Abstract
Cell migration and invasion are very characteristic features of cancer cells that promote metastasis, which is one of the most common causes of mortality among cancer patients. Emerging evidence has shown that coagulation factors can directly mediate cancer-associated complications either by enhancing thrombus formation or by initiating various signaling events leading to metastatic cancer progression. It is well established that, apart from its distinct role in blood coagulation, coagulation factor FVIIa enhances aggressive behaviors of breast cancer cells, but the underlying signaling mechanisms still remain elusive. To this end, we investigated FVIIa's role in the migration and invasiveness of the breast cancer cell line MDA-MB-231. Consistent with previous observations, we observed that FVIIa increased the migratory and invasive potential of these cells. We also provide molecular evidence that protease-activated receptor 2 activation followed by PI3K-AKT activation and GSK3β inactivation is involved in these processes and that β-catenin, a well known tumor-regulatory protein, contributes to this signaling pathway. The pivotal role of β-catenin was further indicated by the up-regulation of its downstream targets cyclin D1, c-Myc, COX-2, MMP-7, MMP-14, and Claudin-1. β-Catenin knockdown almost completely attenuated the FVIIa-induced enhancement of breast cancer migration and invasion. These findings provide a new perspective to counteract the invasive behavior of breast cancer, indicating that blocking PI3K-AKT pathway-dependent β-catenin accumulation may represent a potential therapeutic approach to control breast cancer.
Keywords: β-catenin, breast cancer, coagulation factor, signaling, tissue factor
Introduction
An increasing body of experimental evidence suggests a significant role of factor VIIa (FVIIa)2 and tissue factor (TF) complex in breast cancer progression and metastasis (1). TF, a 47-kDa glycosylated transmembrane protein, is a cellular receptor for plasma clotting factor VII/VIIa with the primary function of supporting FVII activation and initiating blood coagulation (2–4). TF affects various pathophysiological processes ranging from embryological development to cancer (5, 6). Previous studies reveal that TF-deficient mice die due to abnormal fatal bleeding and improper embryonic development (7–9). Although TF-FVIIa-induced thrombin generation contributes to some of these pathophysiological processes via activation of platelets, fibrin deposition and activation of protease-activated receptor 1 (PAR1)-mediated cell signaling (10), it does not account for all TF-dependent processes. Studies so far on TF-FVIIa signaling from several groups have already established that TF–FVIIa binary complex activates PAR2, which is one of the major contributors of tumor growth in breast carcinoma (11–15).
Although it has already been established that TF-FVIIa interaction induces migratory ability of various cancer cells including breast cancer (16), the detailed signaling mechanism involved still remains elusive. Reports also demonstrate that TF-FVIIa signaling modulates several molecules involved in adhesion, inflammation, proliferation, migration, invasion, and angiogenesis including β3 integrin, cyclin D1, interleukin-1, survivin, and vascular endothelial growth factor in various cancer cells (17). It has also been reported that the prometastatic activity of cells is dependent on the extracellular proteolytic activity of FVIIa (17). Studies have shown that overexpression of tissue factor pathway inhibitor attenuates the migration of cells by inhibiting TF-FVIIa-induced signaling (18). Hence, involvement of TF in the enhancement of cancer aggressiveness is likely.
β-Catenin, a key member in the Wnt signaling pathway, has also been reported to be involved in multitasking from embryonic development to cancer progression (19). Upon activation of Wnt signaling, β-catenin accumulates in the cells and ultimately regulates differentiation and developmental processes. It also leads to several human diseases of connective tissue, epithelial tissue, bone, and cancer including breast cancer (20). A recent report shows that, apart from canonical Wnt signaling, multiple non-canonical pathways also result in cellular accumulation of β-catenin (21).
In this study, we address the accumulation of β-catenin in MDA-MB-231 cells by FVIIa through PAR2 receptor activation followed by PI3K-AKT activation and glycogen synthase kinase-3β (GSK3β) inactivation. TF-FVIIa-induced cell signaling promotes transcriptional activation of proteins such as cyclin D1, c-Myc, COX-2, MMP-7, MMP-14, and Claudin-1 directly by manipulating the β-catenin transcriptional complex. Interestingly, these proteins have been previously designated as being downstream of the β-catenin transcriptional complex in the Wnt signaling pathway (21). Our findings provide strong evidence that FVIIa-mediated enhanced migration and invasion of MDA-MB-231 cells are guided by β-catenin. These results also provide supportive evidence for a novel signaling mechanism through which activation of a G protein-coupled receptor family protein, PAR2, contributes to enhanced aggressiveness of human breast cancer cells and suggest that the pathway can be targeted to achieve improved cancer therapeutics to arrest breast tumor growth and increase longevity.
Results
TF-FVIIa interaction leads to β-catenin accumulation in MDA-MB-231 cells
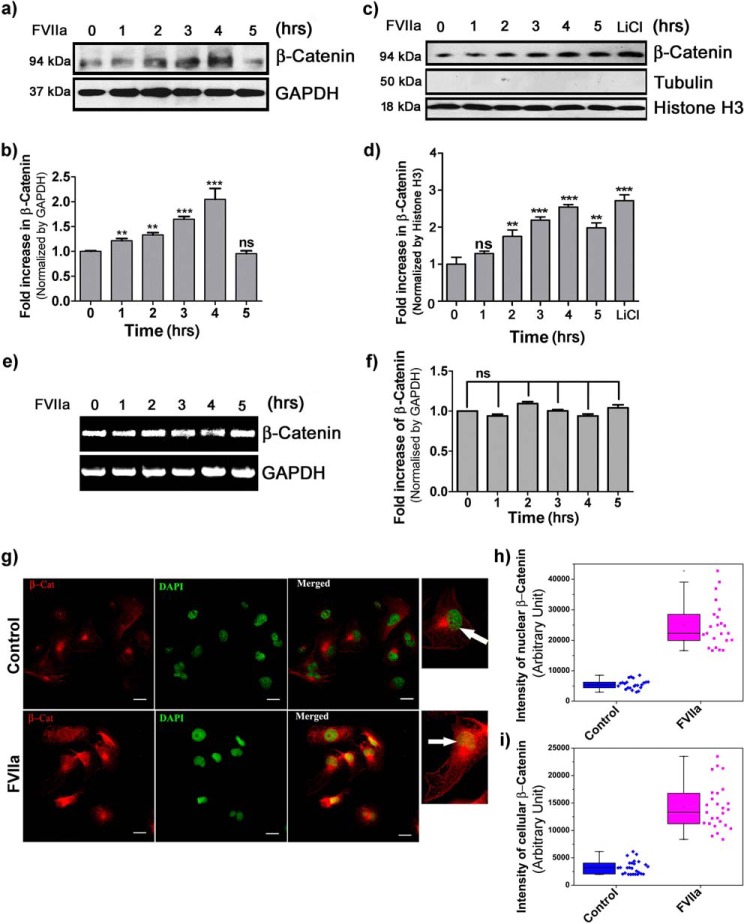
Enhanced migration of cancer cells plays a significant role in cancer metastasis. Among various signaling pathways contributing to the enhancement of migratory potential of cancer cells, Wnt signaling is one of the most prevalent (20, 22). Growing evidence suggests that TF-FVIIa interaction leads to increased breast cancer cell migration and motility (16). Atypical accumulation of β-catenin has been well studied in various complications of cancer, including breast cancer (23). Up-regulation of cytoplasmic/nuclear β-catenin accumulation has been identified in 40–60% of human breast tumors and is correlated with poor prognosis (24, 25). β-Catenin accumulation due to genetic mutations in Axin, APC, or β-catenin has been diagnosed in several other tumor types but is unusual in breast cancer (26). These observations suggest that breast cancer cells adapt an alternate or additional mechanism for β-catenin accumulation. As mentioned earlier, the basic mechanism behind breast cancer cell migration due to FVIIa has yet to be elucidated. Hence, to examine the existence of any correlation between breast cancer cell migration and FVIIa, we challenged MDA-MB-231 human metastatic breast cancer cells with FVIIa for 5 h after 2 h of serum deprivation. Interestingly, we observed a consistent time-dependent increase in the β-catenin level up to 4 h (Fig. 1, a and b). Our data also indicates an increase in the rate of nuclear β-catenin accumulation in a temporal manner (Fig. 1, c and d). Furthermore, semiquantitative PCR analysis revealed an unaltered mRNA level of β-catenin upon subsequent treatment of cells with FVIIa (Fig. 1, e and f). Fluorescence microscopic images further confirmed that β-catenin protein accumulates in a significant level inside the cell nucleus at the 4-h time point after FVIIa exposure (Fig. 1g). Altogether, these findings demonstrate that FVIIa treatment results in β-catenin accumulation in MDA-MB-231 cells without altering its expression level, and this leads to β-catenin translocation inside the cell nucleus.
Figure 1.
Accumulation of β-catenin in MDA-MB-231 cells due to FVIIa treatment. Cells were seeded onto a 12-well plate and allowed to grow. After 2-h serum starvation, cells were treated with FVIIa (100 nm). a, time-dependent accumulation of β-catenin was estimated by standard Western blotting with GAPDH as a loading control. b, band intensity was measured and quantified using ImageJ, and the graph was plotted by GraphPad Prism 5. c, nuclear accumulation of β-catenin was analyzed by Western blotting of nuclear lysate with histone H3 as a loading control. LiCl was used as a positive control. Tubulin was used as a measure of nucleus isolation purity. d, quantification of nuclearly translocated β-catenin was done by ImageJ, and the graph was plotted by GraphPad Prism 5. e, semiquantitative PCR analysis was performed to check the expression status of β-catenin gene in the cells after treatment with FVIIa for various time points. f, quantitative estimation of β-catenin band intensity over GAPDH was done using ImageJ and GraphPad Prism 5. The images of Western blot and RT-PCR analysis are representative of at least three independent experiments. The data are presented as mean ± S.E. Differences are considered to be statistically significant at p < 0.05 using Student's t test. g, cells were grown onto coverglasses followed by treatment with FVIIa. Cells were fixed with 4% paraformaldehyde and permeabilized with 0.01% Triton X-100 followed by immunostaining with β-catenin antibody. DAPI was used for staining nuclei. Scale bars, 25 μm. Quantitative estimation of β-catenin inside the cells and nucleus was performed by ImageJ and MATLAB software; the number of samples (n) per treatment was 23. White arrows indicate nuclear β-catenin. Graphical representations of β-catenin intensity in the nucleus (h) and cells (i) are presented. Error bars represent ±S.E. of the mean. **, p < 0.05; ***, p < 0.001; ns, non-significant using Student's t test; n ≥ 3.
β-Catenin accumulation by FVIIa in MDA-MB-231 cells is PAR2-dependent
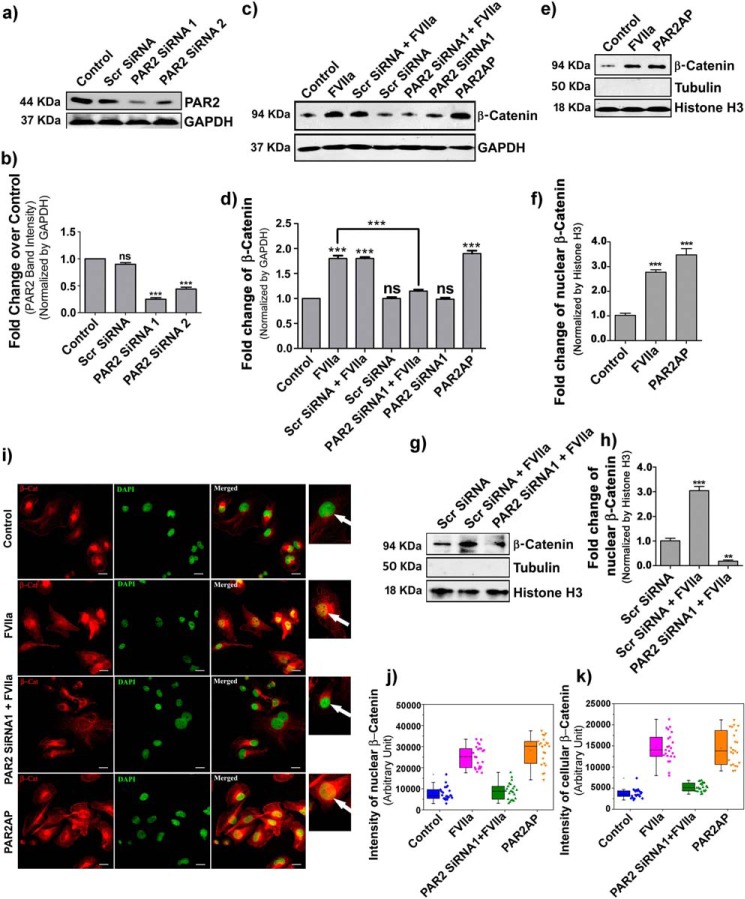
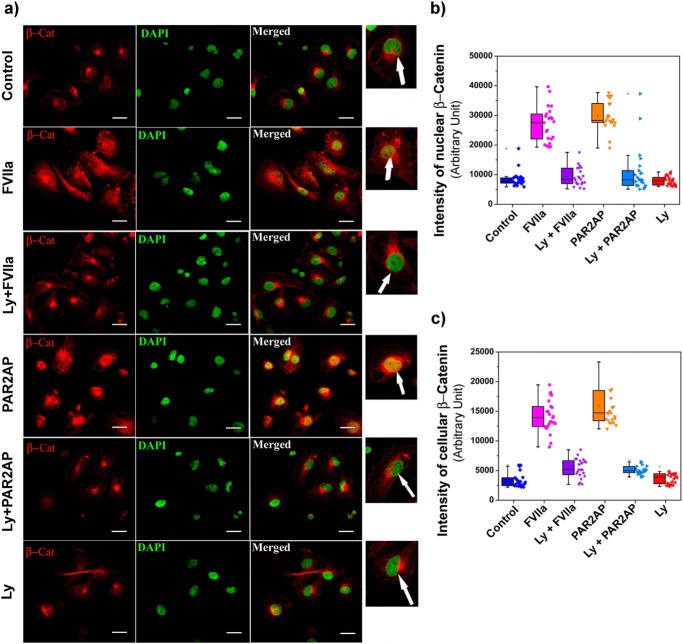
Previous studies have demonstrated that the majority of FVIIa-mediated signaling is dependent on PAR2 (27); hence, we questioned whether FVIIa-modulated β-catenin accumulation in MDA-MB-231 cells is through PAR2 activation. To investigate this, we knocked down PAR2 with PAR2 siRNA and then treated cells with FVIIa and PAR2 activation peptide (PAR2AP; a positive control). The efficiency of PAR2 knockdown with PAR2 siRNA was estimated by Western blotting (Fig. 2, a and b). Interestingly, we found complete reversal of FVIIa-mediated β-catenin accumulation in PAR2-knocked down cells, strongly establishing the involvement of PAR2 in FVIIa-dependent β-catenin accumulation (Fig. 2, c and d). Next, we aimed to examine the status of β-catenin in the nucleus of PAR2-activated cells and observed a significant increment of β-catenin inside the nucleus (Fig. 2, e and f). Here we used histone H3 as a loading control for nuclear lysate and tubulin to measure the purity of nuclear isolation. Consistent with the cellular accumulation, we also observed a suppressed level of β-catenin accumulation in PAR2-deficient cells after FVIIa treatment (Fig. 2, g and h). Fluorescence microscopic images also support less accumulation of β-catenin (red) in the cell cytoplasm as well as in the nucleus (green) in PAR2-silenced FVIIa-challenged cells as compared with treated cells (yellow nuclei due to green DAPI and red β-catenin co-localization) (Fig. 2i). Quantification of the intensity of β-catenin is presented in Fig. 2 for the nucleus (j) and cells (k). As a whole, the findings suggest that PAR2 activation is absolutely required for FVIIa-mediated accumulation of β-catenin in MDA-MB-231 cells.
Figure 2.
β-Catenin accumulation by FVIIa in MDA-MB-231 cells occurs through PAR2 activation. MDA-MB-231 cells were transfected with PAR2 siRNA1 and siRNA2 using Lipofectamine 2000 along with a scrambled (Scr) siRNA as control and allowed to grow for 48 h. a, PAR2 knockdown was verified by Western blotting. b, quantitative estimation of PAR2 band intensity over GAPDH was done using ImageJ and GraphPad Prism 5. c, untreated, scrambled siRNA- and PAR2 siRNA1-treated cells were serum-starved for 2 h and stimulated with FVIIa and PAR2AP (10 μm), and the accumulation of β-catenin was measured after 4 h by Western blotting. d, band intensity of β-catenin over GAPDH was estimated using ImageJ and GraphPad Prism 5. e, nuclear accumulation of β-catenin upon treating the cells with PAR2AP. f, quantification of nuclear β-catenin was done using ImageJ and GraphPad Prism 5. g, nuclear accumulation of β-catenin was verified in PAR2-silenced and control scrambled siRNA-treated cells after challenging with FVIIa. h, quantitative analysis of nuclear β-catenin over histone H3 was performed using ImageJ and GraphPad Prism 5. All the experiments were performed at least three times, and the data are denoted as mean ± S.E. Differences are statistically significant at p < 0.05 using Student's t test. i, cells were transfected with PAR2 siRNA followed by treatment with FVIIa and PAR2AP. β-Catenin accumulation was analyzed by immunostaining with β-catenin antibody. Nuclei were stained with DAPI. Scale bars, 25 μm. White arrows indicate nuclear β-catenin. Quantitative estimation of β-catenin inside the cells and nucleus was performed using ImageJ and MATLAB software. The number of samples (n) per treatment was 23. Graphical representations of β-catenin intensity in the nucleus (j) and cells (k) are presented. Error bars represent ±S.E. of the mean. **, p < 0.05; ns, non-significant using Student's t test; n ≥ 3.
FVIIa-induced β-catenin accumulation also occurs in tissue factor- and PAR2-overexpressing MCF-7 cells
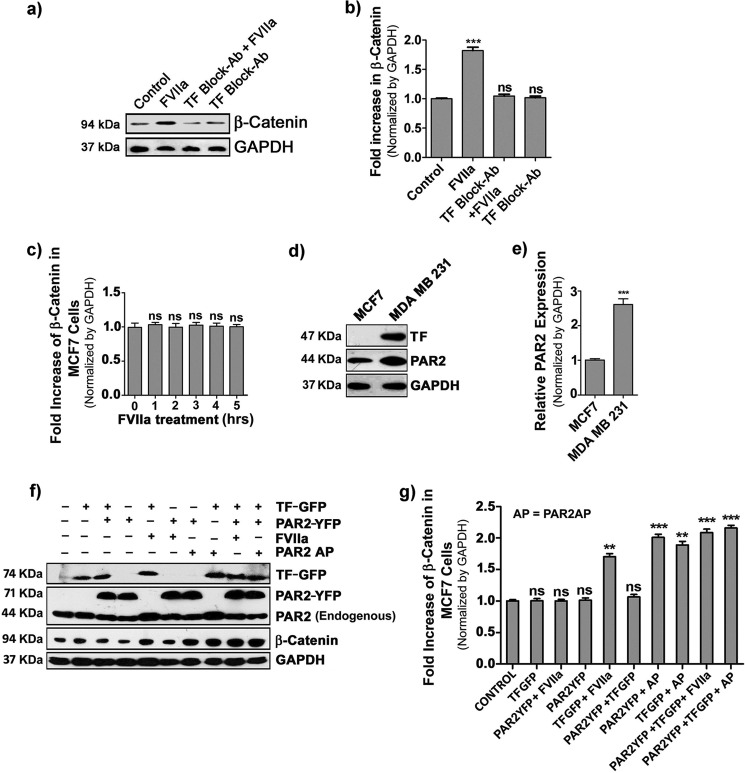
Next, we examined the involvement of TF in the context of FVIIa-mediated β-catenin accumulation. To analyze the importance of TF, we treated MDA-MB-231 cells with TF-blocking antibody prior to FVIIa addition and observed complete attenuation of β-catenin accumulation (Fig. 3, a and b), which confirms that FVIIa-induced β-catenin accumulation is solely TF-dependent. To explore this further, we used another human breast cancer cell line, MCF-7. We estimated β-catenin accumulation in MCF-7 cells after treatment with FVIIa for 5 h and observed no alteration in the β-catenin level (Fig. 3c). Interestingly, we found that MCF-7 cells are devoid of any TF and express lower levels of PAR2 as compared with MDA-MB-231 (Fig. 3, d and e). Therefore, we transfected MCF-7 cells with TF-GFP and PAR2-YFP plasmid constructs, either one or both, followed by FVIIa treatment, and cellular β-catenin accumulation was analyzed (Fig. 3, f and g). TF-overexpressing MCF-7 cells showed increased levels of cellular β-catenin upon FVIIa treatment. Further elevation in β-catenin accumulation was observed in cells co-transfected with both TF and PAR2 followed by FVIIa treatment. Overall, the data reflect that TF is absolutely needed for FVIIa to cleave PAR2 and induce accumulation of cellular β-catenin. In addition, this result also establishes that PAR2-mediated β-catenin accumulation is not specific for MDA-MB-231 cells but also can take place in other breast cancer cells.
Figure 3.
Involvement of tissue factor in FVIIa-induced accumulation of β-catenin. a, MDA-MB-231 cells were treated with TF-blocking antibody (Ab) for 2 h followed by challenging with FVIIa, and β-catenin accumulation was analyzed after 4 h by Western blot analysis. b, quantification of β-catenin accumulation was performed using ImageJ and GraphPad Prism 5. c, quantitative estimation of β-catenin accumulation by Western blotting was checked in MCF-7 cells upon treatment with FVIIa at various time intervals (0–5 h). d, relative levels of TF and PAR2 were measured in MCF-7 and MDA-MB-231 cells by Western blot. e, relative PAR2 band intensity was quantified over GAPDH in MCF-7 and MDA-MB-231 cells using ImageJ and GraphPad Prism 5. f, TF, PAR2, and β-catenin levels in control, TF-GFP-, and PAR2-YFP-overexpressing MCF-7 cells were analyzed by Western blotting after FVIIa or PAR2AP treatment. g, band intensity of β-catenin over GAPDH in control, TF-GFP-, and PAR2-YFP-overexpressing MCF-7 cells was quantified using ImageJ and GraphPad Prism 5. Error bars represent ±S.E. of the mean. **, p < 0.05; ***, p < 0.001; ns, non-significant using Student's t test; n ≥ 3. AP, agonist peptide.
Both TF-FVIIa and PAR2AP modulate β-catenin accumulation in MDA-MB-231 cells via AKT/GSK3β-dependent pathway
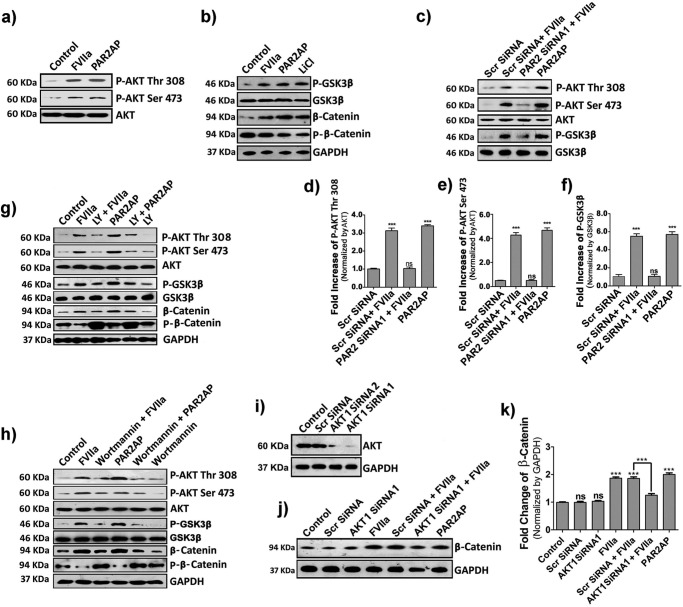
GSK3β is a ubiquitously expressed serine/threonine kinase that synchronizes various cellular functions such as gene expression, apoptosis, cytoskeletal rearrangement, and cell proliferation (28). Its activity is perturbed by phosphorylation, which leads to alterations in downstream targets (28). In the absence of any upstream stimulus, β-catenin is secluded in an idle state by a multimeric “destruction complex” comprising GSK3β, Axin, and APC (29, 30). APC and Axin act as scaffolding proteins, allowing GSK3β-mediated phosphorylation of β-catenin, which leads to its ubiquitination and subsequent proteasomal degradation (30). AKT, an upstream regulator of GSK3β, has been reported to play an important role in cellular migration and cancer (31); therefore, we determined whether PAR2 activation in MDA-MB-231 cells triggers AKT activation via phosphorylation as well as GSK3β inactivation. We found that PAR2 activation by TF-FVIIa or PAR2AP leads to increased phosphorylation of both AKT (Thr-308 and Ser-473) and GSK3β (Ser-9), which remain unaffected upon PAR2 knockdown (Fig. 4, a–f). We also observed that FVIIa or activation peptide not only cause AKT/GSK3β phosphorylation but also decrease phosphorylation of β-catenin, which leads to its subsequent accumulation inside the cell (Fig. 4, a and b). To further elucidate the importance of AKT, we blocked the PI3K/AKT pathway using two different chemical inhibitors, LY294002 and wortmannin, separately followed by ligand treatment. As expected, inhibition of PI3K dramatically reduced the phosphorylation of AKT and GSK3β and increased β-catenin phosphorylation, resulting in less β-catenin accumulation due to enhanced degradation of β-catenin (Fig. 4, g and h). We further confirmed the involvement of AKT by silencing AKT1 in MDA-MB-231 cells (Fig. 4i) followed by ligand addition (Fig. 4, j and k). Results suggest complete reduction of β-catenin accumulation in AKT1-silenced cells. These findings clearly indicate that β-catenin accumulation due to PAR2 activation occurs through PI3K/AKT pathway followed by GSK3β inactivation. The nuclear accumulation of β-catenin in the presence/absence of LY294002 in control and PAR2-activated cells was also evident in fluorescent microscopic images (Fig. 5a). Nuclei with DAPI stain (green) revealed no β-catenin accumulation in LY294002-treated or control cells, whereas FVIIa- or PAR2AP -treated cells with yellow nuclei (due to co-localization of β-catenin and DAPI) indicate significant β-catenin accumulation inside the nucleus. LY294002 addition also reduced nuclear β-catenin accumulation even after FVIIa or PAR2AP treatment. Fig. 5, b and c, present the intensity of the presence of β-catenin inside the nucleus and cells, respectively. Random fields were chosen for imaging and analysis. The number of images taken for every treatment or control analysis was 23 (n = 23).
Figure 4.
TF-FVIIa or PAR2AP modulates β-catenin accumulation in MDA-MB-231 cells via AKT/GSK3β-dependent pathway. a, AKT phosphorylation (both at Thr-308 and Ser-473) at 10 min. b, cells were challenged with FVIIa, PAR2AP, and the positive control LiCl (40 mm), and the phosphorylation status of GSK3β (at Ser-9) and β-catenin and phospho-β-catenin levels were checked at 1, 4, and 4 h, respectively by Western blotting. c, phospho-AKT (at Thr-308 and Ser-473) and -GSK3β (at Ser-9) were checked in PAR2 siRNA1- and scrambled (Scr) siRNA-treated cells after challenging with FVIIa. d, e, and f, quantitative band intensities of phospho-AKT at Thr-308 and Ser-473 and phospho-GSK3β at Ser-9 normalized by respective proteins were measured using ImageJ and GraphPad Prism 5. Cells were treated with either LY294002 (20 μm) (g) or wortmannin (50 μm) (h) 1 h before FVIIa or PAR2AP treatment, and the cell lysates were analyzed for phospho-AKT (both at Thr-308 and Ser-473), phospho-GSK3β (at Ser-9), β-catenin, and phospho-β-catenin by Western blotting. i, AKT1 knockdown was performed by two siRNAs (1 and 2) and analyzed by Western blotting. j, β-catenin levels were estimated in scrambled and AKT1 knockdown cells after FVIIa treatment by Western blotting. k, accumulated β-catenin levels (normalized by GAPDH) were quantified using ImageJ and GraphPad Prism 5. Error bars represent ±S.E. of the mean. ***, p < 0.001; ns, non-significant using Student's t test; n = 3.
Figure 5.
β-Catenin accumulation was assessed by fluorescence microscopy upon inhibiting PI3K with LY294002 followed by PAR2 activation. a, cells were treated with LY294002 (Ly) followed by treatment with FVIIa and PAR2AP. β-Catenin (β-Cat) accumulation was analyzed by immunostaining with β-catenin antibody. Nuclei were stained with DAPI. Scale bars, 25 μm. White arrows indicate nuclear β-catenin. Quantitative estimation of β-catenin inside the cells and nucleus was performed using ImageJ and MATLAB software. The number of samples (n) per treatment was 23. Graphical representations of β-catenin intensity in the nucleus (b) and cells (c) are presented. Error bars represent ±S.E. of the mean; n = 23.
PAR2 activation leads to β-catenin-induced transcriptional activation of downstream metastatic proteins
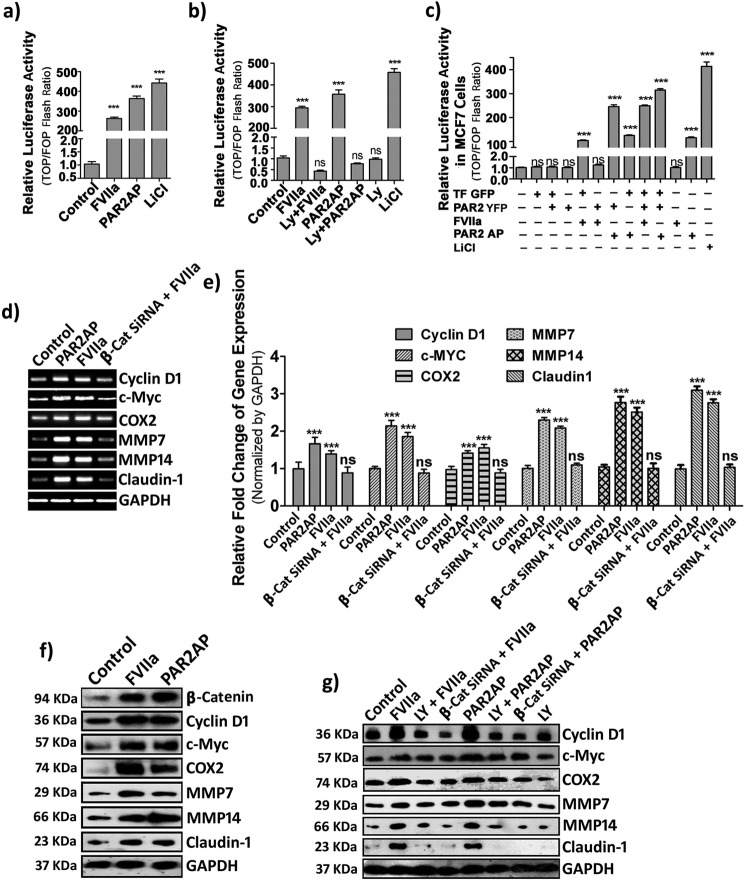
It is well documented that, once stabilized, β-catenin translocates to the nucleus and participates in transcriptional activation of responsive genes critical for tumor cell proliferation and migration via interaction with TCF/LEF (29, 32). To study the fate of nuclearly translocated β-catenin, a TCF/LEF luciferase assay was performed to measure the transcriptional efficiency of β-catenin. We observed a significant increase of luciferase activity in FVIIa- and PAR2AP-treated cells (Fig. 6a). This luciferase activity was abolished by introducing PI3K inhibitor (LY294002) (Fig. 6b), which is consistent with our previous observations. To further establish the role of TF and PAR2 in the context of β-catenin-mediated transcriptional activation, we overexpressed PAR2-YFP and TF-GFP in MCF-7 cells and performed the TCF-LEF luciferase assay (Fig. 6c). We observed that activation of PAR2 also leads to β-catenin accumulation followed by transcriptional activation in MCF-7 cells. Furthermore, we investigated the expression levels of downstream genes responsive to the β-catenin transcriptional complex. Our semiquantitative and real-time PCR analysis revealed a significant rise in the mRNA levels of cyclin D1, c-Myc, COX-2, MMP-7, MMP-14, and Claudin-1 during FVIIa and PAR2AP treatment. To establish the specific role of β-catenin, we knocked down β-catenin by β-catenin siRNA and observed complete attenuation of the above mentioned transcripts in PAR2-activated cells, indicating that the elevated levels of these transcripts by PAR2 activation are mediated via β-catenin (Fig. 6, d and e). Consistent with the transcriptional status, an increase in cyclin D1, c-Myc, COX-2, MMP-7, MMP-14, and Claudin-1 protein levels was also observed during PAR2 activation that remained unaltered by β-catenin knockdown or inhibition of PI3K (Fig. 6, f and g). These findings clearly indicate that β-catenin translocation inside the nucleus leads to the formation of active β-catenin transcriptional complex, which further up-regulates several downstream metastatic proteins.
Figure 6.
PAR2 activation leads to β-catenin-induced transcriptional activation of downstream proteins. a, MDA-MB-231 cells transfected with a TCF/LEF-sensitive or -insensitive reporter vector were seeded in a 96-well plate, grown to full confluence, and subjected to 2-h serum starvation followed by FVIIa, PAR2AP, and LiCl (used as a positive control) treatment for 6 h. Luciferase activity of the cell lysates was analyzed using the Steady-Glo luciferase assay system, and the graph was plotted using GraphPad Prism 5. b, similarly, luciferase activity was quantified for LY294002 (Ly)-pretreated FVIIa/PAR2AP-challenged cells. c, like MDA-MB-231 cells, normal, TF-GFP-, and/or PAR2-YFP-overexpressing MCF-7 cells were transfected with TCF/LEF-sensitive or -insensitive reporter vector, relative luciferase activity was measured after treatment with FVIIa or PAR2AP, and the graph was plotted using GraphPad Prism 5. Transcriptional level of cyclin D1, c-Myc, COX-2, MMP-7, MMP-14, and Claudin-1 were analyzed by semiquantitative PCR (d) and real-time PCR (e) in control and β-catenin-knocked down cells after challenging with FVIIa. f, the protein expression profile of downstream targets after 8 h of ligand addition was analyzed by Western blotting. g, protein expression of these genes was checked in either LY294002-pretreated or β-catenin-knocked down cells after treatment with FVIIa or PAR2AP. Error bars represent ±S.E. of the mean. ***, p < 0.001; ns, non-significant using Student's t test; n = 3.
PAR2 activation promotes migration and invasion of MDA-MB-231 cells through PI3K-AKT-dependent β-catenin accumulation
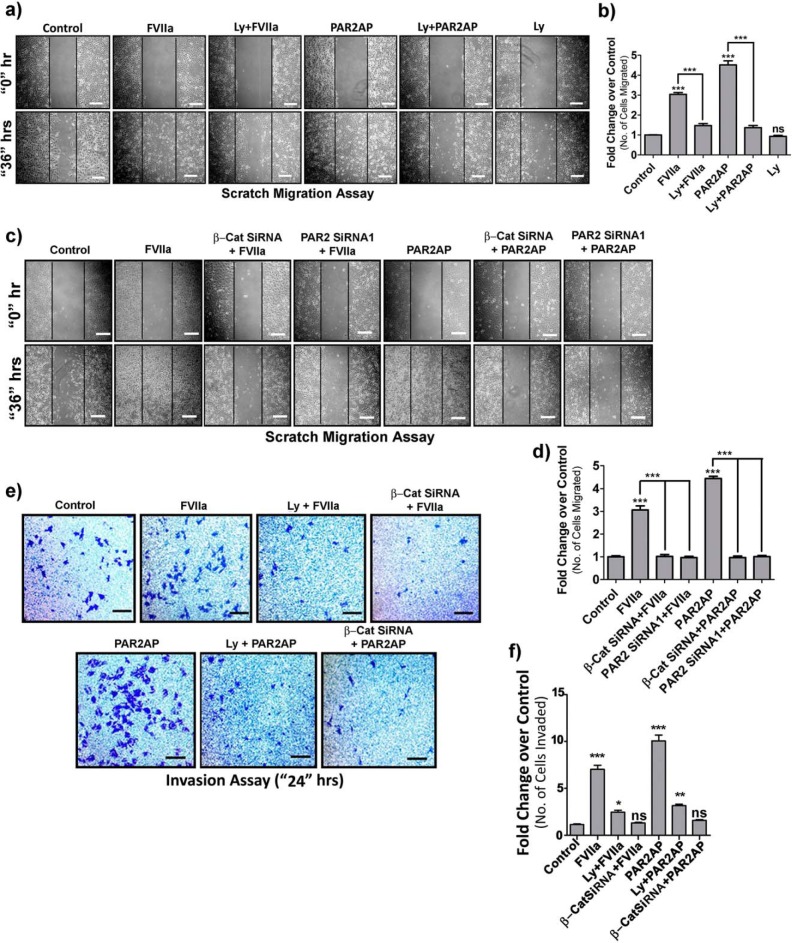
Previous studies have demonstrated that PAR2-mediated signaling induces metastatic behavior of breast cancer both in vitro and in vivo (17, 33–35). Therefore, to elucidate the signaling molecules involved in this transition, we assessed the metastatic potential by migration (Fig. 7, a–d) and invasion assays (Fig. 7, e and f). FVIIa- and PAR2AP-induced enhancement of MDA-MB-231 cell migration and invasion was perturbed by either inhibiting PI3K or silencing β-catenin, suggesting the active participation of the AKT-β-catenin axis in PAR2-dependent cancer metastasis. As a whole, these data clearly demonstrate that PAR2 activation by TF-FVIIa or PAR2AP leads to increased migration and invasion of MDA-MB-231 cells through the interplay of AKT, β-catenin, and related molecules.
Figure 7.
PAR2 activation promotes migration and invasion of MDA-MB-231 cells through PI3K-AKT-dependent β-catenin accumulation. a, MDA-MB-231 cells were seeded on coverglasses and grown to full confluence, and a single scratch line was created followed by FVIIa or PAR2AP treatment. LY294002 (Ly) was added 1 h before ligand addition. Cells were allowed to migrate to the scratched area for 36 h, and images were taken at 0 and 36 h. Vertical black lines indicate the boundary of the edges of the wound at 0 h. Scale bars, 200 μm. b, the number of cells migrated to the scratched area was counted and graphically represented using GraphPad Prism 5. c, similarly, a scratch migration assay was performed on control, β-catenin- or PAR2-knocked down cells after challenging with FVIIa or PAR2AP. Scale bars, 200 μm. d, the number of cells migrated to the scratched area was counted and graphically represented using GraphPad Prism 5. e, control or β-catenin-knocked down cells were seeded on a Transwell membrane previously coated with Matrigel. Cells were treated with FVIIa or PAR2AP and incubated for 24 h. LY294002 was added 1 h prior to ligand addition. The extravasated cells were stained with crystal violet and borate solution after scraping off the cells from the upper chamber (as mentioned under “Materials and methods”). Images were taken by a bright field microscope. Scale bars, 200 μm. f, the number of invaded cells were counted and graphically represented using GraphPad Prism 5. Error bars represent ±S.E. of the mean. *, p < 0.05; **, p < 0.05; ***, p < 0.001; ns, non-significant using Student's t test; n = 3.
β-Catenin and its downstream targets remain well elevated in human breast cancer tissues as compared with normal breast tissues
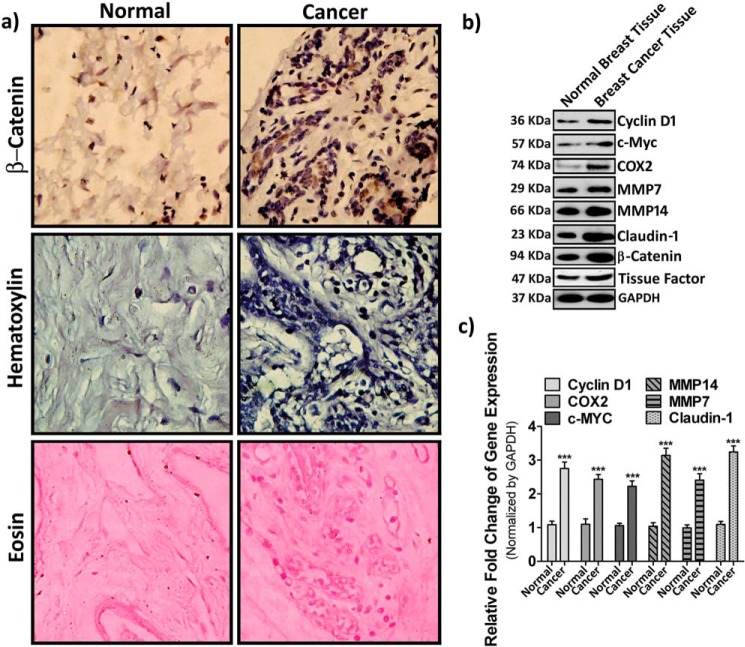
Our present in vitro study indicates that an intrinsic correlation exists between β-catenin accumulation and metastatic potential of breast cancer cells in response to PAR2 activation. Henceforth, to compare the level of β-catenin and its downstream proteins in human breast cancer tissue with respect to normal tissue, samples were collected according to human ethical regulations. Immunohistochemistry data depicts a significant level of β-catenin accumulation in cancer tissue as compare with normal tissue (Fig. 8a). Consistent with immunohistochemistry, Western blot data also revealed an elevated level of β-catenin and its downstream targets in breast cancer tissue (Fig. 8b). Interestingly, we also observed a considerable increment of the TF level in tumor tissue samples. To further analyze the expression profile of β-catenin downstream targets, we also performed real-time PCR analysis of mRNA isolated from cancer and normal breast tissues (Fig. 8c). Consistent with our previous observations, we found up-regulation of β-catenin target gene expression at the mRNA level. In conclusion, the enhanced pattern of signature molecules in cancer tissue observed was similar to the results of our in vitro analysis, suggesting the active involvement of a similar pathway operating in both in vivo and in vitro conditions.
Figure 8.
Expression of β-catenin and its downstream targets in human breast cancer and normal breast tissues. Human breast cancer and normal tissue samples were collected from five different human breast cancer patients and subjected to further analyses. a, immunohistochemistry analysis against β-catenin. Hematoxylin and eosin staining was done (as mentioned under “Materials and methods”) to estimate the level of β-catenin in human breast cancer with respect to normal breast tissue. Images were taken by a bright field microscope under 40× resolution. b, the expression profile of cyclin D1, c-Myc, COX-2, MMP-7, MMP-14, and Claudin-1 along with TF and β-catenin was analyzed at the protein level by Western blotting. c, the transcriptional level of these β-catenin downstream targets was analyzed by real-time PCR. Normal tissues were obtained from the same individual patient's amputated breast tissue. Error bars represent ±S.E. of the mean. ***, p < 0.001 using Student's t test; n = 3.
Discussion
Apart from blood coagulation, the role of coagulation proteases in cancer progression has been well established (36). There is evidence that the primary initiator of the main coagulation cascade, TF, is also involved in oncogenic progression, tumor metastasis, and angiogenesis (16). Through in vitro studies showing the contribution of coagulation protease complex TF–FVIIa and its correlation with various cellular signaling pathways, TF-FVIIa is well accepted as a tumor promoter (35, 37, 38). Most human epithelial cancers have high levels of TF and PAR2 (39, 40). Although the distinct in vivo contribution of TF is well documented, the role of its only ligand, FVIIa, remains elusive due to its physical distance from extravascular cells (where TF physically resides) after crossing the endothelial barrier. Identification of endothelial protein C receptor as an auxiliary receptor for FVIIa (41) and its transcytosis by endothelial protein C receptor provides a novel mechanism for the transfer of FVIIa from the blood to extravascular tissues, crossing the endothelial barrier. Its observed redistribution to extravascular tissues with prolonged retention paved the way for accepting FVIIa as an active in vivo PAR2 activator through TF (42).
Several studies regarding breast cancer have portrayed TF-FVIIa as an active inducer of cancer cell migration, invasion, and proliferation; however, the details of the signaling mechanisms leading to this progression are still unexplored. Through our approach, we have addressed the molecular events responsible for the aggressiveness of breast cancer cells in response to FVIIa. In this study, the observed β-catenin accumulation by FVIIa via the AKT pathway appears to be a novel mechanism by which PAR2 contributes to breast cancer cell migration and invasion. Our results support a pivotal role of PAR2 activation in promoting metastatic behavior of breast cancer cells. The eloquent mechanism that we uncovered is that PAR2 activation increases β-catenin stabilization and subsequent nuclear translocation. Furthermore, we showed that PAR2 activation-dependent stabilization of β-catenin is acquired by coordinating the activation of AKT/GSK3β signaling, leading to destruction of the β-catenin phosphorylation complex (Fig. 9). We also provide molecular evidence supporting the regulatory role of PAR2 and AKT in β-catenin stabilization and its nuclear translocation. The significance of our in vitro results is strengthened by the fact that the key component of this pathway, β-catenin, is well detected in elevated levels in human breast tumors.
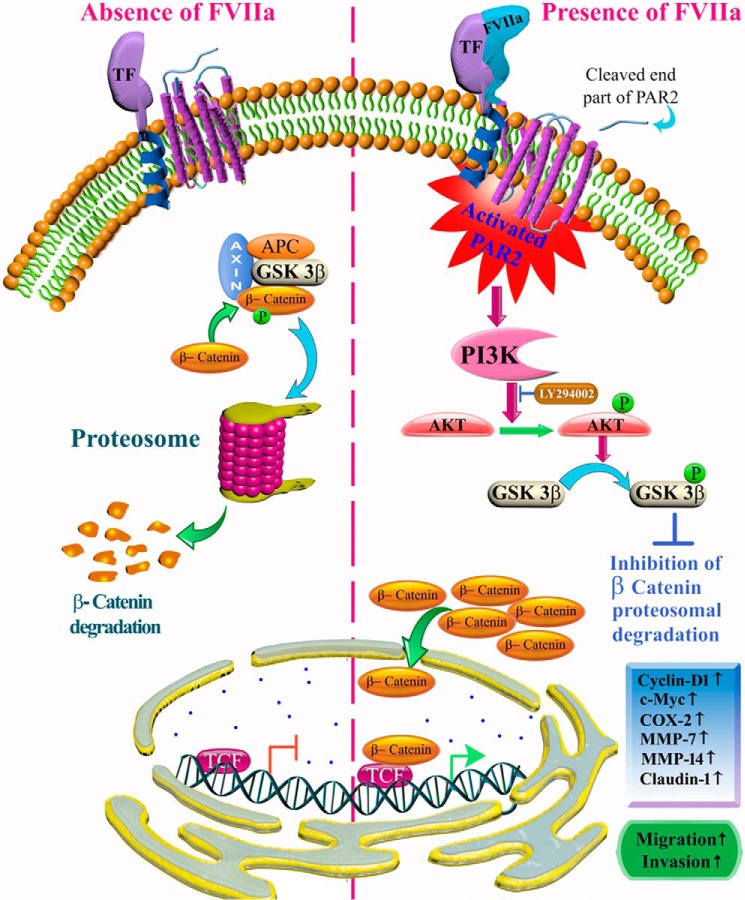
Figure 9.
Schematic representation of TF-FVIIa-dependent PAR2-activated signaling pathway that accounts for β-catenin accumulation and expression of metastatic proteins leading to enhanced migration and invasion of human breast cancer cells. In the normal condition, GSK3β binds to Axin and APC to form an “active phosphorylation complex,” which readily phosphorylates β-catenin, leading to its proteasomal degradation. In the presence of FVIIa, active TF–FVIIa complex is formed and activates PAR2 receptor, leading to intracellular activation of AKT via phosphorylation, which again phosphorylates GSK3β and inactivates it. As a result, β-catenin remains in a non-degradable unphosphorylated state and accumulates inside the cell nucleus to promote the expression of several prometastatic genes like cyclin D1, c-Myc, COX-2, MMP-7, MMP-14, and Claudin-1, leading to enhanced cell migration and invasion of breast cancer cells.
The dual-functioning molecule β-catenin intensifies cell-cell adhesion when bound to E-cadherin complexes and performs a transcriptional co-activator role after nuclear translocation (43). Depletion of E-cadherin–β-catenin complex and nuclear translocation of β-catenin are vital for the development of migratory characteristics of cancer cells (29, 44, 45). These findings suggest that PAR2 activation promotes nuclear translocation of β-catenin and increases the recruitment of β-catenin at TCF/LEF-binding sites. PAR2AP induces overactive β-catenin signaling, which leads to an enhanced co-activation function as evident by higher levels of TCF/LEF-responsive genes (like Cyclin D1, c-Myc, etc.). Up-regulation of cyclin D1 has been implicated in acquisition of higher proliferative potential (46–48). Nuclear β-catenin induces tumor invasion-supporting genes and transition to a metastatic phenotype via recruiting the transcriptional co-activators and cyclic AMP response element-binding protein (21). In the absence of enzymatic activities, β-catenin has been considered “non-targetable.” However, recent findings show that other aspects of this pathway such as β-catenin stabilization or transcriptional activities can be aimed to inhibit its downstream effects. We speculate that disruption of the β-catenin-mediated transcriptional complex assembly could be a productive strategy to inhibit PAR2 activation-induced cell migratory and invasive properties. Consistent knockdown of β-catenin or PI3K inhibition completely attenuates the PAR2-mediated metastatic potential of breast cancer cells. Hence, targeting PAR2 activation-induced hyperactive β-catenin signaling with small molecule inhibitors could represent a novel and effective strategy for therapeutic intervention. We showed that perturbing PAR2 receptor dramatically reduces the phosphorylation of GSK3β. Blocking of GSK3β inhibition may also be a potential target for a therapeutic approach alongside conventional chemo/radiation therapy against breast cancer progression.
Materials and methods
Cell culture
The human breast cancer cell lines MCF-7 and MDA-MB-231 were obtained from the American Type Culture Collection and cultured in standard DMEM (Gibco) supplemented with 10% FBS and 100 units/ml penicillin-streptomycin (Invitrogen). Cells were maintained in a humidified chamber at 37 °C and 5% CO2 level. Cells were seeded on 12-well plates at a density of 0.1 × 106 and allowed to grow to confluence. Serum starvation was applied for 2 h followed by treatment with human recombinant factor VIIa (100 nm; Novo Nordisk) and PAR2 activation peptide (SLIGKV-NH2; 100 μm) (GLBiochem, China). In another set of experiments, cells were treated with PI3K inhibitor LY294002 (20 μm; Calbiochem) and wortmannin (50 μm; Sigma). Antibodies against the following were purchased from the sources indicated: β-catenin, tissue factor, and PAR2 (Abcam); GAPDH and histone H3 (Sigma); p-AKT, AKT, p-GSK3β, GSK3β, cyclin D1, c-Myc, COX-2, MMP-7, MMP-14, Claudin-1, and tubulin (Cell Signaling Technology); antibodies were used at dilutions recommended in the instruction manuals. HRP-conjugated anti-rabbit and anti-mouse secondary antibodies were obtained from Sigma.
Transfection of plasmid constructs
TF-GFP and PAR2-YFP constructs were transfected into MCF-7 cells using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol.
Western blotting
Cells were treated with ligand and lysed with Laemmli buffer at different time points. The lysate was heated at 95 °C for 5 min. Proteins were separated by SDS-PAGE and transferred onto PVDF membrane, which was blocked in 5% nonfat dried milk in TBS and incubated overnight at 4 °C with primary antibodies for specific proteins in 3% BSA in TBS. After washing with TBS-T (TBS, 1% Tween20), secondary antibody was added and incubated for an hour followed by washing and development using the ECL method. Densitometry of the blot was performed using ImageJ, and graphs were prepared using GraphPad Prism 5.
Tissue factor blocking experiment
MDA-MB-231 cells were seeded in 12-well plates and allowed to grow to full confluence. The cell monolayer was incubated at room temperature for 2 h with TF polyclonal blocking antibody, which binds to TF receptors of the cell surface, followed by treatment with FVIIa for 4 h. Cells were lysed thereafter by Laemmli buffer, and Western blot analysis was performed to check the β-catenin accumulation level.
Preparation of subcellular fractions
Equal numbers of MDA-MB-231 cells were seeded in 35-mm dishes and allowed to attain 100% confluence. After 2 h of serum starvation, cells were treated with FVIIa and PAR2 agonist peptide for 4 h. Cytoplasmic and nuclear fractions were prepared by incubating the cells in 50 μl of ice-cold lysis buffer (10 mm NaCl, 10 mm Tris-HCl (pH 7.4), 3 mm MgCl2, 0.5% Nonidet P40, 0.1 mm PMSF, protease inhibitor mixture). Lysate was kept on ice for 7–8 min followed by centrifugation at 8000 ×g for 15 min at 4 °C to precipitate nuclei. The supernatant was stored as the cytoplasmic fraction. The nuclear pellet was incubated in 50 μl of chilled nuclear extraction buffer (20 mm Tris-HCl (pH 7.9), 0.42 m KCl, 0.2 mm EDTA, 10% glycerol, 2 mm DTT, 0.1 mm PMSF, protease inhibitor mixture) for 20 min followed by centrifugation at 21,000 × g for 15 min at 4 °C to precipitate nuclear debris. Equal volumes of lysate were subjected to Western blot analysis.
Immunofluorescence
Cells were seeded onto coverslips etched in 35% hydrofluoric acid for 10 s. Cells were serum-starved for 2 h and treated with FVIIa and PAR2 activation peptide for 4 h followed by washing with PBS, and then cells were fixed with 4% paraformaldehyde for 30 min. Cells were permeabilized with 0.1% Triton X-100 for 10 min, washed thrice with PBS, and then blocked with 5% BSA for an hour. Cells were incubated with β-catenin primary antibody overnight and after washing were treated with Alexa Fluor 555 rabbit secondary antibody for 1 h. After washing, the slides were exposed to DAPI for 10–20 min and again washed with PBS. Cells were imaged with an sCMOS camera (Orca Flash 4.0, Hamamatsu) on an inverted fluorescence microscope from Carl Zeiss (Axio Observer Z1).
Image analysis
Cell images were taken with 63× magnification (numerical aperture, 1.4) using the Axio Observer Z1 microscope. Images were analyzed using ImageJ software. Calculations were performed using MATLAB.
β-Catenin-TCF/LEF-regulated reporter gene assay
MDA cells were seeded in a T-25 flask at a seeding density of 0.7 × 106 cells and incubated to grow for 24 h. On the following day, cells were transfected with 4 μg of either a β-catenin-TCF/LEF-sensitive or -insensitive reporter vector (TOP FLASH/FOP FLASH, Upstate Biotechnology Inc.) using Lipofectamine 2000 reagent according to the manufacturer's instructions. The next day, cells were trypsinized, washed with serum-free medium, counted, and seeded at 40,000 cells/well in a 96-well plate. Cells were allowed to adhere, and then after 2 h of serum starvation, PI3K inhibitor was added to the medium at the required concentration before 1-h FVIIa or PAR2AP treatment with LiCl serving as a positive control. After 6 h of treatment, cells were analyzed for luciferase activity using the Steady-Glo luciferase assay system (Promega) according to the instruction manual.
Migration assay
Equal numbers of MDA-MB-231 cells were seeded onto 35-mm dishes and allowed to reach 60–70% confluence. Cells were transfected with β-catenin or PAR2 siRNA (100 nm) using Lipofectamine 2000 and incubated for 48 h. On the day of the experiment, single scratch lines of equal width were made using a microtip. Both knocked down and untreated cells were serum-starved for 2 h followed by treatment with FVIIa or PAR2AP. LY294002 (PI3K pathway inhibitor) was added 1 h prior to ligand treatment. After 36 h, images were taken, and the number of cells migrated to the scratched area was estimated. The migratory potential of the cells was evaluated by comparing with 0-h treatment.
Invasion assay
MDA-MB-231 cells were grown in a T-25 flask, and β-catenin knockdown was performed. Equal numbers of control and knocked down cells were seeded on the Matrigel (Sigma)-coated upper compartment of a Transwell chamber in serum-free medium. FVIIa or PAR2AP treatment was performed, and LY294001 was added 1 h before ligand addition. FBS-containing DMEM was placed in the lower compartment, and further incubation was carried out for 24 h. Cells that extravasated to the lower surface of the membrane were stained with crystal violet solution after scraping off the upper surface cells. The total number of invaded cells was counted in 10 randomly selected fields under a bright-field microscope, and a comparative graphical analysis was done using GraphPad Prism 5 after repeating the experiments thrice.
Gene knockdown of β-catenin, PAR2, and AKT1 by siRNA
Transfection of siRNAs against human β-catenin, PAR2, and AKT1 and non-target siRNAs into cells was performed at a concentration of 100 nm using Lipofectamine 2000 according to the manufacturer's guidelines. Their respective sequences are as follows: β-catenin siRNA, 5′-GUUAUGGUCCAUCAGCUUU-3′; PAR2 siRNA1, 5′-CUUUGUAUGUCGUGA AGCA-3′; PAR2 siRNA2, 5′-AGUCGUGAAUCUUGUUCAATT-3′; PAR2 scrambled siRNA, 5′-GGACUCUUUAUGGUACGUUUAGAUU-3′; AKT1 siRNA1, 5′-CAGCCC UGAAGUACUCUUU-3′; AKT1 siRNA2, 5′-GGACGGGCACATTAAGATCTT-3′; scrambled siRNA, 5′-AGGUAGUGUAAUCGCCUUGUUTT-3′.
Reverse transcription and quantitative PCR
Extraction of total cellular RNA was performed using TRIzol reagent (Life Technologies). The extracted RNA was used for reverse transcription with the SuperScript III First-Strand cDNA Synthesis System (Invitrogen) according to the manufacturer's protocol. The cDNA was used for quantitative RT-PCR with SYBR Green Master Mix (Applied Biosystems) on the Step One plus Real-Time PCR System (Applied Biosystems). All values were normalized with GAPDH. Primer sequences for real-time PCR are as follows: cyclin D1: forward primer, 5′-ACAAACAGATCATCCGCAAACAC-3′; reverse primer, 5′-TGTTGGGGCTCCTCAGGTTC-3′; c-Myc: forward primer, 5′-AAACACAAACTTGAACAGCTAC-3′; reverse primer, 5′-ATTTGAGGCAGTTTACATTATGG-3′; COX-2: forward primer, 5′-TCAAATGAGATTGTGGAAAAAT-3′; reverse primer, 5′-AGATCATCTCTGCCTGAGTATCTT-3′; MMP-7: forward primer, 5′-TGAGCTACAGTGGGAACAGG-3′; reverse primer, 5′-TCATCGAAGTGAGCATCTCC-3′; MMP-14: forward primer, 5′-TTGGACTGTCAGGAATGAGG-3′; reverse primer, 5′-GCAGCACAAAATTCTCCGTG-3′; Claudin-1: forward primer, 5′-TCTACGAGGGACTGTGGATG-3′; reverse primer, 5′-TCAGATTCAGCTAGGAGTCG-3′; GAPDH: forward primer, 5′-AACGGGAAGCCCATCACC-3′; reverse primer, 5′-CAGCCTTGGCAGCACCAG-3′.
Immunohistochemistry
Breast tumor (n = 10) and normal breast (n = 10) tissues were obtained from Netaji Subhash Chandra Bose Cancer Research Institute, Kolkata, India, following proper norms of ethical human research. Working protocols with tissue samples were also approved by the human ethics committee of Netaji Subhash Chandra Bose Cancer Research Institute (registration number ECR/286/INST/WB/2013), and all the samples were collected obeying the human ethics committee rules. Both cancer and normal tissues were deparaffinized in xylene, dehydrated in various concentrations of ethanol, and again rehydrated. The sections were incubated in wash buffer (0.05 m TBS with 0.01% Tween 20) for 5 min. Heat-induced epitope retrieval was performed in 10 mm citrate buffer (pH 6.0) in a microwave for two sessions of 5 min. After blocking with 3% H2O2 for 10–12 min, the treated tissue slides were rinsed gently with wash buffer and then incubated with β-catenin primary antibody at 4 °C overnight at 1:100 dilutions. Tissue slides were washed with TBS and subsequently incubated with HRP-conjugated secondary antibody. Again after subsequent washes, the slides were treated with a diaminobenzidine and H2O2 mixture followed by counterstaining with hematoxylin. Separate slides for hematoxylin and eosin staining were also prepared.
Statistical analysis
The images of Western blots and other techniques are representative of at least three independent experiments. The error bars of data represent ±S.E of the mean. Differences are considered to be statistically significant at p < 0.05 using Student's t test.
Author contributions
A. R. and P. S. designed the project. A. R. and S. A. A. performed the research work and analyzed data. A. B. cloned TF and PAR2 constructs. K. D., R. P., and S. M. assisted with the experiments. A. M. provided tissue sample for research. A. R. and P. S. wrote the manuscript.
Acknowledgments
We thank the instrument facilities provided by the Department of Biological Chemistry, Indian Association for the Cultivation of Science, Kolkata, India. We pay our gratitude to Dr. Sanghamitra Raha of Saha Institute of Nuclear Physics, Kolkata, India, for providing different constructs including TCF/LEF luciferase constructs, many antibodies, and the reagents. We are thankful to the staff of the Netaji Subhash Cancer Research Institute, Kolkata, India, for humble cooperation and effort in providing breast cancer tissue samples for the study.
This work was supported by Department of Science and Technology, Government of India, Project Grant SR/SO/BB-0125/2012 and a startup grant from the Indian Association for the Cultivation of Science, Kolkata, India. The authors declare that they have no conflicts of interest with the contents of this article.
- FVIIa
- factor VIIa
- PAR
- protease-activated receptor
- PAR2AP
- protease-activated receptor 2 agonist peptide
- TF
- tissue factor
- APC
- adenomatous polyposis coli
- GSK3β
- glycogen synthase kinase-3β
- TCF
- T cell factor
- LEF
- lymphoid enhancer factor
- p-
- phospho-.
References
- 1. Versteeg H. H., Peppelenbosch M. P., and Spek C. A. (2001) The pleiotropic effects of tissue factor: a possible role for factor VIIa-induced intracellular signalling? Thromb. Haemost. 86, 1353–1359 [PubMed] [Google Scholar]
- 2. Sabharwal A. K., Birktoft J. J., Gorka J., Wildgoose P., Petersen L. C., and Bajaj S. P. (1995) High affinity Ca2+-binding site in the serine protease domain of human factor VIIa and its role in tissue factor binding and development of catalytic activity. J. Biol. Chem. 270, 15523–15530 [DOI] [PubMed] [Google Scholar]
- 3. Mann K. G. (1999) Biochemistry and physiology of blood coagulation. Thromb. Haemost. 82, 165–174 [PubMed] [Google Scholar]
- 4. Prasad R., and Sen P. (2017) Structural modulation of factor VIIa by full-length tissue factor (TF1–263): implication of novel interactions between EGF2 domain and TF. J. Biomol. Struct. Dyn. 17, 1–13 [DOI] [PubMed] [Google Scholar]
- 5. Cole M., and Bromberg M. (2013) Tissue factor as a novel target for treatment of breast cancer. Oncologist 18, 14–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carmeliet P., Mackman N., Moons L., Luther T., Gressens P., Van Vlaenderen I., Demunck H., Kasper M., Breier G., Evrard P., Müller M., Risau W., Edgington T., and Collen D. (1996) Role of tissue factor in embryonic blood vessel development. Nature 383, 73–75 [DOI] [PubMed] [Google Scholar]
- 7. Toomey J. R., Kratzer K. E., Lasky N. M., Stanton J. J., and Broze G. J. Jr. (1996) Targeted disruption of the murine tissue factor gene results in embryonic lethality. Blood 88, 1583–1587 [PubMed] [Google Scholar]
- 8. Bugge T. H., Xiao Q., Kombrinck K. W., Flick M. J., Holmbäck K., Danton M. J., Colbert M. C., Witte D. P., Fujikawa K., Davie E. W., and Degen J. L. (1996) Fatal embryonic bleeding events in mice lacking tissue factor, the cell-associated initiator of blood coagulation. Proc. Natl. Acad. Sci. U.S.A. 93, 6258–6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosen E. D., Chan J. C., Idusogie E., Clotman F., Vlasuk G., Luther T., Jalbert L. R., Albrecht S., Zhong L., Lissens A., Schoonjans L., Moons L., Collen D., Castellino F. J., and Carmeliet P. (1997) Mice lacking factor VII develop normally but suffer fatal perinatal bleeding. Nature. 390, 290–294 [DOI] [PubMed] [Google Scholar]
- 10. Ruf W., and Mueller B. (2006) Thrombin generation and the pathogenesis of cancer. Semin. Thromb. Hemost. 32, 61–68 [DOI] [PubMed] [Google Scholar]
- 11. Morris D. R., Ding Y., Ricks T. K., Gullapalli A., Wolfe B. L., and Trejo J. (2006) Protease-activated receptor-2 is essential for factor VIIa and Xa-induced signaling, migration, and invasion of breast cancer cells. Cancer Res. 66, 307–314 [DOI] [PubMed] [Google Scholar]
- 12. Schaffner F., and Ruf W. (2008) Tissue factor and protease-activated receptor signaling in cancer. Semin. Thromb. Hemost. 34, 147–153 [DOI] [PubMed] [Google Scholar]
- 13. Versteeg H. H., Borensztajn K. S., Kerver M. E., Ruf W., Reitsma P. H., Spek C. A., and Peppelenbosch M. P. (2008) TF:FVIIa-specific activation of CREB upregulates proapoptotic proteins via protease-activated receptor-2. J. Thromb. Haemost. 6, 1550–1557 [DOI] [PubMed] [Google Scholar]
- 14. Versteeg H. H., Schaffner F., Kerver M., Petersen H. H., Ahamed J., Felding-Habermann B., Takada Y., Mueller B. M., and Ruf W. (2008) Inhibition of tissue factor signaling suppresses tumor growth. Blood 111, 190–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Versteeg H. H., Schaffner F., Kerver M., Ellies L. G., Andrade-Gordon P., Mueller B. M., and Ruf W. (2008) Protease-activated receptor (PAR) 2, but not PAR1, signaling promotes the development of mammary adenocarcinoma in polyoma middle T mice. Cancer Res. 68, 7219–7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hjortoe G. M., Petersen L. C., Albrektsen T., Sorensen B. B., Norby P. L., Mandal S. K., Pendurthi U. R., and Rao L. V. (2004) Tissue factor-factor VIIa-specific up-regulation of IL-8 expression in MDA-MB-231 cells is mediated by PAR-2 and results in increased cell migration. Blood 103, 3029–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bluff J. E., Brown N. J., Reed M. W., and Staton C. A. (2008) Tissue factor, angiogenesis and tumour progression. Breast Cancer Res. 10, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stavik B., Skretting G., Aasheim H.-C., Tinholt M., Zernichow L., Sletten M., Sandset P. M., and Iversen N. (2011) Downregulation of TFPI in breast cancer cells induces tyrosine phosphorylation signaling and increases metastatic growth by stimulating cell motility. BMC Cancer 11, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang H., and He X. (2008) Wnt/β-catenin signaling: new (and old) players and new insights. Curr. Opin. Cell Biol. 20, 119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clevers H., and Nusse R. (2012) Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 [DOI] [PubMed] [Google Scholar]
- 21. Rao T. P., and Kühl M. (2010) An updated overview on Wnt signaling pathways: a prelude for more. Circ. Res. 106, 1798–1806 [DOI] [PubMed] [Google Scholar]
- 22. Anastas J. N., and Moon R. T. (2013) WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 13, 11–26 [DOI] [PubMed] [Google Scholar]
- 23. Brennan K. R., and Brown A. M. (2004) Wnt proteins in mammary development and cancer. J. Mammary Gland Biol. Neoplasia. 9, 119–131 [DOI] [PubMed] [Google Scholar]
- 24. Lin S. Y., Xia W., Wang J. C., Kwong K. Y., Spohn B., Wen Y., Pestell R. G., and Hung M. C. (2000) β-Catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. U.S.A. 97, 4262–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakopoulou L., Mylona E., Papadaki I., Kavantzas N., Giannopoulou I., Markaki S., and Keramopoulos A. (2006) Study of phospho-β-catenin subcellular distribution in invasive breast carcinomas in relation to their phenotype and the clinical outcome. Mod. Pathol. 19, 556–563 [DOI] [PubMed] [Google Scholar]
- 26. Candidus S., Bischoff P., Becker K. F., and Höfler H. (1996) No evidence for mutations in the α- and β-catenin genes in human gastric and breast carcinomas. Cancer Res. 56, 49–52 [PubMed] [Google Scholar]
- 27. Camerer E., Huang W., and Coughlin S. R. (2000) Tissue factor- and factor X-dependent activation of protease-activated receptor 2 by factor VIIa. Proc. Natl. Acad. Sci. U.S.A. 97, 5255–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jope R. S., and Johnson G. V. (2004) The glamour and gloom of glycogen synthase kinase-3. Trends Biochem. Sci. 29, 95–102 [DOI] [PubMed] [Google Scholar]
- 29. Moon R. T., Bowerman B., Boutros M., and Perrimon N. (2002) The promise and perils of Wnt signaling through β-catenin. Science 296, 1644–1646 [DOI] [PubMed] [Google Scholar]
- 30. Kimelman D., and Xu W. (2006) β-Catenin destruction complex: insights and questions from a structural perspective. Oncogene 25, 7482–7491 [DOI] [PubMed] [Google Scholar]
- 31. Åberg M., and Siegbahn A. (2013) Tissue factor non-coagulant signaling—molecular mechanisms and biological consequences with a focus on cell migration and apoptosis. J. Thromb. Haemost. 11, 817–825 [DOI] [PubMed] [Google Scholar]
- 32. Micalizzi D. S., Farabaugh S. M., and Ford H. L. (2010) Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J. Mammary Gland Biol. Neoplasia. 15, 117–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Versteeg H. H., Spek C. A., Peppelenbosch M. P., and Richel D. J. (2004) Tissue factor and cancer metastasis: the role of intracellular and extracellular signaling pathways. Mol. Med. 10, 6–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Steinhoff M., Buddenkotte J., Shpacovitch V., Rattenholl A., Moormann C., Vergnolle N., and Luger T. A., Hollenberg M. D. (2005) Proteinase-activated receptors: transducers of proteinase-mediated signaling in inflammation and immune response. Endocr. Rev. 26, 1–43 [DOI] [PubMed] [Google Scholar]
- 35. Kocatürk B., and Versteeg H. H. (2013) Tissue factor-integrin interactions in cancer and thrombosis: every Jack has his Jill. J. Thromb. Haemost. 11, Suppl. 1, 285–293 [DOI] [PubMed] [Google Scholar]
- 36. Lima L. G., and Monteiro R. Q. (2013) Activation of blood coagulation in cancer: implications for tumour progression. Biosci. Rep. 33, e00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Degen J. L., and Palumbo J. S. (2012) Hemostatic factors, innate immunity and malignancy. Thromb. Res. 129, Suppl. 1, S1–S5 [DOI] [PubMed] [Google Scholar]
- 38. Sallah S., Wan J. Y., Nguyen N. P., Hanrahan L. R., and Sigounas G. (2001) Disseminated intravascular coagulation in solid tumors: clinical and pathologic study. Thromb. Haemost. 86, 828–833 [PubMed] [Google Scholar]
- 39. van den Berg Y. W., Osanto S., Reitsma P. H., and Versteeg H. H. (2012) The relationship between tissue factor and cancer progression: insights from bench and bedside. Blood 119, 924–932 [DOI] [PubMed] [Google Scholar]
- 40. Su S., Li Y., Luo Y., Sheng Y., Su Y., Padia R. N., Pan Z. K., Dong Z., and Huang S. (2009) Proteinase-activated receptor 2 expression in breast cancer and its role in breast cancer cell migration. Oncogene 28, 3047–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sen P., Clark C. A., Gopalakrishnan R., Hedner U., Esmon C. T., Pendurthi U. R., and Rao L. V. (2012) Factor VIIa binding to endothelial cell protein C receptor: differences between mouse and human systems. Thromb. Haemost. 107, 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nayak R. C., Sen P., Ghosh S., Gopalakrishnan R., Esmon C. T., Pendurthi U. R., and Rao L. V. (2009) Endothelial cell protein C receptor cellular localization and trafficking: potential functional implications. Blood 114, 1974–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cadigan K. M., and Liu Y. I. (2006) Wnt signaling: complexity at the surface. J. Cell Sci. 119, 395–402 [DOI] [PubMed] [Google Scholar]
- 44. Yan D., Avtanski D., Saxena N. K., and Sharma D. (2012) Leptin-induced epithelial-mesenchymal transition in breast cancer cells requires β-catenin activation via Akt/GSK3- and MTA1/Wnt1 protein-dependent pathways. J. Biol. Chem. 287, 8598–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tian X., Liu Z., Niu B., Zhang J., Tan T. K., Lee S. R., Zhao Y., Harris D. C., and Zheng G. (2011) E-cadherin/β-catenin complex and the epithelial barrier. J. Biomed. Biotechnol. 2011, 567305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang K., Hitomi M., and Stacey D. W. (2006) Variations in cyclin D1 levels through the cell cycle determine the proliferative fate of a cell. Cell Div. 1, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu Q., Geng Y., and Sicinski P. (2001) Specific protection against breast cancers by cyclin D1 ablation. Nature 411, 1017–1021 [DOI] [PubMed] [Google Scholar]
- 48. Fredersdorf S., Burns J., Milne A. M., Packham G., Fallis L., Gillett C. E., Royds J. A., Peston D., Hall P. A., Hanby A. M., Barnes D. M., Shousha S., O'Hare M. J., and Lu X. (1997) High level expression of p27(kip1) and cyclin D1 in some human breast cancer cells: inverse correlation between the expression of p27(kip1) and degree of malignancy in human breast and colorectal cancers. Proc. Natl. Acad. Sci. U.S.A. 94, 6380–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]