Abstract
Hemopexin protects against heme toxicity in hemolytic diseases and conditions, sepsis, and sickle cell disease. This protection is sustained by heme-hemopexin complexes in biological fluids that resist oxidative damage during heme-driven inflammation. However, apo-hemopexin is vulnerable to inactivation by reactive nitrogen (RNS) and oxygen species (ROS) that covalently modify amino acids. The resultant nitration of amino acids is considered a specific effect reflecting biological events. Using LC-MS, we discovered low endogenous levels of tyrosine nitration in the peptide YYCFQGNQFLR in the heme-binding site of human hemopexin, which was similarly nitrated in rabbit and rat hemopexins. Immunoblotting and selective reaction monitoring were used to quantify tyrosine nitration of in vivo samples and when hemopexin was incubated in vitro with nitrating nitrite/myeloperoxidase/glucose oxidase. Significantly, heme binding by hemopexin declined as tyrosine nitration proceeded in vitro. Three nitrated tyrosines reside in the heme-binding site of hemopexin, and we found that one, Tyr-199, interacts directly with the heme ring D propionate. Investigating the oxidative modifications of amino acids after incubation with tert-butyl hydroperoxide and hypochlorous acid in vitro, we identified additional covalent oxidative modifications on four tyrosine residues and one tryptophan residue of hemopexin. Importantly, three of the four modified tyrosines, some of which have more than one modification, cluster in the heme-binding site, supporting a hierarchy of vulnerable amino acids. We propose that during inflammation, apo-hemopexin is nitrated and oxidated in niches of the body containing activated RNS- and ROS-generating immune and endothelial cells, potentially impairing hemopexin's protective extracellular antioxidant function.
Keywords: glycoprotein, mass spectrometry (MS), proteomics, reactive nitrogen species (RNS), reactive oxygen species (ROS), covalent modification, hemopexin, tyrosine nitration
Introduction
In several diseases and conditions involving intra- and extravascular hemolysis where heme drives inflammation and vaso-occlusion among other pathology, the heme-binding glycoprotein hemopexin is protective (1, 2). These include genetic and non-genetic hemolytic diseases, e.g. sepsis, sickle cell disease, hemolytic anemias, hemolytic uremic syndrome, as well as hemolytic conditions during stages of dengue and malaria. Sufficient heme is released to put patients at risk from heme toxicity by extracorporeal circulation of blood during cardiac surgery as well as after extensive blood transfusions for cancer chemotherapy, trauma, car accidents, and war injuries. Hemopexin deficiency states develop in clinical sepses that are life-threatening (3, 4). Hemopexin is present in all biological fluids and is relatively high in cerebral spinal fluid where it protects brain cells from heme toxicity. In the plasma, hemopexin binds heme very tightly and in a manner that prevents the oxidative reactions of heme (5). The heme-hemopexin complexes are taken up by endocytosis into liver parenchymal cells where heme is safely degraded and its iron is recycled (6, 7). Normally, hemopexin recycles intact from the liver after delivering heme (6, 8) potentially for up to eight reuses (2).
Inflammation accompanies hemolysis and injury and is associated with increased oxidative stress driven in part via heme-mediated events. The function(s) of many proteins is severely impaired by oxidative modification(s) from reactive oxygen species (ROS).3 Heme-hemopexin complexes resist oxidative damage in vitro (9) and potentially in vivo during inflammation; however, data are accruing indicating that the apo-hemopexin may be vulnerable to inactivation by exposure to ROS that are elevated in vivo in inflammation. Therefore, we considered that the heme binding by hemopexin might become compromised in inflammatory states when ROS, including hypochlorous acid (HOCl), are present. The potential for ROS-mediated damage to apo-hemopexin was revealed by our recent in vitro studies (9). The heme-hemopexin complex was highly resistant to oxidative damage from peroxides and HOCl compared with the apoprotein (9).
In fact, in inflammatory conditions in vivo both ROS and reactive nitrogen species (RNS) are present. For example, activated neutrophils produce their defensive “respiratory burst” of reactive oxygen intermediates; and endothelial cells, macrophages, and astrocytes produce nitric oxide (NO•) after immunological or inflammatory stimuli. NO• is oxidized in vivo to nitrite and nitrate when it becomes a substrate for myeloperoxidase to nitrate proteins. Here, we have investigated whether there is damage to hemopexin from RNS. We have discovered low endogenous levels of nitrated tyrosines (Tyrn) in human (Homo sapiens), rabbit (Oryctolagus cuniculus), and rat (Rattus norvegicus) hemopexin isolated from plasma with standard techniques. Using liquid chromatography-tandem mass spectrometry (LC-MS/MS) and the 3D structure of heme-hemopexin, we identified a predominant endogenously nitrated peptide YYCFQGNQFLR, which is conserved in human, rabbit, and rat hemopexin. Immunoblotting and selective reaction monitoring (SRM) were used to quantitate the extent of nitration of hemopexin from in vivo sources as well as over time after nitrite/myeloperoxidase/glucose oxidase (N/MPO/GO) (10, 11) exposure in vitro. Significantly, we show for the first time that heme binding by hemopexin is increasingly impaired as tyrosine nitration proceeds in vitro. Two of the three nitrated tyrosines that are in the heme-binding site interact directly with heme, one of which resides in YYnCFQGNQFLR. Importantly, nitration is an event requiring a specific environment around the tyrosine residue and lack of steric hindrance because this amino acid is often embedded within the hydrophobic core of the protein. Thus, nitration of tyrosine residues is selective, and such specificity strongly supports the concept that these amino acids are a preferential target for key biological events in vivo (12, 13).
Furthermore, oxidative stress is linked to development of neurodegeneration, and inflammation is considered to contribute to the early stages of Alzheimer's disease (14) to which hemopexin has been linked (15). We have previously shown that three models of ROS, namely H2O2, tert-butyl hydroperoxide, and hypochlorous acid, decrease heme binding by hemopexin, although high molar ratios of ROS/hemopexin were needed, generally supraphysiologically (9). Here, we identify covalent oxidative modifications (COMs)4 of apo-hemopexin after exposure to tert-butyl hydroperoxide and hypochlorous acid. Several tyrosine residues that reside in the heme-binding site were targeted for modification by more than one oxidant and thus constitute susceptible targets for damage. In contrast, we previously showed that the heme-hemopexin complex is fairly resistant to damage by ROS (9). Even after heme-hemopexin is exposed to H2O2 or tert-butyl hydroperoxide, it still delivers heme for heme oxygenase-1 induction that is cytoprotective (9). We propose that in inflammatory conditions inactivation of apo-hemopexin in plasma or other biological fluids by oxidative modification likely occurs close to activated endothelial and immune system cells. Such cells are the sites of production of RNS and ROS in the body during inflammation, and they will be close to activated platelets, which reside at sites of injury and are needed for wound healing. These are conditions in which the protective heme binding function of hemopexin can be life-saving.
Results
Our analytical strategy was to identify endogenously nitrated amino acids in hemopexin isolated from human, rabbit, and rat plasma samples; identify their location in the primary amino acid sequence; determine the extent of conservation among the three species of hemopexin; determine their relative abundance; and map them onto the 3D crystal structure of hemopexin. We then planned to investigate in vitro whether this nitration impaired biological functions of hemopexin such as heme binding. Finally, we wished to assess whether the amino acids that are nitrated are also oxidatively modified by three ROS previously shown to potentially impair heme binding by hemopexin. We reasoned that, if so, there are regions of the apoprotein vulnerable to damage by RNS and ROS leading to loss of function of hemopexin in certain clinically relevant inflammatory conditions.
Identification and semi-quantitation of nitrated tyrosine residues in human hemopexin using LC-MS/MS, which are present also in the rat and rabbit congeners
Human, rabbit, and rat hemopexin are highly conserved proteins (16). Evidence for the occurrence of hemopexin tyrosine nitration in vivo comes from LC-MS/MS data, which show that native isolates of these three species of hemopexin from plasma (16) contain nitrated tyrosines. As expected from non-pathological sera samples, the abundance of nitration is low (see Table 1; for human hemopexin MS/MS data see Fig. 1).
Table 1.
Conserved nitrated tyrosine residues in hemopexin isolates from human, rabbit, and rat plasma
Quantifiable nitration in hemopexin was detected on a few conserved tyrosines as described under “Experimental procedures” and identified using LC-MS/MS of peptides from a trypsin digest of hemopexin from three different species. The predominant nitrotyrosine (Tyrn) identified was a tyrosine in the conserved tryptic peptide YYCFQGNQFLR in human HPX. Using the relative abundance and 3D structural data of heme-hemopexin, the tyrosine most often nitrated in the conserved tryptic peptide YYnCFQGNQFLR is from human (Tyr-199), rabbit (Tyr-201), and rat (Tyr-118) hemopexin. This assignment cannot be unequivocally supported by our current MS/MS spectrum analyses (because of the absence of key diagnostic fragments (b-1, or y-10 using collision induced diffusion, or CID on the ion trap) on different species of hemopexin but, as described under “Results,” the structural data allow only this conclusion. These MS/MS data were obtained from the following hemopexin isolates: eight individual human serum samples, three separate pooled rabbit sera samples, and one pooled rat serum. One human hemopexin isolate lacked detectable nitration.
| HPX species | NCBI gi no. | Peptide sequence | Tyrn site | Charge (+) | Sequest XCorr | Mascot ions score |
|---|---|---|---|---|---|---|
| Homo sapiens | 11321561 | 198YYnCFQGNQFLR208 | 199 | 2 | 3.068 | 77.1 |
| Rabbit | 130500366 | 200YYnCFQGNQFLR210 | 201 | 2 | 4.576 | 67.5 |
| Rat | 123036 | 117YYnCFQGNK124 | 118 | 2 | 2.507 | 46.9 |
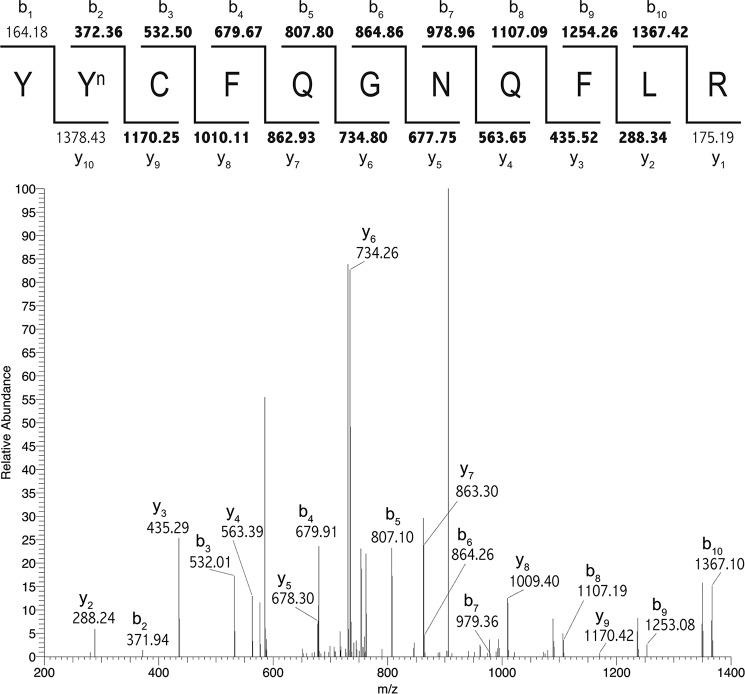
Figure 1.
Nitration of human hemopexin occurs in vivo at Tyr-199. Nitration of human hemopexin occurs in vivo at Tyr-199 detected by LC-MS/MS analysis. SEQUEST was used to identify Tyrn-199 on the tryptic peptide YYnCFQGNQFLR (see Table 1 for XCorr value). Matched b and y ions are shown in bold in the ion diagram (above) and are indicated next to corresponding peaks in the MS/MS spectrum (below). Similar high quality spectra were obtained from rabbit and rat hemopexin (see supplemental Figs. S1 and S2).
Using the relative abundance and 3D structural data of heme-hemopexin, the tyrosine most often nitrated resides in the conserved tryptic peptide YYCFQGNQFLR from human hemopexin (Fig. 1) and is considered to be tyrosine 199 (Tyr-199; YYnCFQGNQFLR). Additional data support that the same peptide is nitrated in other species of hemopexin where rabbit (Tyr-201) and rat (Tyr-118) hemopexin, and Tyr-201 and Tyr-118, respectively, are considered the principal targets (see Tables 1 and 2; for rabbit and rat hemopexin MS/MS data see supplemental Figs. S1 and S2, respectively). This amino acid numbering follows the precursor peptide sequences (derived from the gene) of human, rabbit, and rat hemopexin from the NCBI Protein resource.
Table 2.
Identification using LC-MS/MS of peptides with nitrated tyrosine residues in hemopexin isolates from rabbit plasma
Quantifiable nitration in hemopexin was detected on a few conserved tyrosines as described under “Experimental procedures.” Shown here are four nitrated tyrosines that were unambiguously identified in rabbit hemopexin. These data were obtained from three separate pooled rabbit sera samples (see scans 2592, 2715, 2720, and 2837 in RbtHPXTIIYn files in supplemental data).
| HPX species | NCBI gi number | Peptide sequence | Tyrn site | Charge (+) | Sequest XCorr | Mascot ions score |
|---|---|---|---|---|---|---|
| Rabbit | 130500366 | 118VWVYnTSEK125 | 121 | 2 | 2.100 | 34.7 |
| Rabbit | 130500366 | 228DYnFLSCPGR236 | 229 | 2 | 2.225 | 50.8 |
| Rabbit | 130500366 | 316LYnLIQDTK323 | 317 | 2 | 2.796 | 68.6 |
| Rabbit | 130500366 | 324VYnVFLTK330 | 325 | 2 | 2.601 | 52.5 |
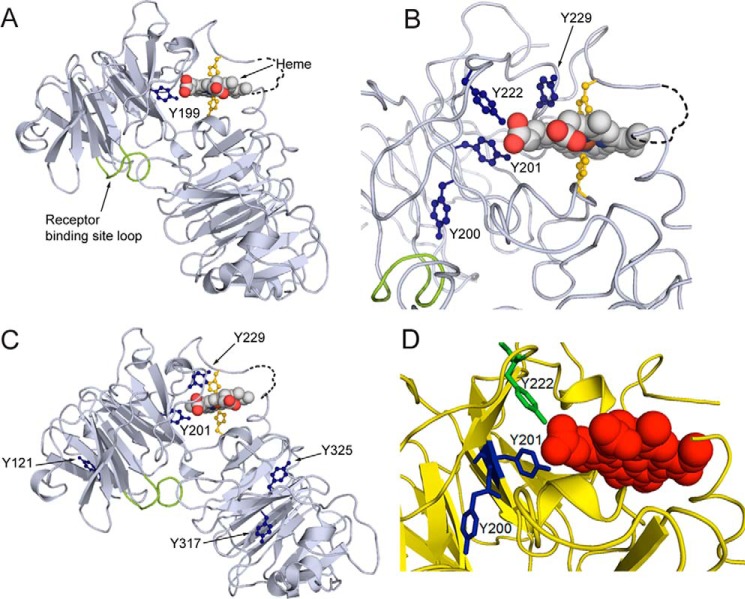
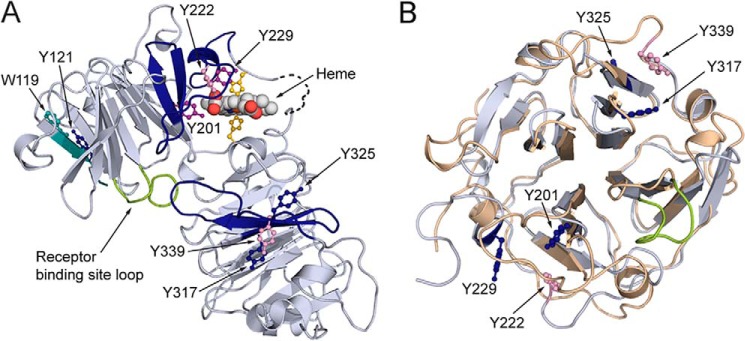
The COBALT multiple sequence alignment tool reveals that each molecule of human hemopexin has 16 tyrosine residues of which 13 are conserved with hemopexin from Norway rat and 15 with rabbit hemopexin, including Tyr-199 of human HPX. Mapping the human hemopexin sequence onto the rabbit 3D crystal structure revealed that this tyrosine (labeled Yn199 in Fig. 2A) interacts directly with the heme propionate in the D ring suggesting that heme binding is likely to be decreased or inactivated by this nitration. Although the spatial distribution of these tyrosine residues may be different in apo-hemopexin and the heme complex, Tyr-201 in the rabbit structure is near the surface of the apoprotein for heme binding. In contrast, Tyr-200 is far from the heme pointing away and buried deeply in the protein interior of the N-domain (Fig. 2B). This is even more apparent in the close up view in Fig. 2D. In rabbit hemopexin, in addition to this conserved nitrated Tyr-201, four other endogenously nitrated tyrosines were unambiguously identified in the MS/MS analyses of rabbit hemopexin isolates (Tyr-121, Tyr-229, Tyr-317, and Tyr-325, Table 2; and the spectra are in supplemental data). Tyr-229 is also close to the heme propionate in the heme-binding site (see Fig. 2D); whereas Tyr-121 is near the N terminus; and Tyr-317 and Tyr-325 are in the C-domain (Fig. 2C).
Figure 2.
Covalent oxidatively modified residues modeled on the rabbit hemopexin crystal structure show a cluster of amino acids in the functionally important heme-binding site. A, when the human hemopexin amino acid sequence is threaded on the rabbit hemopexin crystal structure, the nitrated tyrosine Tyrn-199 in the human sequence (labeled and marine blue; equivalent to Tyrn-201 in rabbit, B) resides in the heme-binding site. Nitration of this tyrosine after incubation of hemopexin with N/MPO/GO in vitro is conserved in human, rabbit, and rat proteins. Also shown are the heme-iron coordinating histidine residues (yellow), the receptor binding site (loop only shown of the Jen 14 epitope (lime)) (17), and the linker peptide modeled on the amino acid backbone (dashed black line). B, in the heme-binding site, the endogenously nitrated Tyr-201 and Tyr-222 interact directly with the propionate group on the D ring of the heme and contribute with other aromatic resides to the hydrophobic environment around the heme. Tyr-201 and Tyr-222 help surround this heme propionate together with Arg-199 and Arg-210 from the N-domain and three histidine residues from the C-domain (data not shown). C shows the endogenously nitrated tyrosine residues in rabbit hemopexin identified by LC-MS/MS and summarized in Table 1. D, in this close-up view, Tyr-200 clearly points away from the heme in contrast to Tyr-201 and Tyr-222 (green). These structures were generated using PyMOL (Molecular Graphics System, version 1.3).
Because hemopexin is not exposed to nitrating reagents during isolation, these observations on endogenous hemopexins from three species, including human, support that tyrosine nitration occurs in vivo, normally with low abundance in the absence of any pathology, and the variation observed, which we recognize is from a small preliminary set of plasma samples, reflects events in vivo. Significantly, key tyrosine residues in the heme-binding site of hemopexin that help stabilize and orient the heme are vulnerable to nitration in vivo.
Enzymatic nitration of human hemopexin in vitro increases the abundance of Tyr- n199 in a population of hemopexin molecules
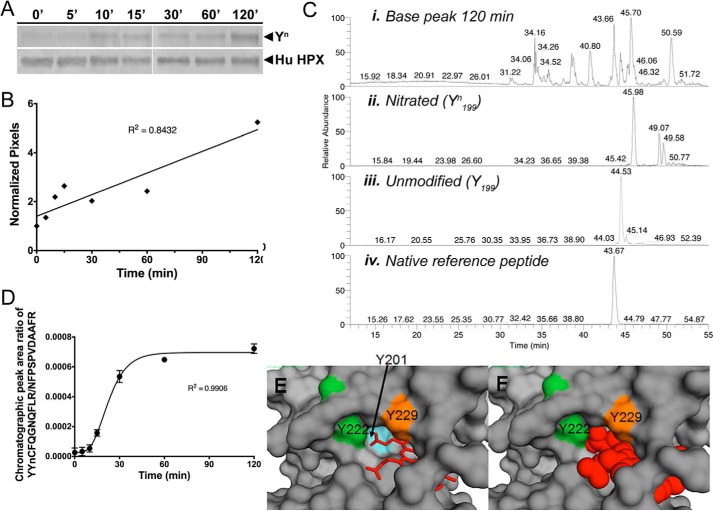
Because hemopexin is likely to be exposed to nitric oxide and other nitrating species in inflammatory conditions that can also accompany hemolysis in the presence and absence of infection, we wanted to determine the abundance of site-specific nitrated tyrosines of human hemopexin. For this we used a gentle, controlled myeloperoxidase/glucose oxidase (N/MPO/GO)-mediated enzymatic nitration system that produces physiological concentrations of oxidants in vitro (10). There is an ∼5-fold increase in nitration of human apo-hemopexin after a 2-h incubation with N/MPO/GO, detected by immunoblotting with anti-3′-nitrotyrosine antibody (Fig. 3, A and B). Similar results were found for rabbit hemopexin (data not shown). Tyrosine nitration in the tryptic peptide YYCFQGNQFLR from native human hemopexin is also nitrated and in vitro detected and identified by LC-MS/MS analyses (Fig. 3, C and D, and supplemental Fig. S3). MASCOT with the site analysis feature cannot unequivocally identify Tyr-118 in rat hemopexin, Tyr-199 in human hemopexin, or Tyr-201 in rabbit hemopexin. However, as mentioned above, our identification of HuTyr-199/RbtTyr-201 as the nitrated tyrosine in this sequence is bolstered by the fact that the 3D structure of hemopexin, albeit with heme bound, supports the sole conclusion that nitration occurs at human hemopexin Tyr-199. This is because this residue side chain is more accessible on the surface of the N-domain, and the four-bladed propeller domains are very stable entities. Significantly, Tyr-199 is clearly blocked by heme in the complex (see Fig. 3, E and F). Selective reaction monitoring (SRM) of human apo-hemopexin at various times after in vitro nitration shows that the extent of nitration of the YYCFQGNQFLR peptide, which we propose is Tyr-199 in the YYnCFQGNQFLR peptide of human hemopexin, increases with the length of exposure time to N/MPO/GO (Fig. 3, C–F). Similar SRM data have also been obtained for the extent of nitration of Tyr-201 of rabbit hemopexin in vitro (data not shown).
Figure 3.
Time course of in vitro nitration of Tyr-199 in human hemopexin. Time course of in vitro nitration of Tyr-199 in human hemopexin. A and B, Western immunoblotting and quantitation using UN-SCAN-IT software show the relative rate of appearance of nitrotyrosine (Tyrn) in human hemopexin compared with total hemopexin (normalized pixels) after incubation with N/MPO/GO. C, chromatographic peaks from SRM analysis for human hemopexin after 120 min of incubation with N/MPO/GO are shown for the base peak (panel i), nitrated peptide, YYnCFQGNQFLR (panel ii), unmodified peptide, YYCFQGNQFLR (panel iii), and internal reference peptide, NFPSPVDAAFR, for normalizing (panel iv). They were quantified by integration using the native reference peptide method (11). The two minor peaks in the unmodified peptide mass chromatogram that appear to the right of the major peak (panel ii) are unrelated ions with the same m/z, coincidentally. These did not interfere with and were not included in the peak area integration for quantitative analysis. D, time course of nitration using SRM data was quantitated using calibration curves (see supplemental Fig. 4) to determine the mole fraction of in vitro nitrated Tyr-199 in human hemopexin over the period of 120 min. Average peak ratio (nitrated/internal reference) of three separate gel excisions (biological replicates) and analysis per time point is shown ± S.D. Standard curves were calibrated using synthetic peptide chromatographic peak area ratios at each mole fraction point. Experimental chromatographic peak area ratios were obtained by dividing the reference peptide peak area by that of either the YYnCFQGNQFLR (E) or the YYCFQGNQGLR (F) peptide. Mole fractions were calculated as described under “Experimental procedures.” The mole ratio for nitration increased with time of incubation with the N/MPO/GO-nitrating system. E and F show a surface representation of the atoms of rabbit hemopexin with a close-up view of the three tyrosine residues that interact directly with the heme (shown as a stick representation in E and space-filling model in F). Tyr-201 is barely visible when heme is bound, consistent with the vulnerability to oxidative modification of this amino acid residue in the apoprotein. The label for Tyr-222 is positioned over the ring side chain, and this amino acid is protected by a loop (see also Fig. 2D).
Overall, these data reveal that nitration in vitro of human hemopexin increases with time, and the abundance of Tyrn-199, as a representative and proposed principal target, increases as nitration proceeds. This demonstrates that the population of hemopexin molecules with one or more nitrated tyrosines in the heme-binding site may increase in biological fluids in vivo depending upon the duration of exposure to and the concentration of nitrating species present in inflammatory conditions.
Apo-hemopexin, the circulating form in plasma, is more susceptible to nitration than the heme-hemopexin complex
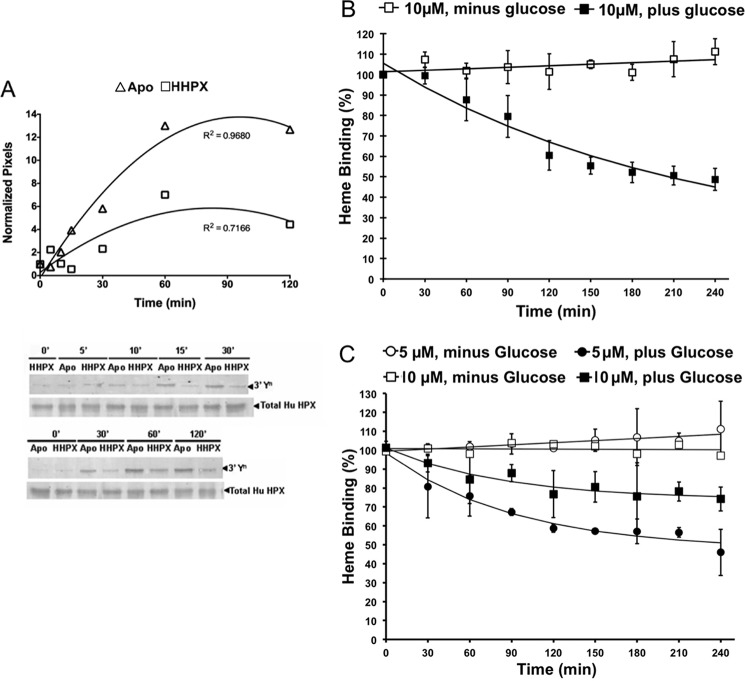
As shown in Fig. 2, hemopexin is composed of two four-bladed β-propeller domains linked by a hinge region (17, 18). Heme binding causes several conformational changes in hemopexin as follows: a compaction in size, enhanced inter-domain interactions, and protection of the hinge region from proteolysis (18, 19). Therefore, we expected that apo-hemopexin, which normally predominates in human plasma and other biological fluids including CSF, would be more vulnerable to nitration than the heme complex. The immunoblot data in Fig. 4A show that nitrated human apo-hemopexin is more extensively nitrated than the heme-hemopexin complex (Fig. 4A). Significantly higher levels of nitration of apo-hemopexin are apparent within 10–15 min of incubation with N/MPO/GO at 37 °C and continue to increase for 1–2 h. Similar results have been obtained with rabbit hemopexin (data not shown). The nitration of the heme complex detected by immunoblotting is likely due to nitration of three tyrosines in the C-domain (see Fig. 4A).
Figure 4.
Human apo-hemopexin is more susceptible to nitration than the heme-hemopexin complex and tyrosine nitration inhibits heme binding by hemopexin. Human apo-hemopexin is more susceptible to nitration than the heme-hemopexin complex, and tyrosine nitration inhibits heme binding by hemopexin. A, apo- and heme-human hemopexin were nitrated in vitro. The signal intensity of nitrated protein relative to total protein in the apo-hemopexin (triangles) and heme complex (squares) was quantitated using UN-SCAN-IT and normalized to the time 0 samples. The higher nitration of apo-hemopexin is apparent within 10–15-min incubation at 37 °C. After 2 h, apo-hemopexin was nitrated ∼2.6-fold more than heme-hemopexin. B, data show the amount of heme bound to nitrated hemopexin (with glucose, filled squares) determined (after 5 min at 25 °C) from the absorbance in the Soret region using our standard procedures (43) and compared with control hemopexin (minus glucose, open squares) treated in parallel with N/MPO/GO. As nitration of human hemopexin increased, heme binding was progressively impaired. Data shown are the average and S.D. of three independent experiments, with R2 values of 0.306 (linear fit) and 0.9478 (monoexponential fit) for the plus and minus glucose data sets, respectively. Heme binding of hemopexin in each of the aliquots was carried out initially by the addition of 1.0 molar eq of heme and after 90 min with 0.5 molar eq of heme to minimize spurious absorbance in the Soret region due to non-bound heme. C, rabbit hemopexin is slightly more resistant to N/MPO/GO nitration than the human congener. Furthermore, the extent of inactivation is greater when the ratio of nitrating species to hemopexin is increased (i.e. by decreasing the hemopexin concentration). Data shown are from 5 and 10 μm rabbit hemopexin (circles and squares) from three and six independent experiments, respectively. For the minus glucose controls, R2 values were 0.7737 and 0.0078 (linear fit) for 5 and 10 μm hemopexin (open symbols), respectively, and R2 values were 0.9663 and 0.9042 (monoexponential fit) for the 5 and 10 μm nitrated hemopexin data sets (filled symbols), respectively. The data in B and C were analyzed in GraphPad Prism and plotted in Excel. Overall, these data support that the conformational changes that occur upon heme binding directly and indirectly protect amino acid residues from N/MPO/GO oxidative modifications, and significantly, tyrosine nitration impairs the ability of hemopexin to bind heme.
Tyrosine nitration in vitro decreases the ability of human hemopexin to bind heme
The most clinically relevant heme complex is protoheme-human hemopexin. The absorbance spectrum of the heme-hemopexin complex is very characteristic, and this is a sensitive, quantitative assay of hemopexin function. Absorbance spectroscopy revealed that protoheme binding by human hemopexin was rapidly impaired during a 4-h incubation in vitro with the N/MPO/GO-nitrating system at 37 °C (Fig. 4B), consistent with the location of Tyrn-199 and other nitrated tyrosines close to the bound heme. The extent of heme binding by human hemopexin decreased as the time of incubation with MPO/GO increased, following a mono-exponential decay (Fig. 4B). The best fit to the data shows a progressive decrease with a level of ∼50% inactivation for 3.5–4 h. Rabbit hemopexin was more resistant to oxidation than the human congener with a slower rate of decline, ∼10% loss of heme binding within 30 min and ∼20–25% loss of heme binding leveling off between 3 and 4 h. The heme-hemopexin complex remains stable for up to 2 h (data not shown). The extent of hemopexin inactivation is greater as the ratio of nitrating species to hemopexin is increased, i.e. by decreasing the rabbit hemopexin concentration from 10 to 5 μm (see Fig. 4C) or conversely damage is reduced by decreasing the enzyme concentration (data not shown). These levels of hemopexin are well within the physiological range. Plasma concentrations of hemopexin vary with age, but adults (20–40 years) average 77 mg/100 ml plasma (∼13.5 μm) and range from 66 to 100 mg/100 ml (20), although more recent data employ a wider range from 0.4 to 1.5 mg/ml (21). Overall, our in vivo and in vitro data support that nitration of hemopexin occurs in vivo, that nitration decreases the ability of hemopexin to bind heme; and if, as we propose, that nitration is significantly increased in inflammation and has the potential to inactivate this important protective function of hemopexin as an extracellular antioxidant against heme toxicity.
ROS also covalently modify apo-hemopexin and impair heme binding
We anticipated that nitrated tyrosine residues of hemopexin would also be susceptible to damage by ROS and also that by comparing results from several different conditions (i.e. type of ROS and ratio of hemopexin: ROS) a pattern showing a hierarchy of susceptible amino acids would become apparent by their COMs. The details of the peptides and their modified amino acids identified by LC-MS/MS in rabbit hemopexin are presented in Tables 3 and 4. The extent and type of modification of eight targeted amino acids is summarized in Table 5 and Fig. 5. Several covalently oxidatively modified peptides were also identified using MASCOT, but at a lower level of confidence because the amino acid was not unambiguously identified (Table 3). Although exposure of apo-hemopexin to high, supra-physiological molar ratios of H2O2 impairs heme binding (9), no amino acid modifications were unambiguously identified by MASCOT searches in hemopexin exposed to H2O2 at 1:1, 1:2.5, or 1:10 molar ratios.
Table 3.
Identification of chlorinated and butylated tyrosines and an oxidized tryptophan in peptides of ROS-treated rabbit hemopexin using LC-MS/MS
These modified amino acids in five peptides were unambiguously detected and identified using MASCOT (see supplemental data). HPX/ROS ratios were 1:10 HOCl (VWoxVYTSEK, FNPVSGEVPPGYClPLDVR, and GGYTLVNGYClPK); 1:2.5 t-BuOOH (YYtBuCFQGNQFLR); and 1:1 t-BuOOH (DYtBuFLSCPGR). Superscripts indicate residues modified by oxidation (ox), t-butylation (t-Bu), or mono-chlorination (Cl).
| HPX species | NCBI gi number | Peptide sequence | COM site | Charge (+) | Mascot ions score |
|---|---|---|---|---|---|
| Rabbit | 130500366 | 118VWoxVYTSEK125 | 119 | 2 | 39.3 |
| Rabbit | 130500366 | 200YYt-BuCFQGNQFLR210 | 201 | 2 | 51.1 |
| Rabbit | 130500366 | 211FNPVSGEVPPGYClPLDVR227 | 222 | 2 | 69.8 |
| Rabbit | 130500366 | 228DYt-BuFLSCPGR236 | 229 | 2 | 25.1 |
| Rabbit | 130500366 | 331GGYTLVNGYClPK341 | 339 | 2 | 47.6 |
Table 4.
Peptides with additional oxidative modifications of tyrosine residues identified by LC-MS/MS after incubation of rabbit hemopexin with t-BuOOH and HOCl
These modified peptides were unambiguously identified by MASCOT with the tyrosine modifications shown, based on mass difference, but the particular amino acid was not validated by MS/MS fragmentation. HPX, ROS ratios were 1:10 HOCl (FNPVSGEVPPGYdi-ClPLDVR, DYClFLSCPGR, and DYdi-ClFLSCPGR), and 1:1, 1:2.5, and 1:10 t-BuOOH (FNPVSGEVPPGYt-BuPLDVR). Mascot scores for FNPVSGEVPPGYt-BuPLDVR from 1:1 and 1:2.5 t-BuOOH searches were 39.2 and 32.8, respectively. Superscripts indicate residues modified by t-butylation (t-Bu), mono- (Cl), or di-chlorination (di-Cl).
| HPX species | NCBI gi number | Peptide sequence | COM site | Charge (+) | Mascot ions score |
|---|---|---|---|---|---|
| Rabbit | 130500366 | 211FNPVSGEVPPGYdi-ClPLDVR227 | 222 | 2 | 63.3 |
| Rabbit | 130500366 | 211FNPVSGEVPPGYtBuPLDVR227 | 222 | 2 | 39.3 |
| Rabbit | 130500366 | 228DYClFLSCPGR236 | 229 | 2 | 30.6 |
| Rabbit | 130500366 | 228DYdi-ClFLSCPGR236 | 229 | 2 | 31.8 |
Table 5.
Summary of covalent oxidative modifications of amino acids of rabbit hemopexin
Data summary: the endogenous nitration of Tyr-201 (rabbit) is conserved on Tyr-199 (human) and Tyr-118 (rat). Significantly, in the 3D crystal structure of hemopexin, Tyr-201 and Tyr-222 interact with the propionate oxygen of heme ring D, and Tyr-229 also resides near the bound heme (17) and see Fig. 2B. There were three additional nitration sites, Tyr-121, Tyr-317, and Tyr-325 (see Table 2). The amino acids and their modifications that were not unambiguously identified by MASCOT (summarized in Table 3), are shown here in square brackets. t-Bu, t-butylation.
| Amino acid | Covalent oxidative modifications | ||||
|---|---|---|---|---|---|
| Tyr-121 | Nitration | ||||
| Tyr-201 | Nitration | t-Bu | |||
| Tyr-222 | Mono-Cl | (di-Cl) | (t-Bu) | ||
| Tyr-229 | Nitration | t-Bu | (Mono-Cl) | (di-Cl) | |
| Tyr-317 | Nitration | ||||
| Tyr-325 | Nitration | ||||
| Tyr-339 | Mono-Cl | ||||
| Trp-119 | Oxidation | ||||
Figure 5.
Distribution of covalently oxidatively modified amino acids after hemopexin is exposed to RNS and ROS. The distribution of covalently and oxidatively modified amino acids after rabbit hemopexin is exposed to RNS and ROS. A, solely endogenously nitrated amino acids are in dark blue; those modified by ROS (treated with HOCl or tert-butyl hydroperoxide, see Table 2) are in pink, and those modified by both nitration and ROS are in purple. Notably, tyrosines 201, 222, and 229 reside in the heme-binding site consistent with the impaired heme binding and are clearly vulnerable to oxidative modification unless protected by heme binding. Also identified were tryptophan 119 and tyrosine 121 in the N-domain and tyrosines 317, 325, and 339 in the C-domains (see Tables 2 and 3). The peptides in which the three modified tyrosines in the N- and C-domains reside are contiguous; however, they are in different blades of the two homologous β-propellers. The relative conformation of the two domains in the open apoprotein structure is unknown. Certain modified residues are no longer solvent-accessible when heme is bound to hemopexin (shown in Fig. 3, E and F), although the receptor-binding site is. B, superimposing the N-domain's (wheat) and the C-domain's (blue/gray) homologous four-bladed β-propellers of hemopexin in the N to C orientation, the three oxidatively modified tyrosine residues in the heme-binding site reside on blade 4, whereas the three on the C-domain reside on blade 6, numbering the blades sequentially from the N to C terminus of hemopexin.
As summarized in Fig. 5A, both Trp-119 and Tyr-121 have single modifications and are located in peptide VWVYTSEK at the N terminus. Trp-119 is oxidized by HOCl (1:10) and Tyr-121 is endogenously nitrated. The six remaining tyrosines, of which four were endogenously nitrated, fall into two groups of three. Those in the heme-binding sites sustained more than one COM, whereas those in the C-domain only had one COM. Tyr-201 and Tyr-229 that were nitrated endogenously were also modified after rabbit hemopexin was incubated with t-BuOOH; in addition, Tyr-229 was modified by hypochlorous acid (HOCl). Tyr-222 was susceptible to HOCl and to t-BuOOH (but apparently not to nitration). In the C-domain of hemopexin, Tyr-317 and Tyr-325 were nitrated, and Tyr-339 was chlorinated by HOCl (1:10). The six peptides containing these modified amino acids form two contiguous regions: one on each domain (Tyr-200 to Arg-236 and Leu-316 to Lys-341). The N- and C-domains of hemopexin are highly homologous four-bladed β-propellers (16). When these domain structures are superimposed (N- to C-orientation), the three endogenously nitrated tyrosines in the N-domain reside in a different HPX repeat from the three in the C-domain (Fig. 5C).
Discussion
As reviewed recently (22), several of the diseases in which hemopexin plasma levels increase are associated with inflammation, increased activity of nitric-oxide synthase, and oxidative stress. Thus, hemopexin is expected to be exposed in vivo to high levels of oxidative species. Of note, however, hemopexin is not an acute-phase protein in humans as it is in rodents (23, 24). Because proteins can be inactivated by oxidative modifications from exposure to RNS and ROS, we reasoned that the increased levels of covalently modified hemopexin might be a physiological response to damage, in which case hemopexin protein levels per se would not necessarily reflect the extent to which hemopexin is functional, i.e. capable of binding to heme or to hemopexin receptors. In proteins, generally, histidine, lysine, cysteine, and methionine residues are oxidized, whereas tyrosine is nitrated. The most common and stable oxidation product of proteins generates the carbonyl derivative of lysine, threonine, arginine, and proline; and carbonylated hemopexin had been detected in plasma from patients with Alzheimer's disease (AD) (25) and in transgenic AD mice (26). In fact, hemopexin was proposed to be a marker of AD (25). Carbonylated hemopexin has been considered a marker of oxidative stress in diabetic rats (27) and in humans with aortic aneurysm (28). However, carbonylation has not yet been shown to compromise the biological function of hemopexin. In contrast, we have identified several key amino acid residues on hemopexin that are particular targets for oxidative damage by RNS and ROS and show that they reside in the heme-binding site. Consequently, heme binding is impaired, which is an important means whereby hemopexin in its capacity as an extracellular antioxidant protects all cells from heme toxicity in a variety of hemolytic conditions, including sickle cell disease (29, 30), sepsis (3), and likely also malaria (31). Hemopexin also protects barrier tissues, for example brain neurons and other brain cells in mouse models of ischemic stroke (32) and intracerebral hemorrhage (33) as well as clinical subarachnoid hemorrhage (34).
The data presented here show that native human, rabbit, and rat hemopexin from plasma are nitrated endogenously (i.e. in vivo), and significantly, the prevalent nitrated peptide sequence is YYCFQGNQFLR conserved in these three species of hemopexin and with one tyrosine residue (Tyrn-199 in human hemopexin) that interacts directly with the heme ligand. The natural low abundance of this nitrated peptide in human blood bank plasma shows that the majority of hemopexin molecules in human plasma from healthy individuals are normally not nitrated. Consistent with this location in the heme site, incubation of hemopexin with an enzymatic nitrating system (N/MPO/GO) in vitro significantly decreased heme binding, which was progressive as nitration proceeded. Additional in vitro studies showing that the longer the exposure of human hemopexin to nitrating species, the greater the relative increase in the abundance of nitrated peptide (which we consider Tyrn-199 the principal target) in a population of hemopexin molecules. Overall, these in vivo and in vitro data bolster our hypothesis that nitration reflects specific events that drive pathology in vivo and that if hemopexin encounters RNS at sufficient concentrations in biological fluids, like plasma and CSF, it could be damaged by nitration of tyrosines in the heme-binding site and lose its ability to bind heme. Importantly, these studies with human hemopexin were confirmed by similar analyses using rabbit hemopexin. Our data support that the human hemopexin is more vulnerable to nitration than rabbit perhaps because of two additional surface-exposed tyrosines (data not shown).
The identification of the nitrated tyrosine in the peptide YYnCFQGNQFLR as Tyrn-201 in rabbit hemopexin, rather than the N-terminal Tyr-200 cannot be unequivocally supported by our current MS/MS spectrum analyses. This is because of the absence of key diagnostic fragments (b-1 or y-10 using collision-induced diffusion on the ion trap) on different species of hemopexin; however, the structural data allow only this conclusion. Inspection of the hemopexin 3D structure, reveals that Tyr-200 is buried deeply in the N-domain away from the heme-binding site and is also not close to the proposed heme-docking site at the end of the tunnel in the N-domain (16). There is little, if any, flexibility in this region because each hemopexin domain is a very stable protein structure. Stability is intrinsic to the four-bladed propellers, which are constrained in part by several disulfide bonds as well as by coordination of the ions in the central tunnel with carbonyl and amino groups of the peptide backbone (17). We are currently extending our MS analyses with additional fragmentation techniques using matrix-assisted laser desorption/ionization with high energy collision-induced dissociation and electron transfer dissociation to identify the nitrated tyrosine in the peptide YYCFQGNQFLR of human hemopexin.
We also wanted to determine whether the damage to apo-hemopexin by, albeit high, supraphysiological levels of H2O2 and HOCl that impair heme binding by ∼30–40% (9) also modified the tyrosine residues that were nitrated and whether there were other oxidant-susceptible amino acid residues in different regions of the protein. Using MASCOT, eight amino acids were identified with covalent oxidative modifications. Our principal findings show that five tyrosines were endogenously nitrated, the same three Tyr-201, Tyr-222, and Tyr-229 in the heme-binding site and another in the C-domain, and together with a tryptophan residue in the N terminus were covalently modified by ROS, including t-BuOOH. The three tyrosines in the heme-binding site sustained more than one COM, two with 3–4 COMs, whereas the three tyrosines in the C-domain only had one COM. The N terminus of rabbit hemopexin was another vulnerable region where Trp-119 and Tyr-121 are located. In humans, these two N-terminal residues may be protected from oxidation because there is an O-linked oligosaccharide chain attached to the N-terminal threonine of human hemopexin (35). Rabbit protein lacks this N-linked oligosaccharide. In vitro studies support that heme can catalyze protein nitration in the presence of H2O2 and nitrite (NO2˙̄), the stable end products of NO and O2˙̄ degradation (36, 37). Based on these published studies with heme bound to bovine serum albumin, heme in vivo (such as that bound to the weak, not the tight, binding site on human serum albumin) may well help catalyze nitration as part of heme toxicity in hemolytic conditions.
The clustering of modified amino acids with the greatest number and variety of modifications in the heme-binding site of hemopexin is clear and quite remarkable given the different chemical reactivity of the RNS and ROS employed. Ligand binding often protects the key amino acids involved in the interaction, and this seems to be the case with hemopexin. Heme binding protects the heme-iron coordinating histidine residues from chemical modification (38). Thus, apo-hemopexin, the predominant form normally present in biological fluids appears potentially vulnerable to inactivation in pathological conditions in which inflammation develops in the presence or absence of heme toxicity, recently shown to drive the inflammatory response in sepsis (3) and sickle cell disease (29, 30).
The modified amino acids identified here constitute a hierarchy susceptible to damage, none of which are close to the region implicated in receptor recognition (39), or in the proposed heme-docking site at the wide end of the tunnel of the N-domain (16). The lack of detectable modifications of amino acids upon exposure to H2O2 was unexpected (a ratio of 1:10 hemopexin/H2O2 reduced binding by ∼30%) (9). This may be due to the fact that in the absence of reduced metals (ferrous and cuprous ions) H2O2 is a relatively weak oxidizing agent (40) or, alternatively, that the carbohydrate chains of hemopexin may be altered by H2O2 as shown for haptoglobin (41).
We draw several conclusions from these observations. First, nitration of the peptide YYCFQGNQFLR of hemopexin occurs in vivo, albeit at low levels, and is the predominant nitration in human, rabbit, and rat hemopexin. Second, we have identified in hemopexin a hierarchy of eight amino acids that are targeted by RNS and physiologically relevant ROS, and three of these particularly vulnerable tyrosines cluster in the heme-binding site of hemopexin. Third, the progressive nitration of human hemopexin in vitro diminishes its ability to bind heme and the proportion of nitro-3-tyrosines, especially Tyr-199 that interacts directly with heme, increases with extent of exposure to nitrating conditions. This supports that the protective heme binding by hemopexin may be decreased in vivo by nitration in certain pathological inflammatory states. Fourth, the large conformational change that hemopexin undergoes driven by heme binding protects the vulnerable tyrosine residues in the heme-binding site from damage; consistent with this, the heme-hemopexin complex is essentially stable in the presence of N/MPO/GO nitration and ROS (9) emphasizing the paramount importance of the extracellular antioxidant role of hemopexin.
Proteins are oxidized and nitrated in oxidative stress when activated neutrophils produce their defensive “respiratory burst” generating superoxide, which reacts with nitric oxide (NO•) forming peroxynitrate (ONOO−), and also by peroxidase activity in the presence of NO2˙̄ and hydrogen peroxide (42). NO• is produced by four members of nitric-oxide synthase (NOS): neuronal, endothelial (e), inducible (i), and mitochondrial. Endothelial NOS is present in cells that line all blood vessels, and NO• causes vasodilation. Following immunological or inflammatory stimuli, macrophages and brain astrocytes produce NO• at high levels for hours or days. Hemopexin helps restrain systemic inflammatory responses in part by sequestering heme from immune cells to prevent heme-mediated activation of TLR4 (29) with release of inflammatory cytokines (44–46), to prevent toxic effects of heme (5), and to safely transport heme to tissues (6, 32). These COMs of hemopexin that we have identified may represent attacks on hemopexin in niches of the body where there is inflammation and oxidative stress with activated endothelial and immune system cells, including neutrophils and astrocytes that generate a variety of RNS and ROS. Hemopexin 3D structure is protected due to many conserved disulfide bonds, which confer stability to the protein in an oxidizing environment. Furthermore, hemopexin lacks free sulfhydryl groups to which redox-active metals can bind. Another mitigating factor protecting apo-hemopexin in vivo is that in biological fluids, including plasma and CSF, there are abundant preferential protein targets such as albumin (9). In fact, in plasma albumin is present at ∼80-fold molar excess over hemopexin.
One clinical condition where we expect that the extent of hemopexin nitration in vivo will increase as inflammation progresses is in sepsis; and tyrosine nitration of proteins has been documented in sepsis (47, 48). Hemopexin deficiency states develop in clinical sepsis and two independent studies have shown that low levels of hemopexin at diagnosis are associated with high morbidity (3, 4). Furthermore, in mouse models of severe sepsis, hemopexin supplementation is life-saving (3), and hemopexin replenishment therapy provides an evidence-based approach to treat clinical sepsis (1). We speculate that if the oxidatively modified hemopexin is recognized as a non-native protein, it would be targeted to the lysosomes for catabolism, rather than the recycling endosomes (6), thus contributing to the development of hemopexin deficiency states documented in sepsis. Hepatic heme levels regulate hemopexin metabolism, and as heme levels rise, the rate of hemopexin catabolism increases thus decreasing plasma hemopexin levels (1).
Hemopexin acts during inflammation that accompanies ischemic (32) and hemorrhagic (49) stroke by diminishing the chemical activity of heme (5) and by delivering it safely to cells. We anticipate that hemopexin may be inactivated in the CSF in AD, the neurodegeneration of which has been linked to oxidative stress and reactive nitrogen species (50). In fact, increased levels of nitrated proteins have been detected in brain tissue and CSF of AD subjects (50). Hemopexin levels are elevated in CSF in inflammation (51) and in AD subjects compared with age-matched controls (15). Although this may be due to its known induction by interleukin-6 (16), hemopexin may be inactivated because increased levels of 3-nitrotyrosine have been detected in the inferior parietal lobe and hippocampus in mild cognitive impairment (MCI) (52). Such nitrosative damage occurs early in the course of MCI, and protein nitration was proposed to be important for the conversion from MCI to AD.
To summarize, we observed that in normal healthy individuals, i.e. in the absence of pathology, nitration of the YYCFQGNQFLR hemopexin peptide is low or undetectable in vivo and that in vitro exposure of the peptide to RNS increases the level of this peptide with one nitrated tyrosine. Of note, this increased tyrosine nitration decreased heme binding by the peptide, supported by our evidence that it is the second tyrosine in this peptide (Tyr-199 in human hemopexin), which directly interacts with the propionate in heme ring D. On the basis of this in vitro evidence, we hypothesize that during inflammatory states hemopexin nitration will be increased in vivo because of elevated RNS produced by activated cells and that increased nitration decreases hemopexin's ability to bind to heme, thus impairing its ability to protect cells from any accompanying heme toxicity due to hemolysis. Because a normal half-life has been recorded for hemopexin with two chemically modified histidines in the heme-binding site that coordinate the heme-iron, we also hypothesize that the observed nitration in the heme-binding site does not decrease hemopexin's circulating half-life because it does not sufficiently alter the protein conformation from the native state. If it were recognized as non-native, it would be rapidly removed from the circulation. Taken together, we caution that when inflammation is present, the levels of hemopexin in biological fluids may not per se accurately reflect the protective capacity of this system against heme toxicity.
Overall, our data establish a strong foundation for analyzing hemopexin in biological fluids such as plasma and CSF to determine its potential as a biomarker for conditions of oxidative stress in which nitration has been detected, including neurodegeneration (e.g. MCI and AD) and systemic inflammatory responses (e.g. acute lung injury, sepsis, and multiple organ failure syndrome). We propose that the type and extent of nitration will reflect key biological processes and, together with other oxidative modifications, COMs of hemopexin as detailed here may provide a more specific biomarker of certain disease states than hemopexin levels alone. Such modifications of hemopexin could also provide evidence-based guidance to physicians when heme toxicity is driving pathology to assess the timing and extent of hemopexin replenishment therapies and the patient responses to such treatment. In the near future, more sophisticated analyses are expected to be developed to detect and quantitate COMs of hemopexin using LC-MS/MS immunoassays and multiplexing immunoassays of biological fluids. Our current research is directed at detecting and quantitating the levels and COMs of hemopexin in relevant diseases and clinical conditions with inflammation.
Experimental procedures
Materials
Sodium nitrite, sodium phosphate monobasic/dibasic, sodium chloride, and glucose were obtained from Thermo Fisher Scientific (Rockford, IL); glucose oxidase, diethylenetriaminepentaacetic acid (DTPA), and wheat germ lectin-agarose were from Sigma; and myeloperoxidase was from EMD Millipore Chemicals (Billerica, MA). Heme (ferric iron-protoporphyrin IX chloride) and mesoheme (ferric iron-mesoporphyrin IX chloride) were obtained from Frontier Scientific (Logan, UT).
Hemopexin isolates
Human, rat, and rabbit hemopexin were isolated and purified from plasma using our published procedures and plasma sources (53, 54), but with some modifications of the liquid chromatography. After several differential precipitation steps, rabbit hemopexin was obtained by ion-exchange chromatography on Q-Sepharose Fast Flow (50 mm sodium acetate buffer, pH 5.2), and the hemopexin fractions were then eluted with a 0–0.2 m NaCl gradient from a Q-Sepharose Fast Flow column (15 mm sodium phosphate butter, pH 7.4). Hemopexin was further purified by gel permeation chromatography using a Superdex 200 column (10 mm sodium phosphate buffer, pH 7.4, containing 0.15 m NaCl). After PEG precipitation and ion-exchange chromatography on Q-Sepharose Fast Flow, human and rat hemopexin were purified using affinity chromatography on wheat germ lectin resin and eluted with N-acetyl d-glucosamine as published previously (55). The purity of hemopexin (>98%) was assessed by SDS-PAGE and Coomassie Gel code Blue staining. Stoichiometric 1:1 heme-hemopexin complexes (>95% saturated) were prepared and characterized as published (53) using heme/DMSO and keeping the final DMSO concentration less than 5% (v/v). Extinction coefficients (A·m−1·cm−1) used were 1.1 × 105 at 280 nm for apo-hemopexin and for heme binding by both rabbit and human hemopexin 1.2 × 105 at 280 nm and 1.4 × 105 at 405 nm. Both protoheme (termed heme here) and mesoheme (iron-mesoporphyrin IX) were used in this work as indicated in the figure legends. Protoheme is the heme in hemoglobin, and we have also used mesoheme, which is more stable; furthermore, the mesoheme-hemopexin complex is chemically and biologically equivalent to protoheme-hemopexin (6).
Experimental design and analyses, nitration by in vitro myeloperoxidase/glucose oxidase treatment and assessment of heme binding after nitration
Purified apo-hemopexin or heme-hemopexin complexes (10 μm) in 0.1 m sodium phosphate solution, pH 7.4, containing 2 mm NaCl were incubated with 53 nm myeloperoxidase, 2 nm glucose oxidase, 0.1 mm DTPA, and 50 μm sodium nitrite (NaNO2) at 37 °C for up to 4 h (10, 11). This reaction mixture was kept on ice until the addition of glucose (0.56 mm), the glucose oxidase substrate, generating H2O2. Myeloperoxidase utilizes H2O2 and NO2− as substrates to catalyze the nitration of amino acid residues (e.g. tyrosine) in proteins. During this incubation, aliquots of hemopexin (6 μl; 3.42 μg) were removed at the indicated times for immunoblotting (250 ng), described below, and for LC-MS/MS (3 μg) and denatured in Laemmli buffer. To more readily distinguish between the extent of nitration of apo- and heme-hemopexin by immunoblotting, the enzyme concentrations were reduced by 75%.
To assess heme binding of MPO/GO-treated apo-hemopexin, aliquots (30 μl) were taken at the times indicated from control and experimental reaction mixtures (after the addition of either glucose or an equal volume of water, respectively) into 15 μl of salt to obtain 7 μm protein in a final concentration of 10 mm sodium phosphate buffer, pH 7.4, with 0.15 m NaCl. Stock heme/DMSO was added at 1.0 molar eq, and the heme binding was assessed after incubation for 5 min at 25 °C by the ratio of absorbance at 280 nm and 404 nm as published (56), from the absorbance spectrum using the “Proteins and Labels” function on a Nanodrop 1000 UV-visible spectrophotometer. For most accurate determination of heme binding (because high levels of unbound heme can distort the absorbance spectrum of hemopexin), additional spectra were obtained with 0.5 molar eq of heme as indicated in the figure legends for samples after extensive nitration causing, for example, a loss of ∼50% heme binding.
Nitrotyrosine quantitation by immunoblotting
The extent of nitration of the hemopexin samples was determined after electrophoresis (10% Tris-HCl gels) by immunoblotting after transfer to Sequi-blot PVDF. Anti-3′-nitrotyrosine rabbit polyclonal antibody (3 μg/ml; antigen-peroxynitrite-treated keyhole limpet hemocyanin (Cayman Chemical, Ann Arbor, MI) was used with horseradish peroxidase-conjugated goat anti-rabbit IgG (dilution 1:50,000). After stripping, the blot was reprobed for total hemopexin by incubating with a goat anti-human HPX polyclonal primary antibody (raised by one of the authors, A. Smith) at a 1:500 dilution with a rabbit anti-goat IgG secondary antibody (1:25,000 dilution). Signal was detected in both cases with Lumigen PS-3 Amigen as substrate for the enhanced chemiluminescence with ECL Plus (Amersham Biosciences, UK), and the blot was scanned on a Typhoon Imager (GE Biosciences). Quantitation was carried out using UN-SCAN-IT gel digitizing software (Silk Scientific, UT).
LC-MS/MS analysis of trypsin-digested hemopexin and detection of tyrosine nitration
After electrophoresis of samples of the various biological isolates of hemopexin, the gels were rinsed twice with pure water before brief staining with a Coomassie R-250-based ImperialTM Stain (Pierce) and rapid de-staining. Excised gel fragments were excised, and hemopexin was reduced and alkylated with iodoacetamide by standard methods followed by digestion with trypsin (20 μg/ml in 50 mm ammonium bicarbonate) for 6–16 h at 37 °C (11). Trypsin, a pancreatic serine protease, cleaves on the C-terminal side of arginine and lysine residues unless a proline is present on the carboxyl side of the potential cleavage site. The tryptic peptides were then extracted and subjected to reverse phase nano-LC tandem MS with a linear 2–50% acetonitrile gradient over 45 min in 50 mm acetic acid (200 nl/min flow rate), through a C18 column (50-μm inner diameter × 7 cm Phenomenex Jupiter Proteo) as described previously (11). Data were acquired on a Thermo Finnigan LTQ MS system by data-dependent acquisition (one survey MS, and eight dependent MS/MS scans). SEQUEST searches were performed using Xcalibur BioWorks 3.1 and Mascot version 2.1 (Matrix Science, Ltd.) for the initial identification of low basal levels of nitration using human (gi 11321561), rabbit (gi 130500366), and rat (gi 123036) hemopexin sequences from the NCBI Proteins resource. Hemopexin searches were carried out in a custom database, with 1.8-Da peptide mass tolerance and 0.8-Da fragment ion tolerance. Two missed cleavages were allowed. Searches included fixed modification of cysteine (carbamidomethylation, +57 atomic mass units) and allowed differential modification of tyrosine (nitration, +45 atomic mass units), and methionine (oxidation, +16 atomic mass units).
Quantitative analyses of tyrosine nitration using SRM
The relative quantitation of nitrated tyrosine (Tyrn) sites was carried out using SRM with scans for the internal unmodified reference peptide, NFPSPVDAAFR: 611.5, for the unmodified peptide YYCFQGNQFLR: 748.5, and for the modified peptide, YYnCFQGNQFLR: 771.6. For each experiment, three vertical gel slices were excised from each gel band (technical replicates) and processed/analyzed separately (technical replicates). The MS/MS chromatographic peak areas (integrated mass chromatograms) of the peptides NFPSPVDAAFR (mass range 959.2–960.4), YYCFQGNQFLR (mass range 748.0–749.5), and YYnCFQGNQFLR (mass range 734–735) were quantitated using Xcalibur software (Vienna, VA) (11). The chromatographic peak area ratios were graphed versus time of exposure to the nitrating in vitro MPO/GO treatment.
Note that Tyr-199 (underlined in sequence) of human hemopexin in the YYCFQGNQFLR sequence is homologous to Tyr-176 in the published rabbit hemopexin crystal structure of the mature secreted protein (17); the 25-amino acid difference was due to the signal peptide (using Protein Data Bank code 1QHU and rabbit hemopexin precursor sequence UniProtKB accession no. P20058. Also, the sequence 216SHRNSTQ222 is missing from the 1QHU structure, and the following histidine was inadvertently assigned His-222; consequently, from His-222 to the C terminus the difference in numbering between the gene and the mature protein is only 24.).
Quantitation of in vitro nitration of Tyr-199 in human hemopexin by the N/MPO/GO system over time was determined by SRM. Synthetic peptides were synthesized by Celtek Peptides (Nashville, TN) with the cysteines carbamidomethylated. The lyophilized native and nitrated peptide samples were diluted in pure water, whereasthe reference peptide required methanol for solubilization. They were then standardized to the arginine curve determined with the 2,4,6-trinitrobenzenesulfonic acid method of Snyder and Sobocinski (57) as described below. Experimental chromatographic peak area ratios were obtained by dividing the reference peptide peak area by that of either the YYCFQGNQFLR (supplemental Fig. S4A) or the YYnCFQGNQFLR (supplemental Fig. S4B) peptide. Mole fractions were found by dividing the concentration of the NFPSPVDAAFR peptide by concentrations of either the YYCFQGNQFLR or YYnCFQGNQFLR peptides ranging from 10 to 90 μm. The standard curves produced the following equations: y = 1368x − 29.98 (YYnCFQGNQFLR) and y = 1298x − 109.8 (YYCFQGNQFLR), generated by GraphPad Prism 6 linear regression (supplemental Fig. S4). These equations were used to obtain the mole fraction for unmodified and nitrated peptide for each time of in vitro nitration (shown in Fig. 3D).
Oxidation by ROS treatment and identification of COMs in apo-hemopexin by LC-MS/MS
Rabbit apo-hemopexin (10 μm) in PBS was incubated for 15 min at 37 °C with HOCl (1:2.5, 1:10, 1:20), H2O2 (1:1, 1:2.5, 1:10), t-BuOOH (1:1, 1:2.5, 1:10), or PBS alone as control as published (9). Protein samples were fractionated by SDS-PAGE, and bands of interest were in-gel digested, and LC-MS/MS analyses of tryptic peptides were conducted as described above. The Thermo RAW data files were searched using MASCOT Server (version 2.4.1; all files are listed in supplemental Table 1). The rabbit hemopexin sequence (gi 130500366) from the NCBI Protein resource was searched with semi-trypsin selected as the proteolytic enzyme. The precursor mass tolerance was set at ±2 Da, and the fragment mass tolerance was set at ±1 Da. Because MASCOT has the limitation of allowing only nine modifications per search, the searches were conducted as follows. The first pass search was for oxidation of Phe, His, Lys, Met, Arg, Trp, Tyr, and kynurenine and oxolactone modifications of Trp. The second pass search focused on di-oxidation of Phe, Lys, Met, Arg, Trp, and Tyr, tri-oxidation of Trp and Tyr, and modification of Trp to hydroxykynurenine. Samples treated with t-BuOOH were additionally searched for t-butylation and di-t-butylation of Trp and Tyr. Samples treated with HOCl-treated were also searched for chlorination of Tyr, His, Lys, and Trp, di-chlorination of Trp and Tyr, and tri-chlorination of Trp and Tyr was included. Modifications were initially validated manually.
Author contributions
A. Smith conceived the research, coordinated the study, designed the experiments, analyzed data, and wrote the manuscript. P. H., R. H., and E. S. B. carried out the experiments, helped analyze the data, and prepared the figures (Fig. 3 was done by E. H., and Fig. 4, B and C, was done by E. S. B.). A. K. carried out the LC-MS/MS generation and helped P. H., R. H., and A. Skaff with their MASCOT searches and analyses that were summarized in the figures, tables, and supplemental figures by P. H. P. H. and A. Skaff generated the PyMOL representations of covalently modified hemopexin (Figs. 1, 3, and 5). All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
The MS data were generated by the School of Biological Sciences Proteomics Core and analyzed using MASCOT by P. H., A. Smith, and A. Skaff with verification from the core director, Dr. A. Keightley. We acknowledge Dr. Marilyn Yoder (School of Biological Sciences, University of Missouri-Kansas City) for help with the PyMOL images and Dr. Roberto Vanacore (Vanderbilt Medical Center, Nashville, TN) for assistance in the preparation of the manuscript. We also thank Dr. Tamas Kapros (School of Biological Sciences, University of Missouri-Kansas City) for help with generating the final figure files.
This work, with Institutional Review Board/Institutional Animal Care and Use (IRB/ACUC)) approval, was supported in part by National Institutes of Health Grant R21 DK064363 (to A. Smith) and Research Incentive Funds (to A. Smith) from the School of Biological Sciences, University of Missouri-Kansas City, Kansas City, MO. A patent application based upon this research with its predicted diagnostic implications has been filed (A. Smith, Serial No. 14/961,144). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The mass spectrometric raw data and spectral libraries associated with this manuscript are available from ProteomeXchange (https://massive.ucsd.edu) with the accession number PXD005560.
This article contains supplemental Table S1 and supplemental Figs. S1–S4.
We have used this term covalent oxidative modifications (COMs) rather than post-translational modifications (PTMs) because COMs are proposed to affect the mature secreted hemopexin, rather than occurring during the synthesis and secretion of hemopexin, which requires the PTMs of glycosylation and cleavage of the signal sequence.
- ROS
- reactive oxygen species
- AD
- Alzheimer's disease
- CSF
- cerebral spinal fluid
- COM
- covalent oxidative modification
- DTPA
- diethylenetriaminepentaacetic acid
- HPX
- hemopexin
- HOCl
- hypochlorous acid
- MCI
- mild cognitive impairment
- MPO/GO
- myeloperoxidase/glucose oxidase
- NOS
- nitric-oxide synthase
- Tyrn
- nitrotyrosine
- PTM
- post-translational modification
- RNS
- reactive nitrogen species
- SRM
- selective reaction monitoring
- NaNO
- sodium nitrite
- t-BuOOH
- tert-butyl hydroperoxide
- COM
- covalent oxidative modification
- N/MPO/GO
- nitrite/myeloperoxidase/glucose oxidase
- AD
- Alzheimer's disease
- DTPA
- diethylenetriaminepentaacetic acid.
References
- 1. Smith A., and McCulloh R. J. (2015) Hemopexin and haptoglobin: allies against heme toxicity from hemoglobin not contenders. Front. Physiol. 6, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith A., and McCulloh R. J. (2016) Mechanisms of haem toxicity in haemolysis and protection by the haem-binding protein, haemopexin. ISBT Sci. Ser. 12, 119–133 [Google Scholar]
- 3. Larsen R., Gozzelino R., Jeney V., Tokaji L., Bozza F. A., Japiassú A. M., Bonaparte D., Cavalcante M. M., Chora A., Ferreira A., Marguti I., Cardoso S., Sepúlveda N., Smith A., and Soares M. P. (2010) A central role for free heme in the pathogenesis of severe sepsis. Sci. Transl. Med. 2, 51ra71. [DOI] [PubMed] [Google Scholar]
- 4. Jung J. Y., Kwak Y. H., Kim K. S., Kwon W. Y., and Suh G. J. (2015) Change of hemopexin level is associated with the severity of sepsis in endotoxemic rat model and the outcome of septic patients. J. Crit. Care 30, 525–530 [DOI] [PubMed] [Google Scholar]
- 5. Gutteridge J. M., and Smith A. (1988) Antioxidant protection by haemopexin of haem-stimulated lipid peroxidation. Biochem. J. 256, 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith A., and Morgan W. T. (1979) Haem transport to the liver by haemopexin. Receptor-mediated uptake with recycling of the protein. Biochem. J. 182, 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davies D. M., Smith A., Muller-Eberhard U., and Morgan W. T. (1979) Hepatic subcellular metabolism of heme from heme-hemopexin: incorporation of iron into ferritin. Biochem. Biophys. Res. Commun. 91, 1504–1511 [DOI] [PubMed] [Google Scholar]
- 8. Smith A., and Hunt R. C. (1990) Hemopexin joins transferrin as representative members of a distinct class of receptor-mediated endocytic transport systems. Eur. J. Cell Biol. 53, 234–245 [PubMed] [Google Scholar]
- 9. Hahl P., Davis T., Washburn C., Rogers J. T., and Smith A. (2013) Mechanisms of neuroprotection by hemopexin: modeling the control of heme and iron homeostasis in brain neurons in inflammatory states. J. Neurochem. 125, 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kostyuk V. A., Kraemer T., Sies H., and Schewe T. (2003) Myeloperoxidase/nitrite-mediated lipid peroxidation of low-density lipoprotein as modulated by flavonoids. FEBS Lett. 537, 146–150 [DOI] [PubMed] [Google Scholar]
- 11. Willard B. B., Ruse C. I., Keightley J. A., Bond M., and Kinter M. (2003) Site-specific quantitation of protein nitration using liquid chromatography/tandem mass spectrometry. Anal. Chem. 75, 2370–2376 [DOI] [PubMed] [Google Scholar]
- 12. Ischiropoulos H. (2009) Protein tyrosine nitration–an update. Arch. Biochem. Biophys. 484, 117–121 [DOI] [PubMed] [Google Scholar]
- 13. Ischiropoulos H. (2003) Biological selectivity and functional aspects of protein tyrosine nitration. Biochem. Biophys. Res. Commun. 305, 776–783 [DOI] [PubMed] [Google Scholar]
- 14. Zhang R., Miller R. G., Madison C., Jin X., Honrada R., Harris W., Katz J., Forshew D. A., and McGrath M. S. (2013) Systemic immune system alterations in early stages of Alzheimer's disease. J. Neuroimmunol. 256, 38–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Castaño E. M., Roher A. E., Esh C. L., Kokjohn T. A., and Beach T. (2006) Comparative proteomics of cerebrospinal fluid in neuropathologically confirmed Alzheimer's disease and non-demented elderly subjects. Neurol. Res. 28, 155–163 [DOI] [PubMed] [Google Scholar]
- 16. Smith A. (2011) in Handbook of Porphyrin Science. Biochemistry of Tetrapyrroles (Kadish K. M., Smith K. M., and Guilard R., eds) pp. 217–356, World Scientific Publishing Co. Pte. Ltd., Singapore [Google Scholar]
- 17. Paoli M., Anderson B. F., Baker H. M., Morgan W. T., Smith A., and Baker E. N. (1999) Crystal structure of hemopexin reveals a novel high-affinity heme site formed between two β-propeller domains. Nat. Struct. Biol. 6, 926–931 [DOI] [PubMed] [Google Scholar]
- 18. Morgan W. T., and Smith A. (1984) Domain structure of rabbit hemopexin. Isolation and characterization of a heme-binding glycopeptide. J. Biol. Chem. 259, 12001–12006 [PubMed] [Google Scholar]
- 19. Smith A., and Morgan W. T. (1984) in Protides of the Biological Fluids (Peeters H., ed) pp. 219–224, Pergamon Press, Oxford, UK [Google Scholar]
- 20. Hanstein A., and Muller-Eberhard U. (1968) Concentration of serum hemopexin in healthy children and adults and in those with a variety of hematological disorders. J. Lab. Clin. Invest. 71, 232–239 [Google Scholar]
- 21. Delanghe J. R., Philippé J., Moerman F., De Buyzere M. L., Vynckier L. L., Verstraete A. G., Vogelaers D. P., and Vandekerckhove L. (2010) Impaired hemoglobin scavenging during an acute HIV-1 retroviral syndrome. Clin. Chim. Acta 411, 521–523 [DOI] [PubMed] [Google Scholar]
- 22. Smith A. (2013) in Handbook of Porphyrin Science (Ferreira G., ed) pp. 311–388, World Publishing Co., New York [Google Scholar]
- 23. Kushner I., Edgington T. S., Trimble C., Liem H. H., and Muller-Eberhard U. (1972) Plasma hemopexin homeostasis during the acute phase response. J. Lab. Clin. Med. 80, 18–25 [PubMed] [Google Scholar]
- 24. Sears D. A. (1968) Plasma heme-binding in patients with hemolytic disorders. J. Lab. Clin. Med. 71, 484–494 [PubMed] [Google Scholar]
- 25. Yu H. L., Chertkow H. M., Bergman H., and Schipper H. M. (2003) Aberrant profiles of native and oxidized glycoproteins in Alzheimer plasma. Proteomics 3, 2240–2248 [DOI] [PubMed] [Google Scholar]
- 26. Shen L., Chen Y., Yang A., Chen C., Liao L., Li S., Ying M., Tian J., Liu Q., and Ni J. (2016) Redox proteomic profiling of specifically carbonylated proteins in the serum of triple transgenic Alzheimer's disease mice. Int. J. Mol. Sci. 17, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Madian A. G., Myracle A. D., Diaz-Maldonado N., Rochelle N. S., Janle E. M., and Regnier F. E. (2011) Differential carbonylation of proteins as a function of in vivo oxidative stress. J. Proteome Res. 10, 3959–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Spadaccio C., Coccia R., Perluigi M., Pupo G., Schinina M. E., Giorgi A., Blarzino C., Nappi F., Sutherland F. W., Chello M., and Di Domenico F. (2016) Redox proteomic analysis of serum from aortic aneurysm patients: insights on oxidation of specific protein target. Mol. Biosyst. 12, 2168–2177 [DOI] [PubMed] [Google Scholar]
- 29. Belcher J. D., Chen C., Nguyen J., Milbauer L., Abdulla F., Alayash A. I., Smith A., Nath K. A., Hebbel R. P., and Vercellotti G. M. (2014) Heme triggers TLR4 signaling leading to endothelial cell activation and vaso-occlusion in murine sickle cell disease. Blood 123, 377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ghosh S., Adisa O. A., Chappa P., Tan F., Jackson K. A., Archer D. R., and Ofori-Acquah S. F. (2013) Extracellular hemin crisis triggers acute chest syndrome in sickle mice. J. Clin. Invest. 123, 4809–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elphinstone R. E., Riley F., Lin T., Higgins S., Dhabangi A., Musoke C., Cserti-Gazdewich C., Regan R. F., Warren H. S., and Kain K. C. (2015) Dysregulation of the haem-haemopexin axis is associated with severe malaria in a case-control study of Ugandan children. Malar. J. 14, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li R. C., Saleem S., Zhen G., Cao W., Zhuang H., Lee J., Smith A., Altruda F., Tolosano E., and Doré S. (2009) Heme-hemopexin complex attenuates neuronal cell death and stroke damage. J. Cereb. Blood Flow Metab. 29, 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ma B., Day J. P., Phillips H., Slootsky B., Tolosano E., and Doré S. (2016) Deletion of the hemopexin or heme oxygenase-2 gene aggravates brain injury following stroma-free hemoglobin-induced intracerebral hemorrhage. J. Neuroinflammation 13, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garland P., Durnford A. J., Okemefuna A. I., Dunbar J., Nicoll J. A., Galea J., Boche D., Bulters D. O., and Galea I. (2016) Heme-hemopexin scavenging is active in the brain and associates with outcome after subarachnoid hemorrhage. Stroke 47, 872–876 [DOI] [PubMed] [Google Scholar]
- 35. Takahashi N., Takahashi Y., and Putnam F. W. (1984) Structure of human hemopexin: O-glycosyl and N-glycosyl sites and unusual clustering of tryptophan residues. Proc. Natl. Acad. Sci., U.S.A. 81, 2021–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bian K., Gao Z., Weisbrodt N., and Murad F. (2003) The nature of heme/iron-induced protein tyrosine nitration. Proc. Natl. Acad. Sci. U.S.A. 100, 5712–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thomas D. D., Espey M. G., Vitek M. P., Miranda K. M., and Wink D. A. (2002) Protein nitration is mediated by heme and free metals through Fenton-type chemistry: an alternative to the NO/O2- reaction. Proc. Natl. Acad. Sci. U.S.A. 99, 12691–12696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morgan W. T., and Muller-Eberhard U. (1976) Chemical modification of histidine residues of rabbit hemopexin. Arch. Biochem. Biophys. 176, 431–441 [DOI] [PubMed] [Google Scholar]
- 39. Morgan W. T., Muster P., Tatum F. M., McConnell J., Conway T. P., Hensley P., and Smith A. (1988) Use of hemopexin domains and monoclonal antibodies to hemopexin to probe the molecular determinants of hemopexin-mediated heme transport. J. Biol. Chem. 263, 8220–8225 [PubMed] [Google Scholar]
- 40. Stocker R., and Keaney J. F. Jr. (2004) Role of oxidative modifications in atherosclerosis. Physiol. Rev. 84, 1381–1478 [DOI] [PubMed] [Google Scholar]
- 41. Vallelian F., Pimenova T., Pereira C. P., Abraham B., Mikolajczyk M. G., Schoedon G., Zenobi R., Alayash A. I., Buehler P. W., and Schaer D. J. (2008) The reaction of hydrogen peroxide with hemoglobin induces extensive α-globin cross-linking and impairs the interaction of hemoglobin with endogenous scavenger pathways. Free Radic. Biol. Med. 45, 1150–1158 [DOI] [PubMed] [Google Scholar]
- 42. Radi R. (2004) Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. U.S.A. 101, 4003–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shipulina N., Smith A., and Morgan W. T. (2000) Heme binding by hemopexin: evidence for multiple modes of binding and functional implications. J. Protein Chem. 19, 239–248 [DOI] [PubMed] [Google Scholar]
- 44. Liang X., Lin T., Sun G., Beasley-Topliffe L., Cavaillon J. M., and Warren H. S. (2009) Hemopexin down-regulates LPS-induced proinflammatory cytokines from macrophages. J. Leukoc. Biol. 86, 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin T., Kwak Y. H., Sammy F., He P., Thundivalappil S., Sun G., Chao W., and Warren H. S. (2010) Synergistic inflammation is induced by blood degradation products with microbial Toll-like receptor agonists and is blocked by hemopexin. J. Infect. Dis. 202, 624–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin T., Sammy F., Yang H., Thundivalappil S., Hellman J., Tracey K. J., and Warren H. S. (2012) Identification of hemopexin as an anti-inflammatory factor that inhibits synergy of hemoglobin with HMGB1 in sterile and infectious inflammation. J. Immunol. 189, 2017–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lanone S., Manivet P., Callebert J., Launay J. M., Payen D., Aubier M., Boczkowski J., and Mebazaa A. (2002) Inducible nitric oxide synthase (NOS2) expressed in septic patients is nitrated on selected tyrosine residues: implications for enzymic activity. Biochem. J. 366, 399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chatterjee S., Lardinois O., Bonini M. G., Bhattacharjee S., Stadler K., Corbett J., Deterding L. J., Tomer K. B., Kadiiska M., and Mason R. P. (2009) Site-specific carboxypeptidase B1 tyrosine nitration and pathophysiological implications following its physical association with nitric oxide synthase-3 in experimental sepsis. J. Immunol. 183, 4055–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen L., Zhang X., Chen-Roetling J., and Regan R. F. (2011) Increased striatal injury and behavioral deficits after intracerebral hemorrhage in hemopexin knockout mice. J. Neurosurg. 114, 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Castegna A., Thongboonkerd V., Klein J. B., Lynn B., Markesbery W. R., and Butterfield D. A. (2003) Proteomic identification of nitrated proteins in Alzheimer's disease brain. J. Neurochem. 85, 1394–1401 [DOI] [PubMed] [Google Scholar]
- 51. Saso L., Leone M. G., Mo M. Y., Grippa E., Cheng C. Y., and Silvestrini B. (1999) Differential changes in α2-macroglobulin and hemopexin in brain and liver in response to acute inflammation. Biochemistry 64, 839–844 [PubMed] [Google Scholar]
- 52. Butterfield D. A., Reed T. T., Perluigi M., De Marco C., Coccia R., Keller J. N., Markesbery W. R., and Sultana R. (2007) Elevated levels of 3-nitrotyrosine in brain from subjects with amnestic mild cognitive impairment: implications for the role of nitration in the progression of Alzheimer's disease. Brain Res. 1148, 243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eskew J. D., Vanacore R. M., Sung L., Morales P. J., and Smith A. (1999) Cellular protection mechanisms against extracellular heme: heme-hemopexin, but not free heme, activates the N-terminal c-Jun kinase. J. Biol. Chem. 274, 638–648 [DOI] [PubMed] [Google Scholar]
- 54. Morgan W. T., Muster P., Tatum F., Kao S. M., Alam J., and Smith A. (1993) Identification of the histidine residues of hemopexin that coordinate with heme-iron and of a receptor-binding region. J. Biol. Chem. 268, 6256–6262 [PubMed] [Google Scholar]
- 55. Vretblad P., and Hjorth R. (1977) The use of wheat-germ lectin-Sepharose for the purification of human haemopexin. Biochem. J. 167, 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Morgan W. T. (1976) The binding and transport of heme by hemopexin. Ann. Clin. Res. 8, 223–232 [PubMed] [Google Scholar]
- 57. Snyder S. L., and Sobocinski P. Z. (1975) An improved 2,4,6-trinitrobenzenesulfonic acid method for the determination of amines. Anal. Biochem. 64, 284–288 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.