, pIII must partially unfold to separate N1 and N2 domains (8). This event is crucial in the infection process as it unmasks the pIII-N1 domain for subsequent binding to a second receptor, the TolAEc protein located in the cell envelope (8–12). In E. coli, it has been proposed that the pIII-N1Ff/TolAIIIEc interaction triggers conformational modifications permitting the pIII-N3 domain to form a pore in the bacterial IM, allowing the subsequent phage DNA injection into the cell cytoplasm (13). The nature of the force driving the DNA out of the capsid remains unknown.
In V. cholerae, TCP retraction seems central to the phage infection process, as TCP production alone is not sufficient to allow CTXΦ uptake (14). Although TCP parasitism facilitates the introduction of CTXΦ into the host cell, subsequent phage binding to TolAVc appears to be the limiting step of the infection process (5, 6, 15). Thus, Heilpern and Waldor (5) have shown that a chimeric fd phage displaying the pIII-N1CTX domain fused to the pIII-N3fd domain can successfully infect V. cholerae cells. This demonstrates that the pIII-N1CTX domain displayed at the tip of the capsid is critical and sufficient to ensure host-specific recognition in a TCP-independent manner (5).
TolA is the central protein of the Tol-Pal cell envelope system, which is highly conserved in Gram-negative bacteria (16, 17). In addition to TolA, the Tol-Pal complex is composed of two IM proteins, TolQ and TolR, of the OM lipoprotein Pal and of the periplasmic protein TolB. In several species, including E. coli and V. cholerae, two additional proteins complete the system, the periplasmic protein CpoB (previously YbgF) (18) and the cytoplasmic thioesterase YbgC (19). The Tol-Pal complex is suspected to function as a nanomachine, using the proton-motive force of the IM to generate movements and to transfer energy to OM proteins. Multiple interactions connecting the different components of the Tol-Pal system have been identified. TolA, TolQ, and TolR form a complex anchored in the IM. The TolA protein extends in the periplasm thanks to a long α2-helix (TolAII domain), whereas its globular C-terminal domain (TolAIII) interacts with TolB, and with Pal in the presence of proton-motive force (20–22). Moreover, Pal interacts with TolB and with the peptidoglycan (23). Thus, the Tol-Pal system links the IM, the OM, and the peptidoglycan. The system is involved in maintaining OM integrity, conferring pleiotropic phenotypes when one of its genes is mutated: increased sensitivity to detergents, cell filamentation in low and high osmolarity media, and outer membrane hypervesiculation (6, 16). In addition, the Tol system is involved in the late stage of cell division corresponding to the OM constriction (25) and has been found associated with the PBP1B–LpoB complex in E. coli (18). It is also required for proper localization of polar factor in Caulobacter crescentus (26) and of chemoreceptors in E. coli (27).
For both coliphage and CTXΦ, structural studies on isolated protein domains have provided new insights into the complex formed with TolAIII in the bacterial periplasm (Fig. 1). First, the structures demonstrate that although the CTXΦ pIII-N1 and M13Φ pIII-N1 domains have only 15% sequence identity, they are both dominantly composed of β-strands, and multiple disulfide bonds stabilize their structures. On the bacterial side, TolAIIIEc and TolAIIIVc are curved structures mixing α-helices and β-sheets. A high-resolution structure of E. coli TolAIII free in solution has been obtained by heteronuclear NMR (Protein Data Bank PDB code 1S62) (12), whereas the TolAIII in complex with coliphage pIII-N1M13 (residues 11–86) has been obtained by X-ray crystallography (PDB code 1Tol). The structure shows that the pIII-N1M13 domain binds the concave side of TolAIIIEc, forming a continuous interprotein β-sheet (8). In 2012, Ford et al. (15) determined the crystal structure of pIII-N1CTX domain alone and in complex with the V. cholerae TolAIII domain (PDB code 4G7X). Surprisingly, the authors showed that pIII-N1CTX binds on the convex face of TolAIIIVc, resulting in a continuous interprotein β-sheet (Fig. 1A). Thus, interaction between the two partners delineates a distinct interface compared with the coliphage model of infection (15).
Figure 1.
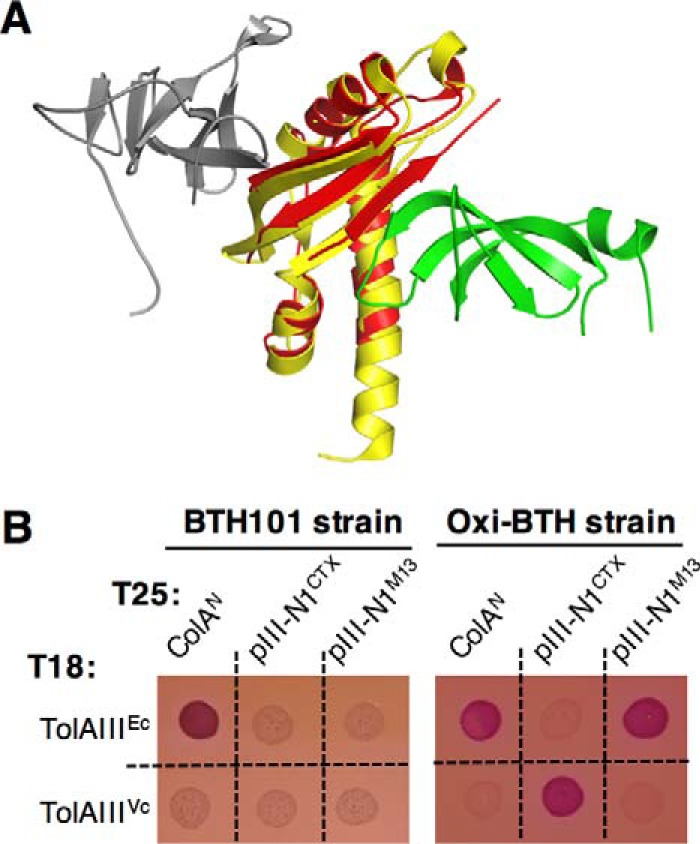
pIII-N1CTX and pIII-N1M13 domains specifically interact with their cognate TolA partner. A, crystal structure of V. cholerae TolAIII (yellow) in complex with CTXΦ pIII-N1 (gray, PDB code 4G7X, and superimposed to E. coli TolAIII (red) in complex with M13Φ pIII-N1 (green, PDB code 1TOL). B, bacterial two-hybrid assay: E. coli BTH101 or Oxi-BTH reporter cells producing the indicated domains fused to the T18 or T25 domain of the Bordetella pertussis adenylate cyclase were spotted on MacConkey plates. The red color of the spot reflects the interaction between tested domains. ColAN (N-terminal colicin A domain), pIII-N1CTX (N-terminal domain of CTXΦ pIII protein), pIII-N1M13 (N-terminal domain of M13-phage pIII protein), and TolAIII (C-terminal domain of E. coli or V. cholerae TolA protein).
Filamentous phages are not the only particles that parasitize the Tol-Pal system to penetrate E. coli cells. Colicins are bacterial toxins comprising various types of lethal activity targeting the IM, the RNA, or the peptidoglycan of its bacterial target. Tol-dependent colicins have been shown to interact with one or more of the Tol proteins during their translocation across the periplasm, showing some similarities with Tol-dependent filamentous phage uptake. In 2012, Li et al. (28) demonstrated that a colicin A peptide (residues 53–107) binds on the convex face of TolAIIIEc, forming an intermolecular antiparallel β-sheet.
It is puzzling to observe that despite the structural similarities between V. cholerae and E. coli TolAIII domains, the molecular binding interfaces with colicin A, pIIICTX, and pIIIM13 differ, illustrating the versatile functioning of TolA as a periplasmic hub protein. In this study, our goal was to investigate the determinants allowing CTXΦ-specific host selection and periplasm transit in vivo thanks to a new oxidative bacterial two-hybrid approach combined to NMR and in vivo studies.
Results
Development of an oxidative bacterial two-hybrid approach dedicated to disulfide bond-folded protein analysis
To gain insights into the mechanism of CTXΦ transit through the periplasm, we first analyzed the interaction that occurs between pIII-N1CTX and TolAIIIVc, compared with the pIII-N1M13 and TolAIIIEc interaction, using a bacterial two-hybrid (BACTH) approach. This system relies on the reconstitution of the signaling cAMP transduction cascade in an endogenous adenylate cyclase-deficient strain (29). The TolAIII domains from V. cholerae and from E. coli were fused to the T18 domain in the pUT18 vector, whereas the pIII-N1 domains from M13 and CTX phages were fused to the T25 domain in the pKT25 vector. Constructs were introduced into the E. coli BTH101 strain, and co-transformants were tested on MacConkey plates. As a control, we first observed that in this assay, the T18-TolAIIIEc construct gives a positive interaction signal with the colicin A N-terminal domain (ColAN) as described previously (10, 22, 30), which validated our approach (Fig. 1B, left panel). We also observed that a T18-TolAIIIVc construct is unable to bind T25-ColAN, attesting the specificity for partner recognition between the two bacterial species. Then, we tested the TolAIIIVc and pIII-N1CTX constructs together, but we did not detect interaction between these different domains. We obtained the same negative result when we tried to detect the TolAIIIEc/pIII-N1M13 interaction (Fig. 1B, left panel). We hypothesized that this result could arise from improper folding of disulfide-bonded TolAIII, pIII-N1M13, or pIII-N1CTX domains when expressed in the cell cytoplasm (Fig. 2, A and B).
Figure 2.
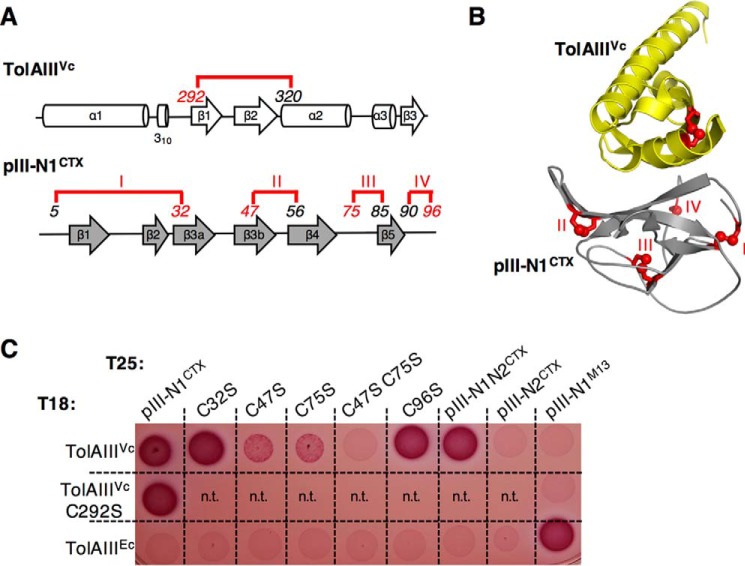
pIII-N1CTX disulfide bonds II and III are required for TolAIIIVc interaction. A, schematic representation of disulfide bond localization (15) on the secondary structure of TolAIIIVc and pIII-N1CTX; B, representation of disulfide bonds on the crystal structure of the complex. Disulfide bonds in pIII-N1 are numbered I–IV and colored in red. Italic numbers refer to cysteine residue positions. C, oxidative bacterial two-hybrid assay: Oxi-BTH reporter cells producing the T25 domain fused to pIII-N1CTX, pIII-N1N2CTX, or pIII-N2CTX domains and the T18 domain fused to V. cholerae TolAIII were spotted on MacConkey plates. Variants bearing substitutions aimed to abolish each of the four disulfide bonds in the pIII-N1CTX domain (C32S, C47S, C75S, and C96S) or in TolAIIIVc (C292S) are presented. TolAIIIEc and pIII-N1M13 are used as a controls. n.t., not tested.
Thus, we envisioned that a bacterial two-hybrid assay in an oxidative environment would allow the proper folding of proteins with disulfide bonds. Several E. coli strains, such as Origami (Novagen) or SHuffle (New England Biolabs), have been engineered to optimize the purification of proteins with disulfide bonds and are commercially available. We chose the SHuffle T7 strain as a chassis because it is deleted for glutaredoxin reductase (gor) and thioredoxin reductase (trxB) genes, allowing disulfide bond formation in the cytoplasm. In addition, this strain expresses a cytoplasmic version of the disulfide bond isomerase DsbC, promoting correct disulfide bond formation and proper oxidative folding of proteins containing multiple cysteines (31). We transduced the cya mutation in the SHuffle strain to make the Oxi-BTH strain (for Oxidative Bacterial Two-Hybrid). The resulting strain was co-transformed with the pUT18 and pKT25 vectors expressing domains of interest. As shown in Fig. 1B (right panel), the Oxi-BTH strain allowed the detection of interaction between TolAIIIVc and pIII-N1CTX, as well as between TolAIIIEc and pIII-N1M13. Thus, we concluded that the Oxi-BTH strain is a powerful tool to apply the BACTH system to the study of disulfide-bonded proteins. Indeed, our data suggest that in both E. coli and V. cholerae, disulfide bond-dependent folding of the TolAIII domain and/or the phage minor capsid domain is required to allow binding of the two partners. Conversely, TolAIII Ec is able to interact with the colicin A N-terminal domain, which does not contain cysteines, in both reducing and oxidizing conditions (Fig. 1B). Finally, we did not observe cross-interaction between the two species (i.e. TolAIIIVc/pIII-N1M13 or TolAIIIEc/pIII-N1CTX) despite strong structural conservation between E. coli and V. cholerae TolAIII domains.
Role of disulfide bonds in TolAIIIVc/pIII-N1CTX interaction
Because the TolAIIIVc/pIII-N1CTX interaction is only seen in our oxidative two-hybrid assay, we hypothesized that one or more disulfide bonds might be essential for bacterial and phage domain recognition. To test this hypothesis, each disulfide bond (one in TolAIIIVc and four in pIII-N1CTX, Fig. 2, A and B) was sequentially abolished by introducing substitutions of individual cysteine to serine in the BACTH constructs. The resulting mutants were then tested in the Oxi-BTH assay. As shown in Fig. 2C, TolAIIIVc (C292S) construct was still able to interact with pIII-N1CTX. This suggests that disulfide bond formation in TolAIIIVc is not required for CTX phage binding. We then targeted each of the four disulfide bonds present in pIII-N1CTX. None of the individual mutations we performed was able to totally abolish binding to TolAIIIVc. Interestingly, mutation of the second or the third S–S bond (mutations C47S and C75S, respectively) of the phage pIII-N1 domain resulted in a faint interaction signal, suggesting a weaker binding affinity between TolAIIIVc and these two pIII-N1CTX mutants compared with the wild-type construct. In agreement with this observation, a pIII-N1CTX(C47S/C75S) double mutant was not able to interact with TolAIIIVc. These observations are unlikely to result from stability defects of the cysteine variants compared with the native pIII-N1 protein. Indeed, inserting the same mutations on the His-tagged pIII-N1CTX domain expressed from the pIN vector resulted in equivalent expression of the different constructs (supplemental Fig. S1). Moreover, as the pIII-N1CTX/TolAIII interaction is not seen in the regular BACTH assay, it is more likely that pIII-N1CTX folding via its 2nd and 3rd disulfide bonds is critical for TolAIIIVc binding.
CTXΦ central domain pIII-N2 does not block phage pIII-N1 accessibility to TolAIIIVc
It has been shown that Ff coliphages require an activation step to become able to infect the host cell. Indeed, in the native conformation of the minor capsid protein pIIIFf, the N2 domain is tightly associated with N1, which buries the phage TolAIII-binding site at the domain interface. Phage activation is processed upon binding of N2 to the primary receptor, the F pilus, which initiates partial unfolding, prolyl cis-to-trans isomerization in the hinge between N1 and N2 and domain disassembly, thereby exposing its binding site for the ultimate receptor TolA (32). It has been proposed that the isomerization sets a molecular timer to maintain the binding-active state long enough for the phage to interact with TolA. Conversely, Craig and co-workers (15) have suggested that the TolA-binding site on pIII-N1CTX is permanently accessible and does not require initial pilus-induced conformational change. We wondered whether a fusion to the two-hybrid T25 domain would allow us to test the influence of pIII-N2CTX on the accessibility of pIII-N1CTX in an Oxi-BTH interaction assay. Indeed, our data show that the T25–pIII-N1N2CTX construct is able to bind T18–TolAIIIVc, whereas the T25–pIII-N2CTX domain alone is not (Fig. 2C). This result demonstrates that pIII-N1CTX exposes a TolAIIIVc-binding motif that is not masked by the pIII-N2 domain.
Specific recognition between the TolAIIIVc and pIII-N1CTX domains relies on two salt bridges
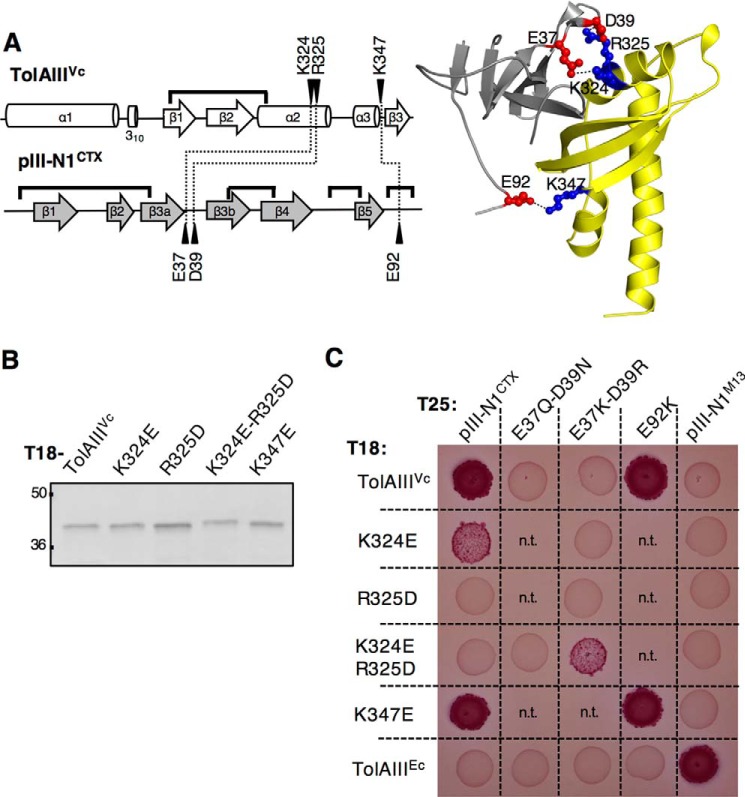
We aimed to identify the residues showing a predominant role in the specificity of binding. Reversible protein/protein interactions involve low energy ionic bonds, hydrogen bonds, and van der Waals interactions. Based on the co-crystal structure of TolAIIIVc and pIII-N1CTX (PDB code 4G7X (15)), we focused our analysis on the intermolecular salt bridges. Residues Lys-324, Arg-325, and Lys-347 on TolAIIIVc, respectively, interact with Glu-37, Asp-39, and Glu-92 on pIII-N1CTX (Fig. 3A). To analyze the importance of these three salt bridges in the complex formation, we abolished them sequentially by introducing single residue substitutions on TolAIIIVc. We confirmed that the different TolAIIIVc variants showed production level and stability equivalent to the wild-type T18-TolAIII construct by immunodetection (Fig. 3B), and we tested their capacity to interact with T25-pIII-N1 in a Oxi-BTH assay (Fig. 3C). The TolAIII(K324E) and TolAIII(R325D) mutants were, respectively, partially and totally impaired in their ability to interact with pIII-N1CTX, whereas the TolAIII(K347E) mutant retained its ability to bind pIII-N1CTX. Conversely, we introduced the mirror mutations on T25-pIII-N1CTX to generate the E37Q/D39N and the E92K variants. We demonstrated that T25-pIII-N1(E37Q/D39N) was unable to bind T18-TolAIIIVc, although T25-pIII-N1(E92K) was. Together, these data suggested that the two intermolecular salt bridges engaged between Lys-324 and Arg-325 on TolAIIIVc and Glu-37 and Asp-39 on pIII-N1CTX are crucial for pIII-N1–TolAIIIVc complex stabilization.
Figure 3.
pIII-N1CTX-TolAIIIVc binding relies on two salt bridges. A, left, schematic representation of the V. cholerae TolAIII and CTX phage pIII-N1 secondary structure. Residues (black arrowheads) engaged into intermolecular salt bridges (dotted lines) and disulfide bonds (black lines) are pointed out. Right, X-ray structure of the complex showing the three key salt bridges. B, Western immunoblot of 0.2 absorbance units of whole-cell lysates of E. coli DH5α strain carrying T18-TolAIIIVc construct, or variants of interest, and probed with anti-T18 antibody. The molecular mass markers (in kDa) are indicated on the left. C, TolAIIIVc and pIII-N1CTX point mutants were tested for their binding ability, in comparison with the wild-type constructs, in an Oxi-BTH assay on MacConkey plates. TolAIIIEc and pIII-N1M13 are used as a controls. n.t., not tested.
To confirm the role of the TolAIII charged patch (Lys-324–Arg-325) in pIII-N1CTX binding, we swapped the charged residues engaged into salt bridges between the two partners. As predicted, the T18-TolAIII(K324E/R325D) double mutant was not able to interact with the wild-type T25-pIII-N1CTX construct but regained interaction with the mutated variant presenting oppositely charged residues T25-pIII-N1(E37K/D39R) (Fig. 3C). Of note, the restored interaction signal appeared weaker than the one observed for wild-type constructs, possibly reflecting that additional local steric effects operate at the binding interface.
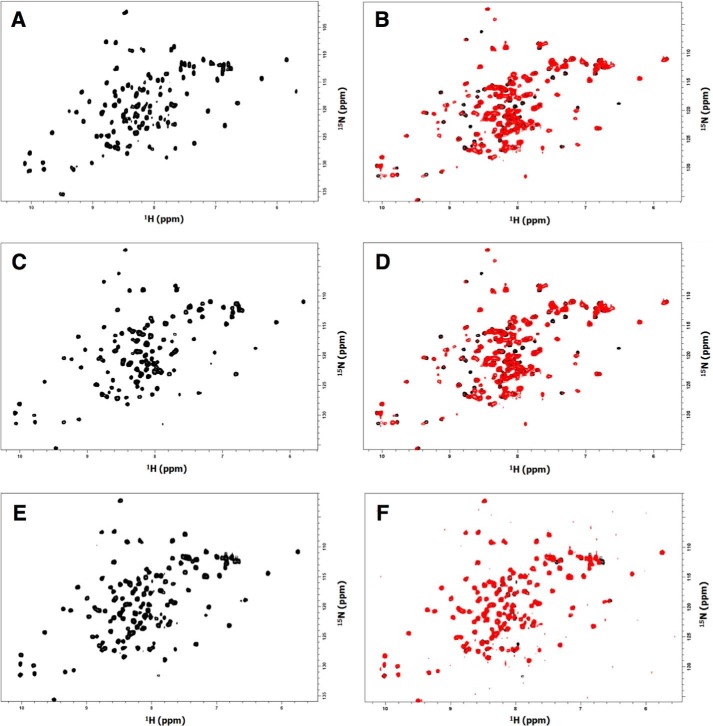
To further establish that our data result from a specificity of interaction rather than a folding defect of the protein variants, we developed an NMR approach. We used the pIN3-ompA2 vector (33) to overproduce 15N-labeled wild-type pIII-N1CTX, pIII-N1CTX(E37Q/D39N), and pIII-N1CTX(E92K) variants fused to a His C-terminal tag. The 1H-15N HSQC spectra of the proteins (Fig. 4, A, C, and E) shows 15N/1H correlations for the NH group of each amino acid of the proteins. The 15N and 1H resonances are associated with the nuclear chemical environment; thus, the 1H-15N HSQC spectrum represents the fingerprint of a protein. In the case of the native and variant pIII-N1CTX, the 1H-15N HSQC spectra are well resolved. This indicates that each residue has a particular environment in agreement with the X-ray structure of the protein (15). The fact that the spectra of the variant proteins are similar to that of the native protein demonstrates the conservation of the folding. Moreover, superimposition 1H-15N HSQC in the presence and the absence of TolAIIIVc showed chemical shift perturbations for pIII-N1CTX (Fig. 4B) and pIII-N1CTX(E92K) variants (Fig. 4D), whereas no perturbation was seen with the pIII-N1CTX(E37Q/D39N) variant (Fig. 4F). These observations demonstrate that pIII-N1CTX and pIII-N1CTX(E92K) could interact with the TolAIIIVc domain, modifying the chemical environment of the nucleus located at the interface, whereas the pIII-N1CTX(E37Q/D39N) variant could not, which confirmed the Oxi-BTH results.
Figure 4.
A, 1H-15N HSQC of 15N-labeled pIII-N1CTX domain. B, 1H-15N HSQC of 15N-labeled pIII-N1CTX domain in the absence (black) and in the presence of TolAIIIVc domain (red). C, 1H-15N HSQC of 15N-labeled pIII-N1CTX (E92K) domain. D, 1H-15N HSQC of 15N-labeled pIIIN1CTX(E92K) domain in the absence (black) and in the presence of TolAIIIVc domain (red). E, 1H-15N HSQC of 15N-labeled pIIIN1CTX(E37Q/D39N) domain. F, 1H-15N HSQC of 15N-labeled pIIIN1CTX(E37Q/D39N) domain in the absence (black) and in the presence of TolAIIIVc domain (red).
In V. cholerae, periplasmic expression of the CTX pIII-N1 domain leads to Tol phenotypes
Oxi-BTH assay and NMR studies allowed us to identify key residues on the CTXΦ minor capsid protein that were involved in TolAIII binding. To confirm these data in vivo, we used a periplasmic expression assay in V. cholerae. In E. coli, it has been previously shown that production of the N-terminal domain of colicin A (34) or of the pIII-N1 domain of coliphage f1 (10) into the periplasm of WT cells perturbs the Tol-Pal system by sequestering the TolA protein, and consequently results in tol phenotypes. This approach is particularly suitable to study the translocation step of phage infection, as it does not require production and assembly of mutated phage particles, their TCP-dependent reception, and their OM transport.
We used the pIN3-ompA2 vector to express the His-tagged pIII-N1CTX wild-type sequence (or variants) fused to a N-terminal ompA signal sequence to allow transport of the produced proteins into the periplasm via the sec pathway. Correct production of the resulting proteins in V. cholerae cells was confirmed by immunodetection, whereas transport of the produced domains into the periplasm was attested by sodium azide inhibition of the sec pathway (supplemental Fig. S2). Cells producing the WT pIII-N1 phage domain targeted to the periplasm presented a 5-fold decrease in susceptibility to phage infection (Fig. 5A) and high sensitivity to deoxycholate (DOC) (Fig. 5B) and to SDS (Fig. 5C), which are characteristic tol phenotypes. This suggests that the exogenous pIII-N1CTX domain interacts with the endogenous TolAVc protein in the V. cholerae cell envelope, competing with the other Tol proteins of the system and preventing adequate functioning that ensures membrane integrity and permits phage uptake. We also observed that exogenous production of pIII-N1M13 in V. cholerae periplasm does not result in tol phenotypes, which demonstrates further that the M13Φ capsid protein pIII does not bind V. cholerae TolA protein. In accordance with the Oxi-BTH results, periplasmic expression of the pIII-N1CTX(E37Q/D39N) construct resulted in phage infection rate, DOC sensitivity, and SDS sensitivity similar to the control cells carrying an empty vector, whereas periplasmic expression of the pIII-N1CTX(E92K) variant showed the same phenotypes as those obtained with the WT pIII-N1CTX construct. In accordance with the Oxi-BTH results, these in vivo data confirm that the pIII-N1CTX negatively charged residues Glu-37 and Asp-39 are required for pIII-N1 interaction with the native TolA protein.
Figure 5.
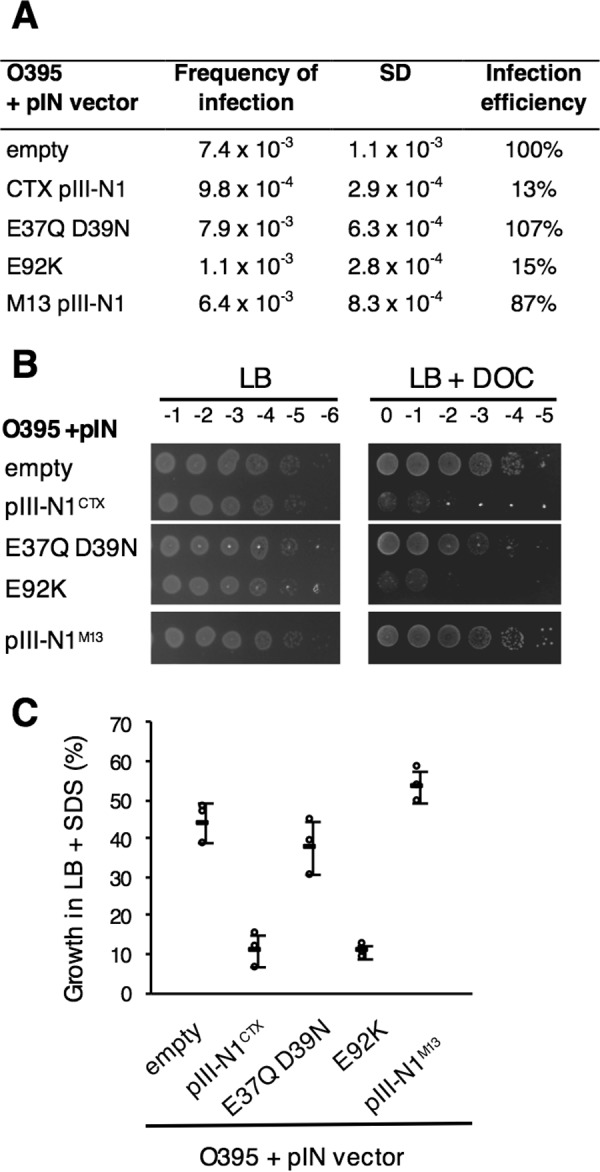
Periplasmic production of CTXΦ pIII-N1 domain or variants in V. cholerae and phenotypic characterization. Precultures of V. cholerae O395 cells carrying various pIN constructs were grown in LB supplemented with MgCl2 to promote OM integrity and cell growth. A, CTX transduction assay was conducted in triplicate, and CFU were counted on LB or LB supplemented with Cm. Frequency of infection is calculated by dividing the number of transductants (CmR colonies) by the number of O395 recipients. The mean and the standard deviation of the triplicate is presented. For each construct, infection efficiency is expressed as the percentage of infection compared with the receiver strain carrying the empty vector. B, membrane integrity assay. 4 μl of 10-fold dilution of normalized cultures (initial A595 = 1) were spotted on LB+amp plates alone or supplemented with 1% DOC. C, percentage of survival to SDS 0.125% of the different strains compared with V. cholerae WT strain carrying a pIN empty vector. For each strain, percentage of growth is calculated as A600 nm of each stain grown in LB+SDS × 100/A600 nm of the WT strain carrying an empty vector and grown in LB. The experiments were conducted in triplicate, and the standard deviation is presented.
Evidence that TolA Arg-325 residue is essential for proper functioning of the Tol-Pal system in V. cholerae
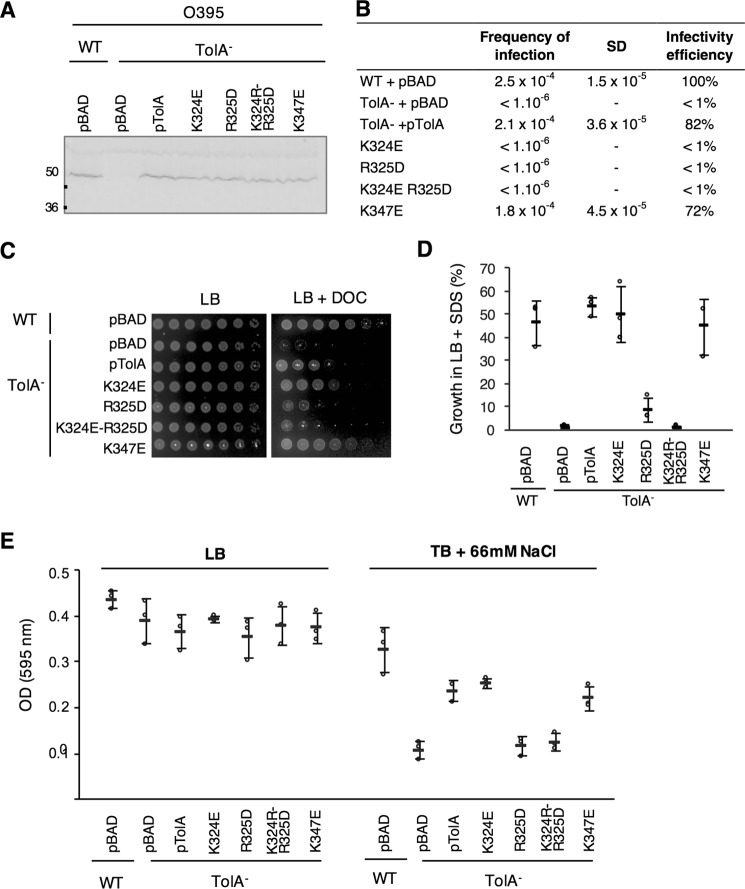
We decided to investigate whether the TolA charged patch (Lys-324–Arg-325) required for phage binding is important for the Tol-Pal system function in V. cholerae. Although multiple attempts to delete the V. cholerae full-length tolA gene by double homologous recombination were unsuccessful, we found that a TolAΔ(41–421) mutant, deleted for its periplasmic domains but retaining its IM domain, is viable. The resulting strain presents characteristic tol phenotypes (6, 35), including resistance to CTXΦ infection, sensitivity to SDS and to DOC, and finally growth defect in a low osmolarity medium (tryptone broth, 66 mm NaCl) compared with LB medium (Fig. 6, A–E).
Figure 6.
Phenotypic characterization of V. cholerae O395 WT and TolA− strains complemented with TolA variants of interest. A, expression of the different pBAD-TolAVc constructs in V. cholerae was assessed by Western immunoblotting. A total of 0.3 absorbance units of whole-cell lysate were loaded onto an SDS-13.5% acrylamide gel and immunodetected using polyclonal antibody raised against E. coli TolA. B, CTX transduction assays were conducted in triplicate, and CFU were counted on LB or LB supplemented with Cm. Frequency of infection is calculated by dividing the number of transductants (CmR colonies) by the number of O395 recipients. The mean and the standard deviation of the triplicate is presented. For each construct, infection efficiency is expressed as the percentage of infection compared with the receiver strain carrying the empty vector. C, membrane integrity assay. 4 μl of 5-fold dilution of normalized cultures (initial A600 = 1) were spotted on LB+kanamycin plates alone or supplemented with 1% DOC. D, percentage of survival to SDS-0.125% of the different strains compared with V. cholerae WT strain carrying an pBAD empty vector. For each strain, percentage of growth is calculated as A600 nm of each stain grown in LB+SDS × 100/A595 nm of the WT strain carrying an empty vector and grown in LB. E, quantification of V. cholerae O395 WT and TolA− mutant growth in LB (407 mosm) and in tryptone broth (TB) supplemented with 66 mm NaCl (123 mosm). The experiment was conducted in triplicate, and the error bars report standard deviations.
We first performed a complementation assay of the TolAΔ(41–421) mutant using a pBAD expression vector. TolA has been reported to be a low abundance protein (400–800 copies in E. coli cells (36)). In our assay, basal level of TolA production from the pBAD-TolA vector was detected even in the absence of induction (Fig. 6A), and it was sufficient to almost fully complement (80–100% rescue) the V. cholerae tolA mutant for phage infection (Fig. 6B), for resistance to SDS and DOC (Fig. 6, C and D), and for hypo-osmotic growth in tryptone broth (Fig. 6E). These results also confirmed that the deletion had no polar effect on the other genes of the Tol-Pal operon.
We then tested V. cholerae strains carrying TolA variants where residues Lys-324, Arg-325, or Lys-347 were mutated for oppositely charged residues. Although the K324E variant behaved similarly to the wild-type pBAD-TolA construct, rescue was no longer observed for the TolA(R325D) variant, despite adequate production of the mutant protein (Fig. 6, A–E). The TolA(K324E/R325D) double mutant showed a slightly stronger phenotype than the individual mutants for membrane integrity assays (Fig. 6, C and D). Finally, the TolA(K347E) variant complemented the mutant strain similarly to the WT TolA construct. Together, these findings support the conclusion that the positively charged residue Arg-325 plays a dominant role in TolA function in V. cholerae, whereas Lys-324 and Lys-347 do not.
Conservation of the Lys-324 and Arg-325 residues in other TolA Vibrio species
The Tol-Pal system has a widespread distribution in Gram-negative bacteria. We first questioned TolA sequence conservation among 128 V. cholerae isolate genomes available on the NCBI database, and we found that the full-length protein is 100% identical (supplemental Fig. S3). We then broadened the study to the Vibrionaceae family, including 82 Vibrio species (supplemental Fig. S4) and 33 non-Vibrio species (six Aliivibrio, three Enterovibrio, five Grimontia, 13 Photobacterium, and six Salinovibrio (supplemental Fig. S5). PSIPRED analysis (37) of the TolAIII amino acid sequences suggests that the secondary structures are conserved among Vibrionaceae (data not shown). Alignments of TolAIII sequences from multiple Vibrio species indicates that Lys-324 is highly conserved (73/82 sequences) with only few variants where it is replaced by another positively charged residue, arginine (6/82). Three exceptions were found with Vibrio breoganii, Vibrio gazogenes, and Vibrio rhizosphaerae that carry an uncharged glutamine residue at position 324. Overall, the positive charge at position 324 appears to be conserved in other Vibrionaceaes tested (31/33), suggesting its importance in TolA function (supplemental Figs. S4 and S5). Conversely, the positively charged residue Arg-325 is less conserved in Vibrio species (51/82), frequently being replaced by the uncharged residue serine (25/82) or less frequently by alanine, threonine, or valine (6/82). Notably, the positively charged residue at position 325 was never observed in other Vibrionaceae species (0/33).
As we previously demonstrated that pIII-N1CTX interaction with the V. cholerae TolA receptor is driven by two intermolecular salt bridges engaging TolA Lys-324 and Arg-325 residues, we expected orthologous TolAIII sequences carrying the charged patch (Lys-324–Arg-325) to also be able to bind CTX pIII-N1. As shown in Fig. 7A, the TolAIII domain from Vibrio anguillarum, Vibrio tasmaniensis, and Vibrio alginolyticus carry the (KR) patch, whereas Vibrio harveyi has a (KS) motif and the Vibrionaceae Aliivibrio fischeri has a (KT) motif. The TolAIII coding sequences from the different species were cloned into the pUT18 vector, and correct expression of the different constructs was assessed by immunodetection (Fig. 7B) before testing their interaction ability with T25-pIII-N1CTX in the Oxi-BTH assay. As shown on Fig. 7C, T18-TolAIII from V. anguillarum, V. tasmaniensis, and V. alginolyticus showed positive interaction signal with T25-pIII-N1CTX. In contrast, T18-TolAIIIV. harveyi and T18-TolAIIIV. fischeri, lacking the positively charged patch (KR), were not able to bind pIII-N1CTX. We analyzed the TolAIII sequences that were able to bind pIII-N1 to define a consensus sequence for partner recognition. We found that TolAIII tolerates multiple sequence variation in the β2-strand, likely because this region of the protein interacts with pIII-N1 β1-strand through backbone hydrogen bond interactions. Conversely, TolAIII α2-helix alignments allowed the definition of a consensus sequence (GD(S/T)R(L/V)CAA(T/A)KRA(V/I)AQ) surrounding the (KR) residues. We noticed that V. harveyi TolAIII carry the defined consensus sequence, apart from Arg-325. However, point mutation to generate T18-TolAIIIV. harveyi S325R and reconstitute a positively charged patch (KR) was not sufficient to restore interaction with pIII-N1CTX (Fig. 7C).
Figure 7.
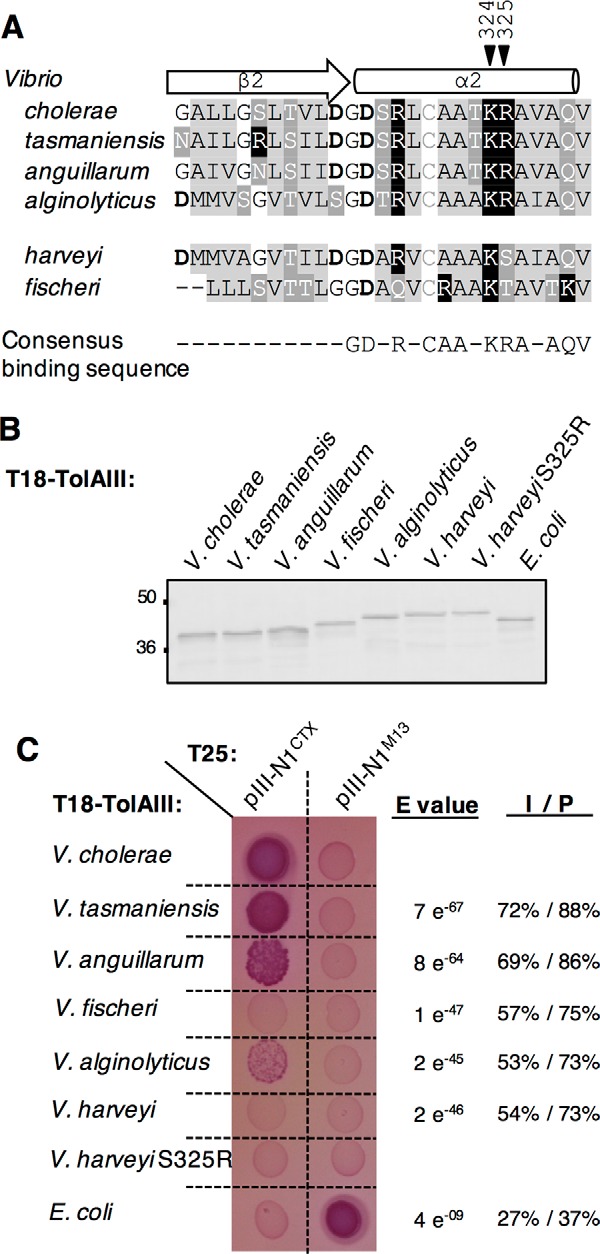
Conservation of TolA Lys-324 and Arg-325 residues in other Vibrio species. A, sequence alignment between V. cholerae, V. tasmaniensis, V. anguillarum, V. alginolyticus, V. harveyi, and V fischeri TolAIII β2-α2 domain. Residues are colored as follows: basic (black squares), acidic (bold), hydrophobic (light gray),and polar uncharged (dark gray). Black arrowheads point the Lys-324–Arg-325 motif. B, Western immunoblot of 0.2 absorbance units of whole-cell lysates of E. coli DH5α strain carrying T18-TolAIII construct from selected Vibrionaceae and from E. coli and probed with anti-Cya antibody. The molecular mass markers (in kDa) are indicated on the left. C, T18-TolAIII constructs from selected species were tested for their binding ability to the T25-pIII-N1CTX in an Oxi-BTH assay on MacConkey plates. E. coli TolAIII and pIII-N1M13 are used as a controls. Sequence comparison between each TolAIII of interest and the V. cholerae TolAIII amino acid sequences using Blast2 is reported on the right of the panel as E value, identity (I) and positive (P) values.
Discussion
New in vivo protein–protein interaction assay dedicated to disulfide-oxidized proteins
For many proteins functioning in the periplasm, exposed at the cell surface, or secreted in the extracellular environment, stability and/or activity require the formation of disulfide bonds. For example, in V. cholerae, S–S bond proteins include the cholera toxin (38), the pilin subunits that assemble in different type IV pili (TCP, MshA, and PilA) operating into the various aspects of Vibrio ecology (39, 40), the TolA protein in the Tol-Pal complex involved in envelope integrity and cell division (15), and sensors such as TcpP (41, 42) and ToxR (43) operating signal transduction to regulate virulence pathways. Thus, identification and characterization of physiologically relevant interactions between these S–S bond proteins in vivo are crucial tasks for the understanding the molecular processes within the cell.
The original BACTH approach is an easy yet efficient technique that has become a common laboratory tool used to identify and dissect protein/protein interactions (29, 44). As the interaction between candidates of interest is tested in the cell cytoplasm, studies were restricted so far to proteins in their reduced state. Recently, the development of new BACTH plasmids inserting a transmembrane segment downstream of the T25 and T18 fragments was shown to allow the detection of interactions occurring within the periplasmic space of the cell (45).
In this study, we extended the range of BACTH application to the study of disulfide-oxidized proteins directly in the reporter cell cytoplasm thanks to a new genetic background that we named Oxi-BTH. In this strain, correct disulfide reshuffling is catalyzed by a cytoplasmic version of the disulfide bond isomerase DsbC (31). The Oxi-BTH assay was validated by the visualization of interactions previously described between proteins with unique (TolAIIIEc and TolAIIIVc) and multiple disulfide bonds (pIII-N1M13 and pIII-N1CTX), which could not be observed in the regular BACTH assay. As a protein with eight cysteines, pIII-N1CTX has a probability of less than 1% to form the correct four disulfide bonds, and our results also attest the efficiency of the cytoplasmic DsbC proofreading activity (Fig. 1). Finally, this tool was suitable to individually test the importance of each disulfide bond and of targeted residues in protein/protein interactions (Figs. 2 and 3). Many BACTH pUT18 and pKT25 constructs have already been published, and genome-wide libraries are available for E. coli (46) and Pseudomonas aeruginosa (47) allowing the constitution of a large collection of potential partners that can be directly tested in the Oxi-BTH assay without requiring new cloning steps. In this context, we believe that Oxi-BTH constitutes a robust and versatile tool to test the importance of oxidative folding in protein/protein interaction, to identify key amino acids involved, and to investigate the specificity of binding between two proteins of interest.
Refining the model of CTXΦ uptake
Filamentous phage infection is a two-step process, requiring a primary receptor at the surface of the cell (the TCP pilus in the case of CTXΦ) and a secondary receptor, TolA, inside the cell envelope. In this study, our aim was to gain insight into the second step of the infection process by investigating in vivo the phage/TolA interaction. In contrast to E. coli, we did not manage to obtain the V. cholerae TolA clean deletion mutant by homologous recombination, although we were able to delete the periplasmic domains of the protein. It is noteworthy that the only other V. cholerae TolA− mutant published in the literature is a tolA::pGP704 disruption mutant (obtained by single crossover) that, according to the primer design, also resulted in a truncated TolA protein somewhere in the periplasmic domain (6). This suggests that, under laboratory conditions, TolA is essential in V. cholerae, as reported previously in E. coli O7:K1 (48), P. aeruginosa (49), and C. crescentus (26), but is dispensable in the E. coli K12 strain (35).
We observed that pIII-N1CTX domain is sufficient for TolA binding and that pIII-N2CTX does not impede the interaction between the two partners (Fig. 2). This result is in agreement with previous observations made by Ford et al. (15). Indeed, the authors previously showed that CTXΦ incubation with an excess of purified TolAIIIVc domain before infection of V. cholerae O395 cells reduced the infection efficiency. However, in this study, binding between pIII-N1-N2CTX and TolAIIIVc had not been formally tested because pIII-N1-N2CTX could not be purified in a soluble form (15). Together, our data further demonstrate that the CTXΦ naturally exposes a TolA-binding site on the N-terminal domain of pIII, which is reminiscent to IF1 and IKe coliphage infection strategy (50). It implies that CTXΦ binding to the primary receptor TCP might be responsible for phage recruitment from the environment and possibly active pulling to the cell surface through retraction (14), but it is not required to unmask the pIII-N1 TolA-binding site.
The crystal structure of the complex (PDB code 4G7X) suggested that multiple interactions (ionic, hydrophobic, polar contacts, and van der Waals forces) contribute to the binding interface between TolAIIIVc and pIII-N1CTX. The TolAIIIVc β2-strand folds around pIII-N1CTX, resulting in a continuous intermolecular β-sheet that involves multiple hydrogen bonds between the backbone chains of TolAIII β2-strand and pIII-N1 β1-strand. TolAIIIVc residues Lys-324, Arg-325, and Lys-347 form three salt bridges with pIII-N1 Glu-37, Asp-39, and Glu-92 residues, respectively (Fig. 3). However, the contribution of each of the individual interactions in the TolAIII–pIIICTX complex formation cannot be assessed from the crystal structure. By combining in silico analysis and in vivo experimental data, we defined that the consensus sequence for CTX binding on TolAIIIVc is restricted to the α2-helix, whereas the β2-strand tolerates multiple residues variations (Fig. 7). Moreover, our work clearly demonstrates that the two salt bridges engaged between TolAIIIVc residues Lys-324 and Arg-325, and CTX phage pIII-N1 residues Glu-37 and Asp-39, respectively, provide the driving forces of the interaction, forming ion locks required for the complex to form, whereas the TolAIII Lys-347–pIII-N1 Glu-92 bond is dispensable. It is noteworthy that mutating the TolAIII Arg-325 residue alone had a stronger negative impact on phage binding than mutating the TolAIII Lys-324 residue (Fig. 3). We used the 2P2I database (http://2p2idb.cnrs-mrs.fr/2p2i_inspector.html)4 (51, 52) to gain insights into the protein–protein interface parameters. Results showed a total of 88 non-bonded contacts (distance cutoff 4 Å) between pIII-N1CTX and TolAIIIVc (supplemental Fig. S4). Among these 88 contacts, TolAIII Arg-325 was engaged in 20 different contacts with pIII-N1 surface, whereas TolAIII Lys-324 was engaged in six (supplemental Fig. S6). These observations might explain the more predominant role of the TolAIII Arg-325 residue in pIII-N1 binding, compared with Lys-324. Overall, this is reminiscent of what was previously observed for M13 coliphage pIII-N1 binding on the concave side of TolAIIIEc, where the interaction makes an antiparallel continuous β-sheet stabilized by two salt bridges (TolAIIIEc Asp-210, Lys-212 interacting with pIII-N1M13 Arg-29 and Asp-28, respectively (PDB code 1Tol (8)). It would be interesting to test the importance of each of these ionic interactions for coliphage–TolAEc complex formation.
TolA is involved in the fitness of V. cholerae, in particular in low osmolarity conditions, such as those found when the bacterium reaches low salinity environments (fresh water), or in response to detergent components, such as the bile acid deoxycholate during its transit through the gastrointestinal track (Figs. 5 and 6) (6). We found that the TolA(R325D) variant was functionally unable to complement the V. cholerae tol mutant, whereas the TolA(K324E) mutant was able to do so (Fig. 6). We questioned the sequence conservation of TolAIII in other Vibrio species, but we did not observe strict correlation between conservation of the Lys-324 and Arg-325 residues and their importance in TolA function in the tested conditions. Interestingly, we noticed that in the 33 Vibrionaceae species studied, the TolAIII Arg-325 residue is absent (supplemental Fig. S3), whereas additional positive charges are found at more or less one pitch of the α2 helix: Arg-321 (occurrence 33/33) and Lys-329 (occurrence 24/33). Because TolA is a hub protein, and part of a multicomponent complex, the patch of positive residues on the α2 helix might be involved in partners binding, either with an already known partner of the TolAIII domain such as TolB or Pal (16, 22, 53) or an unknown partner that still has to be identified.
We demonstrated that the KR patch on TolA is crucial but not sufficient for CTX binding (Fig. 7). Indeed, study of the V. harveyi TolA(S325R) variant suggests that additional local steric effects operate at the binding interface, outside the α2 helix. Although salt bridges provide the forces to drive protein–protein initial attraction, subsequent complex stabilization is usually the result of cooperative interactions that encompass multiple pairwise bonds (54). Our results allow a better understanding of the TolAIII/pIII-N1 interaction interface, with two salt bridges providing the driving forces of the interaction, whereas the formation of the intermolecular anti-continuous β-sheet provides stabilization interactions to the complex. By targeting important functional and/or conserved residues of TolA protein, CTXΦ uses a parasite infection strategy that may help to prevent V. cholerae from escaping the infection.
Implication for the CTX infection host specificity
In this work, we observed that the CTX phage protein pIII-N1 responsible for host selection (5) can bind the TolAIII receptor domain from at least three other Vibrio species: V. alginolyticus, V. anguillarum, and V. tasmaniensis (Fig. 7). However, the host range reported for CTX from environmental sampling studies is surprisingly narrow, even in species carrying a conserved TolA sequence. Thus, although all the epidemic V. cholerae strains belonging to the O1 or O139 serogroups carry the CTX prophage, it is rarely detected (2–5%) in non-O1/non-O139 V. cholerae environmental isolates genomes (55–59). Apart from V. cholerae, CTXΦ has been reported to infrequently infect other Vibrio species, even for very close species such as Vibrio mimicus (60). It has been proposed that the limited distribution of the primary receptor TCP among Vibrio species plays a predominant role in the fate of the entire infection process (55). It is interesting to note that TCP-independent CTXΦ infection has been reported to occur in defined conditions (5). Thus, whereas TolA is a conserved protein, variation in the sequence of the TolAIII domain might serve as a second level of CTXΦ host-specificity checkpoint.
Because bacteriophages, like any other viruses, are obligate intracellular parasites, successful uptake across the bacterial cell envelope is an essential condition to complete their life cycle. However, current knowledge on host/phage interactions is based on a limited number of microbial models. This scientific problem outreaches the question of V. cholerae pathogenic conversion, as other filamentous phages (sharing similarities with CTXΦ) have been demonstrated to affect the virulence and fitness of a large range of bacterial hosts: meningococcal disease-associated (MDA) phage in invasive isolates of Neisseria meningitidis (61); YpfΦ in the plague bacillus Yersinia pestis (62); and CUS-1 in the high-virulence clone E. coli O18:K1:H7 (63). In this context, our work emphasizes the necessity to study CTXΦ infection on its own and to overtake the Ff coliphage model of infection. It also sheds light on the mechanism underlying the initial filamentous phage–bacterial host binding and provides basic knowledge that might serve the understanding of transduction-dependent spreading of virulence factors in bacterial populations.
Experimental procedures
Bacterial strains and growth conditions
Relevant bacterial strains and plasmids used in this study are listed in supplemental Table S1. Bacteria were routinely cultivated in Luria-Bertani broth (LB, 407 mosm) at 37 °C (E. coli) or 30 °C (V. cholerae). When specified, MgCl2 (2 mm) was added to the culture medium to promote OM integrity and cell growth of tol mutants. Tryptone broth (1% (w/v) tryptone, 66 mm NaCl, pH 8.5) was used as a low osmolarity medium (123 mosm (64)). When indicated, antibiotics were added to the medium at the following concentrations: streptomycin (100 μg/ml); ampicillin (50 or 100 μg/ml); kanamycin (50 μg/ml); chloramphenicol (30 μg/ml for E. coli and 1 μg/ml for V. cholerae). V. cholerae tolA Δ(41–421) in-frame deletion mutant was constructed as described previously using the primers listed in supplemental Table S2 and the suicide plasmid pWM91 (65, 66). The absence of the TolA protein in the mutant strain was confirmed by Western blotting using polyclonal antibodies directed against E. coli TolAII-III protein (30) and cross-reacting with TolAIIIVc.
Plasmid construction
Polymerase chain reactions (PCR) were performed using Q5 High Fidelity DNA polymerase (New England Biolabs). Primer sets required to generate genetic constructs were synthesized by Sigma (supplemental Table S2). Enzymes (New England Biolabs) were used according to the manufacturer's instructions. Plasmids have been constructed either by standard restriction/ligation protocol, by Sequence and Ligase Independent Cloning (SLIC) (67) as modified by Jeong et al. (68), or by restriction-free cloning as described previously (69). Briefly for restriction-free cloning, genes of interest were amplified with oligonucleotides introducing extensions annealing to the target vector. The double-stranded product of the first PCR has then been used as oligonucleotides for a second PCR using the target vector as template and Pfu Turbo polymerase (Stratagene, La Jolla, CA). For BACTH plasmid constructs, the V. cholerae El Tor N16961 genome was used as a template to PCR-amplify the pIIICTX-encoding gene (orfU, at loci vc1460). The tolA sequence was amplified from V. cholerae O395 (locus VC0395_A1430) or from E. coli W3110 (locus BL257_RS03625) genomes. The pG3 vector (10) was used as a template to amplify pIIIM13. Amplified products were cloned into a pUT18c or a pKT25 expression vector (29) to generate fusions with the adenylate cyclase T18 and T25 domains. For construction of the pBAD-TolA rescue plasmid, the native sequence of V. cholerae O395 TolA, retaining the start and the stop codons, was amplified by PCR. The forward primer was designed to introduce a Shine-Dalgarno consensus sequence GAAGGAGATATACATACCC directly upstream of the start codon. The amplification product was then introduced into the pBAD18-Kan vector (70). For periplasmic expression, pIN-pIII-N1CTX plasmid was constructed by PCR-amplifying the pIII-N1CTX sequence (without start codon) with an upstream oligonucleotide encoding Strep-tag II (WSHPQFEK) and a downstream oligonucleotide encoding a C-terminal His6 sequence. Digestion of the PCR product and the pIN-ompA2 vector (33) with EcoRI and BamHI enzymes allowed subsequent ligation of the PCR product into the vector. The pIN-PIII-N1M13 plasmid (previously named pG3) has been described previously (10).
Mutations on pUT18-TolA, pKT25-pIII-N1, pIN-pIII-N1, and pBAD-TolA plasmids were performed by QuikChange site-directed mutagenesis using complementary pairs of oligonucleotides (listed in supplemental Table S2) and Pfu Turbo polymerase. All constructs were confirmed by DNA sequencing (Eurofins, MWG).
Construction of the Oxi-BTH strain
To conduct bacterial two-hybrid experiments in an oxidative environment, the E. coli K12 SHuffle T7 strain (New England Biolabs) was engineered further. This initial strain is deleted for glutaredoxin reductase (gor) and thioredoxin reductase (trxB) genes, which allows disulfide bond formation in the cytoplasm. Moreover, cytoplasmic expression of the disulfide bond isomerase DsbC acts on proteins with multiple disulfide bonds, promoting correct disulfide bond formation and proper folding. In this genetic background, the cya° mutation was transduced using a P1 lysate of an E. coli K12 cya° strain. The resulting mutant strain, named Oxi-BTH, was unable to ferment sugars, and consequently it grew as white colonies when plated on MacConkey plates.
Bacterial two-hybrid assay in E. coli BTH101 and Oxi-BTH strains
The adenylate cyclase-based bacterial two-hybrid technique was used as published previously (29), with the following modifications. Pairs of proteins to be tested were fused to the isolated T18 and T25 catalytic domains of the Bordetella adenylate cyclase. After transformation of the two plasmids producing the fusion proteins into the reporter BTH101 or Oxi-BTH strains, plates were incubated at 37 °C for 24 h. Three colonies for each transformation were inoculated into 600 μl of LB medium supplemented with ampicillin, kanamycin, and IPTG (0.5 mm). After overnight growth at 30 °C, 5 μl of each culture were dropped onto MacConkey plates supplemented with ampicillin, kanamycin, and IPTG (0.5 mm). Plates were incubated for 16–24 h (BTH101) and 2–3 days (Oxi-BTH) at 30 °C. The experiments were done at least in triplicate, and a representative assay is shown.
SDS-PAGE and immunoblotting
Protein samples resuspended in 2× loading buffer were subjected to SDS-PAGE. When specified, 2-mercaptoethanol (5% final) was added to the samples. For detection by immunostaining, proteins were transferred onto nitrocellulose membranes, and immunoblots were probed with primary antibodies and goat secondary antibodies coupled to alkaline phosphatase and developed in alkaline buffer in the presence of 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium. The anti-TolAIIIEc polyclonal antibodies are from our laboratory collection, and the anti-5 His monoclonal antibody (Qiagen), the anti-CyaA monoclonal antibody (3D1, Santa Cruz Biotechnology), and alkaline phosphatase-conjugated goat anti-rabbit and anti-mouse antibodies (Millipore) have been purchased as indicated.
V. cholerae phenotypic analysis
Sensitivity test to SDS
V. cholerae cells harboring the empty plasmid as a control or the plasmid encoding the constructs of interest were grown in LB medium at 30 °C until stationary phase, then back-diluted to initial A595 nm = 0.2 in LB supplemented or not with 0.125% SDS, and grown for 7 h at 30 °C with agitation. The percentage of surviving cells was estimated from the turbidity ratio of the SDS-treated cells and the control samples. Experiments were performed in triplicates.
Sensitivity test to deoxycholate
Normalized serial dilutions of strains to be tested were spotted onto 1% DOC-supplemented LB plates. After overnight incubation at 37 °C, survival was reported as the highest dilution of strain able to form colonies.
Growth in low osmolarity conditions
The different strains were grown in LB medium at 30 °C until stationary phase, then back-diluted 100-fold in LB medium (osmolarity 407 mosm) and in tryptone broth (1% (w/v) tryptone, 66 mm NaCl, pH 8.5, 123 mosm (64)), and incubated for 16 h at 30 °C. The percentage of growth was estimated from the turbidity ratio of the tested strains and the control sample. Experiments were performed in triplicates.
CTX-cm phage particle preparation
The dif1− strain BS2 (71) harboring the chloramphenicol-marked CTXEl Tor phage replicative form (pCTX-cm) was used as a donor strain to produce CTX phage particles. The donor strain was streaked onto LB-Cm (1 μg/ml) plates and incubated at 37 °C overnight. A single colony was used to inoculate a 2-ml LB + Sm 100 culture and incubated at 37 °C for 16 h. Cell were pelleted by centrifugation, and the supernatant containing phages was filter-sterilized using a 0.22-μm syringe filter. Phage preparation was checked for sterility by plating on an LB plate.
Susceptibility to CTX phage infection assays
Cell susceptibility to phage infection was conducted as described previously (72). Briefly, for each recipient strain three independent clones were cultivated separately in TCP-inducing conditions (LB 1% tryptone, 0.5% yeast extract, 0.5% NaCl, pH 6.5, 30 °C). After 20 h of growth, 75 μl of cells were mixed with 75 μl of CTX-cm phage suspension. The mixture was incubated during 30 min at room temperature without shaking, then 500 μl of LB was added, and the cell suspension was incubated at 37 °C with vigorous shaking for 45 min to allow cell recovery. Then dilutions of the cell suspension were plated on LB agar plates supplemented with Sm (100 μg/ml) or with Sm (50 μg/ml), and Cm (1 μg/ml) to enumerate total cells and transductant cells, respectively. The frequency of infection was determined by dividing the number of transductants by the number of total recipient cells.
NMR spectroscopy
For NMR studies, a TolA(237–356)Vc (TolAIIIVc) construct was included in a plasmid with the gene of OmpA signal sequence for protein secretion at the N terminus and His6 tag for the purification at the C terminus. For the native and variant pIII-N1CTX, the OmpA signal sequence and His6 tag construct were used, with an additional strep tag at the N terminus of the gene sequence. 15N isotopic labeling of native and mutant pIII-N1CTX was obtained from bacteria grown on M9 medium containing 1 g/liter 15NH4Cl as sole source of nitrogen. The proteins were overexpressed in E. coli BL21 strain. Protein production was obtained after 2 h of IPTG induction at 37 °C. Protein purification was obtained from periplasmic extract pulled on nickel-nitrilotriacetic acid-agarose and eluted with imidazole step gradient.
NMR spectra were recorded on a Bruker 600 MHz spectrometer equipped with a TCI cryoprobe at 300 K. 1H-15N HSQC spectra were processed with TopSpin software. For NMR experiments pIII-N1CTX wild type and (E37Q/D39N) double mutant, the concentration was 0.16 mm in 50 mm NaPO4, 50 mm NaCl buffer at pH 6.9. In the case of the complexes, the final TolAIIIVc concentration was 0.32 mm.
In silico analysis
Search for V. cholerae O395 TolA orthologous sequences was performed with BlastP and restricted to Vibrionaceae species. Multisequence alignments were performed using Clustal Omega (73) and color-coded with JalView2 tool (24). The 2P2I database (51, 52) was used to list all the non-bonded contacts at the complex interface (PDB code 4G7X) with a cutoff distance of 4 Å.
Author contributions
L. H., M. N., F. G., D. D., and R. L. contributed to the design of the study; L. H., R. N., M. N., and F. G. performed experiments. L. H., R. N., M. N., F. G., D. D., and R. L. contributed to data interpretation. L. H. wrote most of the manuscript. All authors approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank François-Xavier Barre, Paula Watnick, Delphine Destoumieux-Gazon, Marie-Stephanie Aschteng, Long-Fei Wu, and Emmanuelle Bouveret for providing useful strains and plasmids. We thank Emmanuelle Bouveret, Eric Cascales, Bérengère Ize, Marlon Sidore, and all the members of the Laboratoire d'Ingénierie des Systèmes Macromoléculaires for helpful discussions and support; Annick Brun, Isabelle Bringer, Moly Ba, Olivier Uderso, and Faniry Raboanarivola for technical assistance; and Octave Uhleure for encouragements. We thank Olivier Bornet at the NMR platform of IMM for NMR time machine necessary for recording NMR spectra.
This work was supported in part by CNRS and the Aix-Marseille Université (to the R. L. and W. G. Laboratories) and Agence Nationale de la Recherche Grant ANR-14-CE09-0023 (to the R. L. research team). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S6 and Tables S1–S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- TCP
- toxin co-regulated pilus
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- CT
- cholera toxin
- PDB
- Protein Data Bank
- OM
- outer membrane
- IM
- inner membrane
- BACTH
- bacterial two-hybrid
- DOC
- deoxycholate
- HSQC
- heteronuclear single quantum coherence
- Sm
- streptomycin
- Cm
- chloramphenicol.
References
- 1. Waldor M. K., and Mekalanos J. J. (1996) Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914 [DOI] [PubMed] [Google Scholar]
- 2. Mai-Prochnow A., Hui J. G., Kjelleberg S., Rakonjac J., McDougald D., and Rice S. A. (2015) Big things in small packages: the genetics of filamentous phage and effects on fitness of their host. FEMS Microbiol. Rev. 39, 465–487 [DOI] [PubMed] [Google Scholar]
- 3. Rakonjac J., Bennett N. J., Spagnuolo J., Gagic D., and Russel M. (2011) Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 13, 51–76 [PubMed] [Google Scholar]
- 4. Holliger P., and Riechmann L. (1997) A conserved infection pathway for filamentous bacteriophages is suggested by the structure of the membrane penetration domain of the minor coat protein g3p from phage fd. Structure 5, 265–275 [DOI] [PubMed] [Google Scholar]
- 5. Heilpern A. J., and Waldor M. K. (2003) pIIICTX, a predicted CTX minor coat protein, can expand the host range of coliphage fd to include Vibrio cholerae. J. Bacteriol. 185, 1037–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heilpern A. J., and Waldor M. K. (2000) CTXΦ infection of Vibrio cholerae requires the tolQRA gene products. J. Bacteriol. 182, 1739–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng L.-W., and Perham R. N. (2002) Delineating the site of interaction on the pIII protein of filamentous bacteriophage fd with the F-pilus of Escherichia coli. J. Mol. Biol. 319, 603–614 [DOI] [PubMed] [Google Scholar]
- 8. Lubkowski J., Hennecke F., Plückthun A., and Wlodawer A. (1999) Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Structure 7, 711–722 [DOI] [PubMed] [Google Scholar]
- 9. Lubkowski J., Hennecke F., Plückthun A., and Wlodawer A. (1998) The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p. Nat. Struct. Biol. 5, 140–147 [DOI] [PubMed] [Google Scholar]
- 10. Pommier S., Gavioli M., Cascales E., and Lloubès R. (2005) Tol-dependent macromolecule import through the Escherichia coli cell envelope requires the presence of an exposed TolA binding motif. J. Bacteriol. 187, 7526–7534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riechmann L., and Holliger P. (1997) The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell 90, 351–360 [DOI] [PubMed] [Google Scholar]
- 12. Deprez C., Lloubès R., Gavioli M., Marion D., Guerlesquin F., and Blanchard L. (2005) Solution structure of the E. coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain. J. Mol. Biol. 346, 1047–1057 [DOI] [PubMed] [Google Scholar]
- 13. Bennett N. J., and Rakonjac J. (2006) Unlocking of the filamentous bacteriophage virion during infection is mediated by the C domain of pIII. J. Mol. Biol. 356, 266–273 [DOI] [PubMed] [Google Scholar]
- 14. Ng D., Harn T., Altindal T., Kolappan S., Marles J. M., Lala R., Spielman I., Gao Y., Hauke C. A., Kovacikova G., Verjee Z., Taylor R. K., Biais N., and Craig L. (2016) The Vibrio cholerae minor Pilin TcpB initiates assembly and retraction of the toxin-coregulated pilus. PLoS Pathog. 12, e1006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ford C. G., Kolappan S., Phan H. T., Waldor M. K., Winther-Larsen H. C., and Craig L. (2012) Crystal structures of a CTX pIII domain unbound and in complex with a Vibrio cholerae TolA domain reveal novel interaction interfaces. J. Biol. Chem. 287, 36258–36272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lloubès R., Cascales E., Walburger A., Bouveret E., Lazdunski C., Bernadac A., and Journet L. (2001) The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152, 523–529 [DOI] [PubMed] [Google Scholar]
- 17. Sturgis J. N. (2001) Organisation and evolution of the tol-pal gene cluster. J. Mol. Microbiol. Biotechnol. 3, 113–122 [PubMed] [Google Scholar]
- 18. Gray A. N., Egan A. J., Van't Veer I. L., Verheul J., Colavin A., Koumoutsi A., Biboy J., Altelaar A. M., Damen M. J., Huang K. C., Simorre J. P., Breukink E., den Blaauwen T., Typas A., Gross C. A., Vollmer W. (2015) Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. Elife 4, e07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gully D., and Bouveret E. (2006) A protein network for phospholipid synthesis uncovered by a variant of the tandem affinity purification method in Escherichia coli. Proteomics 6, 282–293 [DOI] [PubMed] [Google Scholar]
- 20. Cascales E., Gavioli M., Sturgis J. N., and Lloubès R. (2000) Proton motive force drives the interaction of the inner membrane TolA and outer membrane pal proteins in Escherichia coli. Mol. Microbiol. 38, 904–915 [DOI] [PubMed] [Google Scholar]
- 21. Cascales E., Lloubès R., and Sturgis J. N. (2001) The TolQ–TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA–MotB. Mol. Microbiol. 42, 795–807 [DOI] [PubMed] [Google Scholar]
- 22. Walburger A., Lazdunski C., and Corda Y. (2002) The Tol/Pal system function requires an interaction between the C-terminal domain of TolA and the N-terminal domain of TolB. Mol. Microbiol. 44, 695–708 [DOI] [PubMed] [Google Scholar]
- 23. Cascales E., and Lloubès R. (2004) Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box: Pal interacts independently with OmpA, TolA and TolB. Mol. Microbiol. 51, 873–885 [DOI] [PubMed] [Google Scholar]
- 24. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gerding M. A., Ogata Y., Pecora N. D., Niki H., and de Boer P. A. (2007) The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol. Microbiol. 63, 1008–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yeh Y.-C., Comolli L. R., Downing K. H., Shapiro L., and McAdams H. H. (2010) The caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J. Bacteriol. 192, 4847–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Santos T. M., Lin T.-Y., Rajendran M., Anderson S. M., and Weibel D. B. (2014) Polar localization of Escherichia coli chemoreceptors requires an intact Tol-Pal complex: chemoreceptor localization in Escherichia coli. Mol. Microbiol. 92, 985–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li C., Zhang Y., Vankemmelbeke M., Hecht O., Aleanizy F. S., Macdonald C., Moore G. R., James R., and Penfold C. N. (2012) Structural evidence that colicin A protein binds to a novel binding site of TolA protein in Escherichia coli periplasm. J. Biol. Chem. 287, 19048–19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karimova G., Pidoux J., Ullmann A., and Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Derouiche R., Zeder-Lutz G., Bénédetti H., Gavioli M., Rigal A., Lazdunski C., and Lloubès R. (1997) Binding of colicins A and El to purified TolA domains. Microbiology 143, 3185–3192 [DOI] [PubMed] [Google Scholar]
- 31. Lobstein J., Emrich C. A., Jeans C., Faulkner M., Riggs P., and Berkmen M. (2012) SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 11, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martin A., and Schmid F. X. (2003) The folding mechanism of a two-domain protein: folding kinetics and domain docking of the gene-3 protein of phage fd. J. Mol. Biol. 329, 599–610 [DOI] [PubMed] [Google Scholar]
- 33. Ghrayeb J., Kimura H., Takahara M., Hsiung H., Masui Y., and Inouye M. (1984) Secretion cloning vectors in Escherichia coli. EMBO J. 3, 2437–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bouveret E., Rigal A., Lazdunski C., and Bénédetti H. (1998) Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol. Microbiol. 27, 143–157 [DOI] [PubMed] [Google Scholar]
- 35. Meury J., and Devilliers G. (1999) Impairment of cell division in tolA mutants of Escherichia coli at low and high medium osmolarities. Biol. Cell 91, 67–75 [PubMed] [Google Scholar]
- 36. Levengood S. K., Beyer W. F. Jr., and Webster R. E. (1991) TolA: a membrane protein involved in colicin uptake contains an extended helical region. Proc. Natl. Acad. Sci. U.S.A. 88, 5939–5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buchan D. W., Minneci F., Nugent T. C., Bryson K., and Jones D. T. (2013) Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 41, W349–W357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tomasi M., Battistini A., Araco A., Roda L. G., and D'Agnolo G. (1979) The role of the reactive disulfide bond in the interaction of cholera-toxin functional regions. Eur. J. Biochem. 93, 621–627 [DOI] [PubMed] [Google Scholar]
- 39. Craig L., Pique M. E., and Tainer J. A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 [DOI] [PubMed] [Google Scholar]
- 40. Gao Y., Hauke C. A., Marles J. M., and Taylor R. K. (2016) Effects of tcpB mutations on biogenesis and function of the toxin-coregulated pilus, the type IVb Pilus of Vibrio cholerae. J. Bacteriol. 198, 2818–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang M., Liu Z., Hughes C., Stern A. M., Wang H., Zhong Z., Kan B., Fenical W., and Zhu J. (2013) Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc. Natl. Acad. Sci. U.S.A. 110, 2348–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morgan S. J., French E. L., Thomson J. J., Seaborn C. P., Shively C. A., and Krukonis E. S. (2015) Formation of an intramolecular periplasmic disulfide bond in TcpP protects TcpP and TcpH from degradation in Vibrio cholerae. J. Bacteriol. 198, 498–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fengler V. H., Boritsch E. C., Tutz S., Seper A., Ebner H., Roier S., Schild S., and Reidl J. (2012) Disulfide bond formation and ToxR activity in Vibrio cholerae. PLoS ONE 7, e47756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Battesti A., and Bouveret E. (2012) The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58, 325–334 [DOI] [PubMed] [Google Scholar]
- 45. Ouellette S. P., Gauliard E., Antosová Z., and Ladant D. (2014) A Gateway®-compatible bacterial adenylate cyclase-based two-hybrid system. Environ. Microbiol. Rep. 6, 259–267 [DOI] [PubMed] [Google Scholar]
- 46. Handford J. I., Ize B., Buchanan G., Butland G. P., Greenblatt J., Emili A., and Palmer T. (2009) Conserved network of proteins essential for bacterial viability. J. Bacteriol. 191, 4732–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Houot L., Fanni A., de Bentzmann S., and Bordi C. (2012) A bacterial two-hybrid genome fragment library for deciphering regulatory networks of the opportunistic pathogen Pseudomonas aeruginosa. Microbiology 158, 1964–1971 [DOI] [PubMed] [Google Scholar]
- 48. Gaspar J. A., Thomas J. A., Marolda C. L., and Valvano M. A. (2000) Surface expression of O-specific lipopolysaccharide in Escherichia coli requires the function of the TolA protein. Mol. Microbiol. 38, 262–275 [DOI] [PubMed] [Google Scholar]
- 49. Dennis J. J., Lafontaine E. R., and Sokol P. A. (1996) Identification and characterization of the tolQRA genes of Pseudomonas aeruginosa. J. Bacteriol. 178, 7059–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lorenz S. H., Jakob R. P., Weininger U., Balbach J., Dobbek H., and Schmid F. X. (2011) The filamentous phages fd and IF1 use different mechanisms to infect Escherichia coli. J. Mol. Biol. 405, 989–1003 [DOI] [PubMed] [Google Scholar]
- 51. Basse M. J., Betzi S., Bourgeas R., Bouzidi S., Chetrit B., Hamon V., Morelli X., and Roche P. (2013) 2P2Idb: a structural database dedicated to orthosteric modulation of protein/protein interactions. Nucleic Acids Res. 41, D824–D827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Basse M.-J., Betzi S., Morelli X., and Roche P. (2016) 2P2Idb v2: update of a structural database dedicated to orthosteric modulation of protein/protein interactions. Database 2016 baw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bonsor D. A., Hecht O., Vankemmelbeke M., Sharma A., Krachler A. M., Housden N. G., Lilly K. J., James R., Moore G. R., and Kleanthous C. (2009) Allosteric β-propeller signalling in TolB and its manipulation by translocating colicins. EMBO J. 28, 2846–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sinha N., and Smith-Gill S. J. (2002) Electrostatics in protein binding and function. Curr. Protein Pept. Sci. 3, 601–614 [DOI] [PubMed] [Google Scholar]
- 55. Faruque S. M., Asadulghani, Saha M. N., Alim A. R., Albert M. J., Islam K. M., and Mekalanos J. J. (1998) Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXΦ: molecular basis for origination of new strains with epidemic potential. Infect. Immun. 66, 5819–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hasan N. A., Ceccarelli D., Grim C. J., Taviani E., Choi J., Sadique A., Alam M., Siddique A. K., Sack R. B., Huq A., and Colwell R. R. (2013) Distribution of virulence genes in clinical and environmental Vibrio cholerae strains in Bangladesh. Appl. Environ. Microbiol. 79, 5782–5785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ceccarelli D., Chen A., Hasan N. A., Rashed S. M., Huq A., and Colwell R. R. (2015) Non-O1/non-O139 Vibrio cholerae carrying multiple virulence factors and V. cholerae O1 in the Chesapeake Bay, Maryland. Appl. Environ. Microbiol. 81, 1909–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jiang S., Chu W., and Fu W. (2003) Prevalence of cholera toxin genes (ctxA and zot) among non-O1/O139 Vibrio cholerae strains from Newport Bay, California. Appl. Environ. Microbiol. 69, 7541–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sarkar A., Nandy R. K., Nair G. B., and Ghose A. C. (2002) Vibrio pathogenicity island and cholera toxin genetic element-associated virulence genes and their expression in non-O1 non-O139 strains of Vibrio cholerae. Infect. Immun. 70, 4735–4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boyd E. F., Moyer K. E., Shi L., and Waldor M. K. (2000) Infectious CTXΦ and the Vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect. Immun. 68, 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bille E., Zahar J.-R., Perrin A., Morelle S., Kriz P., Jolley K. A., Maiden M. C., Dervin C., Nassif X., and Tinsley C. R. (2005) A chromosomally integrated bacteriophage in invasive meningococci. J. Exp. Med. 201, 1905–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Derbise A., Chenal-Francisque V., Pouillot F., Fayolle C., Prévost M.-C., Médigue C., Hinnebusch B. J., and Carniel E. (2007) A horizontally acquired filamentous phage contributes to the pathogenicity of the plague Bacillus. Mol. Microbiol. 63, 1145–1157 [DOI] [PubMed] [Google Scholar]
- 63. Gonzalez M. D., Lichtensteiger C. A., Caughlan R., and Vimr E. R. (2002) Conserved filamentous prophage in Escherichia coli O18:K1:H7 and Yersinia pestis biovar orientalis. J. Bacteriol. 184, 6050–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goforth J. B., Walter N. E., and Karatan E. (2013) Effects of polyamines on Vibrio cholerae virulence properties. PLoS ONE 8, e60765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Metcalf W. W., Jiang W., Daniels L. L., Kim S. K., Haldimann A., and Wanner B. L. (1996) Conditionally replicative and conjugative plasmids carrying lacZ α for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35, 1–13 [DOI] [PubMed] [Google Scholar]
- 66. Houot L., and Watnick P. I. (2008) A novel role for enzyme I of the Vibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a Biofilm. J. Bacteriol. 190, 311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li M. Z., and Elledge S. J. (2007) Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 [DOI] [PubMed] [Google Scholar]
- 68. Jeong J.-Y., Yim H.-S., Ryu J.-Y., Lee H. S., Lee J.-H., Seen D.-S., and Kang S. G. (2012) One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl. Environ. Microbiol. 78, 5440–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. van den Ent F., and Löwe J. (2006) RF cloning: a restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods. 67, 67–74 [DOI] [PubMed] [Google Scholar]
- 70. Guzman L.-M., Belin D., Carson M. J., and Beckwith J. O. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Das B., Bischerour J., Val M.-E., and Barre F.-X. (2010) Molecular keys of the tropism of integration of the cholera toxin phage. Proc. Natl. Acad. Sci. U.S.A. 107, 4377–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kirn T. J., Lafferty M. J., Sandoe C. M., and Taylor R. K. (2000) Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35, 896–910 [DOI] [PubMed] [Google Scholar]
- 73. McWilliam H., Li W., Uludag M., Squizzato S., Park Y. M., Buso N., Cowley A. P., and Lopez R. (2013) Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.