Abstract
Glycolysis and the pentose phosphate pathway both play a central role in the degradation of glucose in all domains of life. Another metabolic route that can facilitate glucose breakdown is the gluconate shunt. In this shunt glucose dehydrogenase and gluconate kinase catalyze the two-step conversion of glucose into the pentose phosphate pathway intermediate 6-phosphogluconate. Despite the presence of these enzymes in many organisms, their only established role is in the production of 6-phosphogluconate for the Entner-Doudoroff pathway. In this report we performed metabolic profiling on a strain of Schizosaccharomyces pombe lacking the zinc-responsive transcriptional repressor Loz1 with the goal of identifying metabolic pathways that were altered by cellular zinc status. This profiling revealed that loz1Δ cells accumulate higher levels of gluconate. We show that the altered gluconate levels in loz1Δ cells result from increased expression of gcd1. By analyzing the activity of recombinant Gcd1 in vitro and by measuring gluconate levels in strains lacking enzymes of the gluconate shunt we demonstrate that Gcd1 encodes a novel NADP+-dependent glucose dehydrogenase that acts in a pathway with the Idn1 gluconate kinase. We also find that cells lacking gcd1 and zwf1, which encode the first enzyme in the pentose phosphate pathway, have a more severe growth phenotype than cells lacking zwf1. We propose that in S. pombe Gcd1 and Idn1 act together to shunt glucose into the pentose phosphate pathway, creating an alternative route for directing glucose into the pentose phosphate pathway that bypasses hexokinase and the rate-limiting enzyme glucose-6-phosphate dehydrogenase.
Keywords: gene expression, glucose metabolism, pentose phosphate pathway (PPP), secondary metabolism, zinc, NADPH regeneration, gluconate, glucose degradation
Introduction
The Embden-Meyerhof-Parnas pathway (glycolysis) and the pentose phosphate pathway play a central role in the degradation of glucose in organisms from all domains of life. In glycolysis, glucose is broken down to pyruvate to provide energy in the form of ATP, metabolic intermediates, and reduced nicotinamide adenine dinucleotide. In the pentose phosphate pathway, glucose is broken down to provide reducing equivalents in the form of NADPH and pentose sugars that are biosynthetic precursors of nucleic acids and amino acids (1–3). As the processes of glycolysis and the pentose phosphate pathway run in parallel, all cells have mechanisms to tightly regulate the flow of glucose into each pathway. On entering cells, the majority of glucose is phosphorylated to glucose 6-phosphate (Glu-6-P) in the first step of glycolysis. The intermediate Glu-6-P can then be further broken down in the process of glycolysis, or shunted into the pentose phosphate pathway. In the first committed step of the pentose phosphate pathway, Glu-6-P dehydrogenase catalyzes the dehydrogenation of Glu-6-P. This irreversible, rate-limiting step of the pentose phosphate pathway is typically highly regulated within cells, and therefore holds a prominent position in determining the overall flow of glucose into the pentose phosphate pathway (1, 2).
The gluconate shunt is a less studied metabolic route that can facilitate the breakdown of glucose. In the gluconate shunt, glucose is oxidized by glucose dehydrogenase to gluconate, which is then phosphorylated by gluconate kinase to produce 6-phosphogluconate (4, 5). As 6-phosphogluconate is the second intermediate of the pentose phosphate pathway, the gluconate shunt potentially creates a route for directing glucose into the pentose phosphate pathway that bypasses the rate-limiting enzyme Glu-6-P dehydrogenase. However, the only established role for the gluconate shunt is found in plants, algae, cyanobacteria, and some bacteria, which all use the Entner-Doudoroff (ED)2 pathway to degrade glucose or gluconate (4, 6, 7). In the ED pathway 6-phosphogluconate is dehydrated to generate 2-keto-3-deoxygluconate-6-phosphate, which is then cleaved to generate pyruvate and glyceraldehyde 3-phosphate. The gluconate shunt is therefore a metabolic route that can be used to direct glucose and gluconate to the ED pathway (6, 8). Despite this established role for the gluconate pathway, glucose dehydrogenase and gluconate kinase activities have been detected in mammals, fission yeast, and flies, which all lack the key ED pathway enzymes (5, 9–13).
In this report we used metabolomics profiling to identify metabolites that accumulate in a fission yeast strain lacking the transcriptional repressor Loz1. In Schizosaccharomyces pombe Loz1 plays a central role in zinc homeostasis by regulating the expression of genes required for zinc uptake and zinc storage (14, 15). Loz1 also regulates the availability of zinc in cells by controlling the levels of the non-essential, abundant zinc-binding enzyme alcohol dehydrogenase 1 (Adh1). Specifically, under zinc-limiting conditions, inactivation of Loz13 results in the derepression of an adh1 antisense transcript, and the strong antisense transcription in turn inhibits the expression of adh1 (16, 17). Although the regulation of adh1 gene expression has been well characterized at a transcriptional level, it is largely unclear if the lowered levels of Adh1 in zinc-deficient cells also affects cellular metabolism. As Loz1 regulates the expression of adh1, and potentially other abundant zinc-binding proteins, the goal of this study was to determine whether the changes in transcription that are mediated by Loz1 also affect cell metabolism. By using metabolomic analyses to screen for metabolites whose levels were altered in fission yeast mutants with impaired Loz1 function, we found that the metabolite that showed the highest fold-increase in loz1Δ cells was d-gluconate.
Here we show that the higher levels of d-gluconate in loz1Δ cells results from increased expression of gcd1, a gene encoding a novel NADP+-dependent glucose dehydrogenase. We also find that the function of gcd1 overlaps with zwf1, the gene encoding Glu-6-P dehydrogenase. We propose that in fission yeast the gluconate shunt creates an alternative route for directing glucose into the pentose phosphate pathway that bypasses the rate-limiting enzyme Glu-6-P dehydrogenase.
Results
loz1Δ cells have increased levels of gluconate
Our previous studies revealed that cells lacking the transcriptional repressor Loz1, or cells expressing the hypomorphic allele loz1-1, have impaired zinc homeostasis and reduced levels of the enzyme Adh1 (14). To determine whether impaired Loz1 activity affected cell metabolism, wild-type, loz1Δ, and loz1-1 cells were grown to exponential phase in the nutrient-rich YES medium. Cells were then harvested and metabolites identified using both GC-MS and LC-MS. 314 unique metabolites were detected. 11 of these accumulated >2-fold in both loz1-1 and loz1Δ cells relative to the wild-type control, including a variety of lipids and hydrolyzed phospholipids, the amino acid ergothioneine, the organic acids citrate, cis-aconitate, and gluconate, and the alcohol 2,3-butanediol (Table 1). Of these metabolites, gluconate, a naturally occurring derivative of glucose, showed the highest fold-increase in loz1Δ. As little is known about the biological function of gluconate in eukaryotes, we chose to further investigate how and why gluconate levels were regulated by Loz1.
Table 1.
Metabolites that accumulate to significantly higher levels (p ≤ 0.05) in loz1 mutants
Results show the average fold-increase from 5 independent cultures.
| Biochemical name | Fold increase |
|
|---|---|---|
| loz1–1 /WT | loz1Δ/WT | |
| Gluconate | 3.49 | 4.24 |
| Mevalonate | 2.71 | 3.53 |
| Phytosphingosine | 3.77 | 3.34 |
| 2-Palmitoleoylglycerophosphoinositol | 3.19 | 3.07 |
| Ergothioneine | 2.09 | 2.87 |
| Citrate | 2.69 | 2.85 |
| 2,3-Butanediol | 2.54 | 2.81 |
| 1-Stearoylglycerophosphocholine (18:0) | 2.24 | 2.79 |
| cis-Aconitate | 2.40 | 2.22 |
| 2-Myristoylglycerophosphocholine | 2.41 | 2.05 |
| 1-Eicosenoylglycerophosphocholine (20:1n9) | 2.89 | 2.03 |
The Idn1 gluconate kinase is required for the breakdown of gluconate
Studies of glucose metabolism in cell-free extracts of S. pombe have demonstrated the presence of two enzymes involved in gluconate metabolism, an NADP+-dependent glucose dehydrogenase, which catalyzes the oxidation of d-glucose to d-gluconate, and gluconate kinase, which phosphorylates d-gluconate to produce 6-phosphogluconate (Fig. 1) (5, 18). A single hexose transporter named Ght3 has also been identified that facilitates uptake of gluconate when glucose is limiting (19). As the growth medium used for the metabolic profiling contained 3% glucose, which results in the strong repression of the Ght3 gluconate uptake system (supplemental Fig. S1), we predicted that the increased levels of gluconate in loz1Δ cells were likely to be a result of altered expression of one or both of the enzymes involved in gluconate metabolism.
Figure 1.
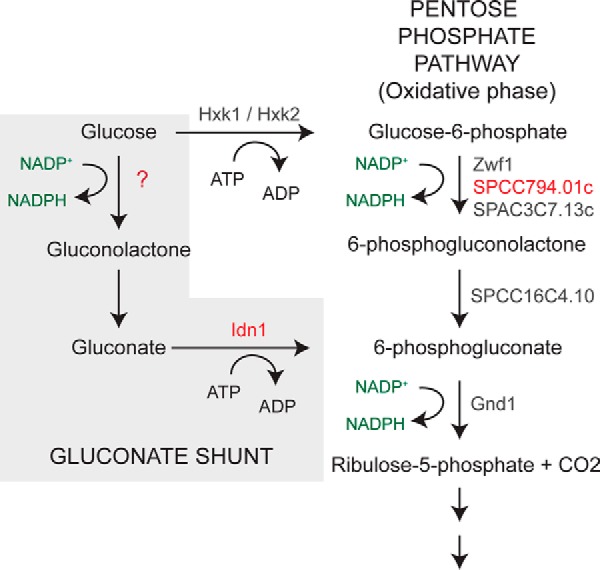
The gluconate shunt and oxidative phase of the pentose phosphate pathway in fission yeast. The names of enzymes predicted to catalyze reactions are shown in gray and red.
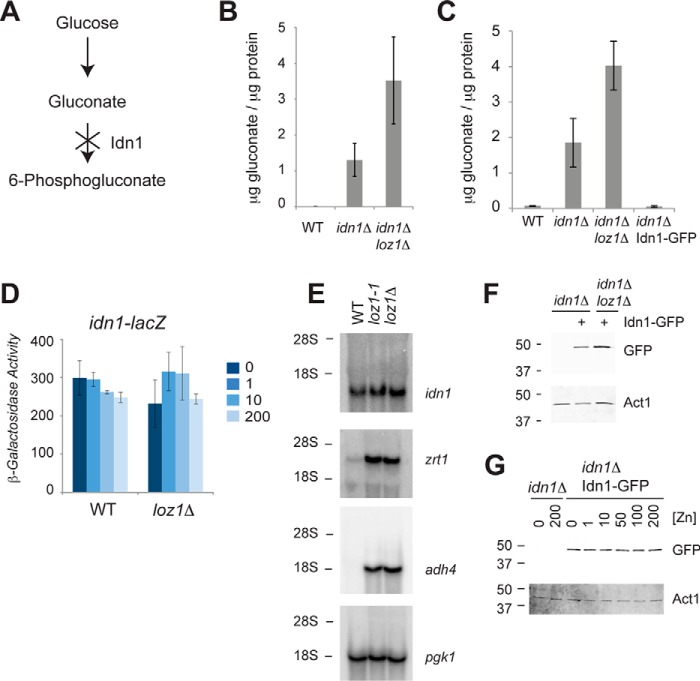
The simplest explanation for the increase in gluconate levels in the loz1 mutants was that the expression or activity of the gluconate kinase was reduced in these strains (Fig. 2A). In S. pombe a single gene named idn1 encodes a protein with high sequence similarity to characterized gluconate kinases (supplemental Fig. S2). To test whether Idn1 was required for the breakdown of gluconate levels in vivo, wild-type and idn1Δ cells were grown to exponential phase in YES medium and gluconate levels were measured by LC-MS/MS. As shown in Fig. 2B, idn1Δ cells accumulated ∼300-fold higher levels of gluconate relative to the wild-type control, consistent with Idn1 catalyzing the phosphorylation of gluconate to 6-phosphogluconate. As a complementary approach to examine Idn1 activity, we employed an enzymatic assay to measure gluconate levels in cell extracts. In these assays, intracellular gluconate levels are coupled to the consumption of 6-phosphogluconate (see “Experimental procedures”). When gluconate levels were measured by this method, idn1Δ cells accumulated ∼28-fold higher levels of gluconate relative to the wild-type control (Fig. 2C). This phenotype was also rescued by the introduction of a plasmid expressing an Idn1-GFP fusion protein from the constitutive pgk1 promoter (idn1Δ Idn1-GFP). These results are consistent with the predicted role of Idn1 in phosphorylating gluconate in vivo.
Figure 2.
idn1 encodes gluconate kinase in fission yeast. A, a schematic diagram to illustrate the effects of deletion of gluconate kinase on gluconate accumulation. B, LC-MS/MS and C, enzymatic assays were used to measure gluconate accumulation in wild-type, idn1Δ, and idn1Δ loz1Δ cells grown to exponential phase in YES medium. Data represents the average values from 5 independent cultures for the LC-MS/MS, and 3 independent experiments for enzymatic assays, with error bars representing standard deviations. Enzymatic assays were also used to confirm that an Idn1-GFP fusion was functional. D, β-galactosidase activity was measured in wild-type and loz1Δ cells expressing an idn1-lacZ reporter following growth overnight in ZL-EMM supplemented with 0, 1, 10, or 200 μm Zn2+. Data represents the average values from 3 independent experiments with error bars representing standard deviations. E, RNA blot analysis of total RNA purified from wild-type, loz1-1, and loz1Δ cells grown to exponential phase in YES medium. Blots were probed for idn1, the zinc-regulated transcripts adh4 and zrt1, and loading control pgk1. The positions of the 28 S and 18 S ribosomal RNAs are shown on the left. F, immunoblot analysis of crude protein extracts prepped from idn1Δ and idn1Δ Idn1-GFP, and idn1Δ loz1Δ Idn1-GFP cells grown to exponential phase in YES medium, or G, in ZL-EMM supplemented with the indicated amount of Zn2+. Immunoblots were probed for GFP and loading control Actin (Act1). The positions of the 50- and 37-kDa molecular mass markers are shown on the left.
As decreased expression of gluconate kinase would potentially lead to gluconate accumulating within cells, we next tested whether idn1 expression was altered in loz1Δ cells. To determine whether the transcription of idn1 was controlled by Loz1, a construct was generated in which ∼1200 bp of the idn1 promoter, extending from the open reading frame, was fused to the lacZ reporter gene. In wild-type and loz1Δ cells expressing this reporter, no significant changes in β-galactosidase activity were apparent following growth overnight in a zinc-limited minimal medium (ZL-EMM) supplemented with 0–200 μm zinc (Fig. 2D). The presence of a fully functional Loz1 also had little effect on idn1 mRNA levels, as similar levels of idn1 transcripts accumulated in wild-type, loz1-1, and loz1Δ cells grown to exponential phase in the zinc-rich YES medium (Fig. 2E). This was in contrast to the Loz1-regulated zrt1 and adh4 mRNA controls, which accumulated to higher levels in loz1 mutants grown in the zinc-replete YES medium. Idn1 protein levels were also not regulated by Loz1 or zinc as similar levels of the functional Idn1-GFP protein were detected in idn1Δ loz1Δ and idn1Δ cells (Fig. 2F), and in idn1Δ cells grown over a range of zinc levels (Fig. 2G). Although the above experiments do not eliminate a model in which Loz1 influences the activity of Idn1, the results indicate that the ability of loz1Δ and loz1-1 cells to accumulate gluconate is not a result of altered idn1 gene expression.
SPCC794.01c encodes an NADP+-dependent glucose dehydrogenase
An alternative explanation for the increased gluconate in the loz1 mutants is that the unknown glucose dehydrogenase is expressed at higher levels in the loz1-1 and loz1Δ cells. To gain insight into whether the first step of the gluconate shunt was regulated by Loz1, gluconate levels were compared in loz1Δ idn1Δ and idn1Δ cells. As shown in Fig. 2, B and C, ∼2-fold higher levels of gluconate accumulated in loz1Δ idn1Δ relative to idn1Δ. These results reveal that the increase in gluconate in loz1Δ cells is independent of Idn1, and are consistent with gluconate synthesis being higher in loz1Δ.
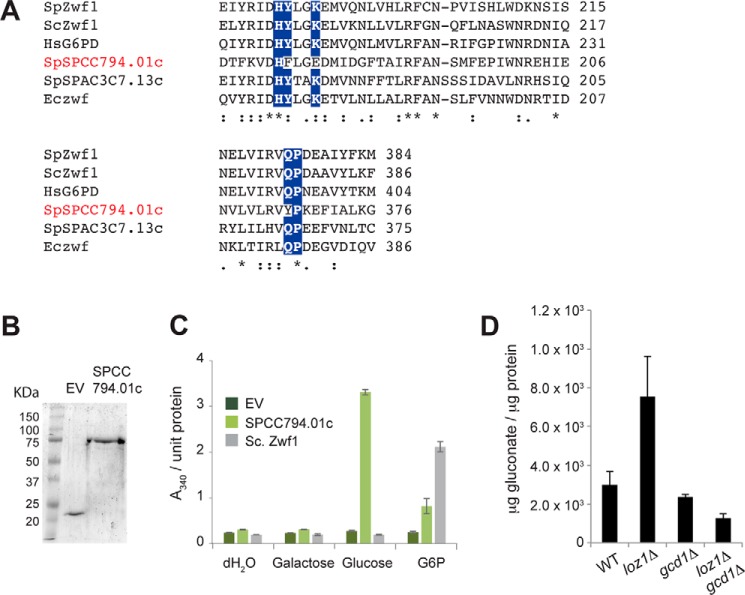
As Loz1 represses its target gene expression when zinc is in excess, we predicted that the gene encoding the unknown glucose dehydrogenase might be expressed at higher levels in zinc-deficient cells. When published microarray data were searched for putative NADP+-dependent oxidoreductases that were regulated by zinc, we noted that SPCC794.01c, a gene encoding a putative NADP+-dependent Glu-6-P dehydrogenase, was induced in response to zinc deficiency in multiple analyses (17, 20). In S. pombe, two additional genes (zwf1 and SPAC3C7.13c) are also predicted to encode NADP+-dependent Glc-6-P dehydrogenase, which catalyzes the first committed step of the pentose phosphate pathway (Fig. 1). Although all of the three putative Glc-6-P dehydrogenases from S. pombe have multiple conserved domains that are common to this family of proteins (supplemental Fig. S3), a notable exception was that the residues predicted to be involved in coordinating the phosphate moiety of Glc-6-P (21, 22) were not conserved in SPCC794.01c (Fig. 3A). This lack of conservation suggested that SPCC794.01c might differ in its substrate specificity relative to other Glc-6-P dehydrogenases.
Figure 3.
Gcd1 is a NADP+-dependent glucose dehydrogenase. A, an alignment of the Glc-6-P-binding domain of Glc-6-P dehydrogenase or putative G6P dehydrogenase family members from S. pombe (SpZwf1, SpSPCC794.01c, and SpSPAC3C7.13c), S. cerevisiae (ScZwf1), Homo sapiens (HsG6PD), and E. coli (Eczwf). Residues that are predicted to coordinate Glc-6-P are highlighted by white text. The alignment was performed using Clustal-Omega (EMBL-EBI). B, SDS-PAGE analysis of purified recombinant His-tagged SpSPCC794.01c, or the His tag (EV). Gels were stained with Coomassie Brilliant Blue. A protein ladder with sizes in kDa is shown on the left (C). Activity of 1 μg of purified His tag (EV), His-tagged SpSPCC794.01c, or 0.5 units of S. cerevisiae Zwf1 in the presence of NADP+ and the indicated substrate. Reactions were allowed to go to completion and enzyme activity was determined by measuring NADPH generation via absorbance at 340 nm (D). The indicated strains were grown to exponential phase in YES medium and gluconate levels were measured by LC-MS/MS. Data represents the average values from 5 independent cultures with error bars representing standard deviations.
To examine the substrate specificity of SPCC794.01c, His-tagged recombinant SPCC794.01c was purified from Escherichia coli (Fig. 3B). The activities of His-SPCC794.01c, the His tag alone (EV), were then assayed by measuring NADPH production in the presence of a given substrate (Fig. 3C). The activity of Glc-6-P dehydrogenase from Saccharomyces cerevisiae (Sc.Zwf1) was also measured as a control. As expected, incubation of the control enzyme (Sc.Zwf1) with each of the substrates led to an ∼8.5-fold increase in NADPH levels in the presence of Glc-6-P relative to the His tag control (EV). This activity was specific to Glc-6-P as no increase in NADPH was observed in the presence of glucose or galactose substrates. In contrast, incubation of SPCC794.01c in the presence of glucose resulted in an ∼12-fold increase in NADPH levels relative to the His tag control, and in the presence of Glc-6-P, a 3-fold increase in NADPH. No increase in NADPH was observed in the presence of a galactose substrate. Together these experiments reveal that SPCC794.01c has different substrate specificity than other Glc-6-P dehydrogenases and that it is able to use glucose as a substrate in vitro. As SPCC794.01c functions as a glucose dehydrogenase, it was named Gcd1 for Glucose dehydrogenase 1.
To test whether Gcd1 is necessary for the increased gluconate in loz1Δ cells in vivo, wild-type, loz1Δ, and loz1Δ gcd1Δ cells were grown to exponential phase in YES medium and gluconate levels were measured by LC-MS/MS (Fig. 3D). As expected, higher levels of gluconate accumulated in loz1Δ cells relative to the wild-type control. This increase was not apparent in loz1Δ gcd1Δ indicating that Gcd1 is required for the increased levels of gluconate in loz1Δ cells.
Gluconate accumulates in zinc-limited cells in a manner that is dependent upon Gcd1
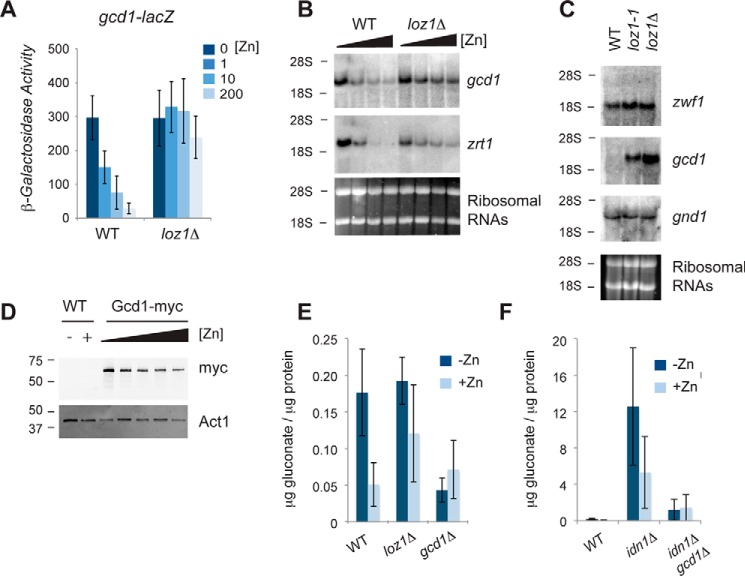
As Gcd1 was necessary for the Loz1-dependent increase in gluconate we next tested whether gcd1 was a Loz1 target gene. To determine whether the expression of gcd1 was dependent upon Loz1, a reporter construct containing ∼1450 bp of the gcd1 promoter fused to the lacZ gene was integrated into the genome of wild-type and loz1Δ cells. As Loz1 represses gene expression when zinc is in excess, β-galactosidase activity was examined following growth in ZL-EMM or ZL-EMM supplemented with 1–200 μm Zn2+. An ∼10-fold increase in β-galactosidase activity was observed in zinc-limited wild-type cells, whereas high levels of β-galactosidase activity were observed under all conditions in loz1Δ (Fig. 4A). Cellular gcd1 mRNA levels were also dependent upon zinc and Loz1 (Fig. 4, B and C). To examine the effects of zinc on Gcd1 protein levels, a strain was generated that expressed the endogenous Gcd1 protein fused to 13 myc epitope tags. As shown in Fig. 4D, there was an ∼2–3-fold increase in levels of Gcd1-Myc protein in cells grown overnight under zinc-limiting conditions relative to the levels of Gcd1-Myc that accumulated in zinc-replete cells. Thus, Gcd1 accumulates in both zinc-limited and zinc-replete cells, yet higher levels accumulate in zinc-starved cells consistent with the increased expression of gcd1 under this condition.
Figure 4.
gcd1 expression is regulated by zinc in a Loz1-dependent manner. A, β-galactosidase activity was measured in wild-type and loz1Δ cells expressing the gcd1-lacZ reporter following growth overnight in ZL-EMM or ZL-EMM supplemented with 0, 1, 10, or 200 μm Zn2+. Results show average values from 3 independent experiments. B, RNA blot analysis of total RNA purified from wild-type and loz1Δ cells grown overnight in ZL-EMM or ZL-EMM supplemented with 1, 10, or 100 μm Zn2+, or C, to exponential phase in YES medium. Blots were probed for the indicated transcripts. Ribosomal RNAs are shown as loading controls. D, immunoblot analysis of crude protein extracted prepped from wild-type cells and cells expressing Gcd1–13×Myc from its endogenous promoter (Gcd1-myc). Immunoblots were probed for c-Myc and loading control Actin (Act1). The positions of the 50- and 37-kDa molecular mass markers are shown on the left. E and F, the indicated strains were grown overnight in ZL-EMM or ZL-EMM supplemented with 200 μm Zn2+ and gluconate levels were measured using an enzymatic assay. Data represents the average values from 3 independent experiments with error bars representing standard deviations.
To determine whether increased expression of gcd1 in zinc-limited cells also resulted in gluconate accumulation, gluconate levels were measured in strains grown in ZL-EMM with or without a 200 μm zinc supplement. Zinc-limited wild-type cells accumulated ∼3-fold higher levels of gluconate relative to zinc-replete cells (Fig. 4E). This fold-change was reduced in loz1Δ mutants, whereas deletion of gcd1 resulted in lower levels of gluconate accumulating under zinc-limiting conditions. Although these results are consistent with the Loz1-dependent derepression of gcd1 leading to increased gluconate accumulation in zinc-limited cells, the levels of gluconate that accumulated were small relative to the large increase in gluconate seen in idn1Δ (Fig. 4F). The increases in gluconate in idn1Δ cells were largely dependent upon Gcd1 as lower levels of gluconate were detected in gcd1Δ idn1Δ cells compared with idn1Δ. Taken together these results are consistent with Gcd1 and Idn1 acting in the same pathway. They also suggest that most of the gluconate generated within cells is rapidly phosphorylated by Idn1.
The function of Gcd1 overlaps with Zwf1
Why would the expression of gcd1 be increased in response to zinc limitation? As Gcd1 is a NADP+-dependent enzyme and the phosphorylation of gluconate by Idn1 requires 1 ATP, there is no obvious energetic advantage of using the gluconate shunt to generate 6-phosphogluconate instead of hexokinase and Glc-6-P dehydrogenase (Fig. 1). However, as these reactions could run in parallel, a potential explanation for the increased expression of gcd1 is that it allows increased carbon flux into the pentose phosphate pathway to increase NADPH regeneration and/or the levels of needed biosynthetic intermediates. In support of this hypothesis, there are precedents for the regulation of NADPH levels by zinc in other yeast (23). loz1Δ cells also accumulate ergothioneine (Table 1), which is potentially generated from precursors supplied by the pentose phosphate pathway (24). To test whether the enzymes of the gluconate shunt influenced total cellular NADPH regeneration, NADP+ and NADPH levels were measured in wild-type and gcd1Δ cells following growth to exponential phase in YES medium. No significant changes in the NADP+/NADPH ratio were observed in gcd1Δ compared with wild-type cells (supplemental Fig. S4). We also observed no obvious growth defect of gcd1Δ cells in zinc-limiting medium, zinc-replete medium, or in the presence of strong oxidants such as H2O2 (Fig. 5 and data not shown).
Figure 5.
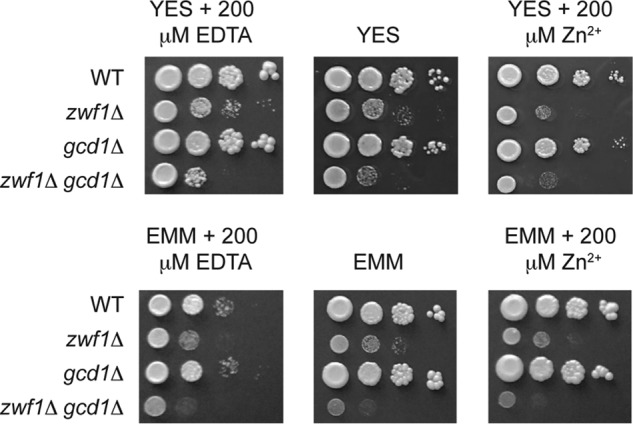
Gcd1 functions overlap with Zwf1. The indicated strains were grown overnight in the nutrient-rich YES medium, or minimal medium (EMM). Cells were diluted to an A600 of 1.0 and 10-fold serial dilutions of each strain plated onto YES or EMM medium, with or without 200 μm EDTA or Zn2+. Plates were incubated at 31 °C for 3–6 days before photography.
As we were unable to find any phenotype that was a result of loss of gcd1, we next tested whether there was redundancy between the gluconate shunt and the first steps of the pentose phosphate pathway. As two genes (zwf1 and SPAC3C7.13c) encode Glc-6-P dehydrogenase in S. pombe, we initially performed RNA blot analysis to examine their expression in exponentially growing cells. We also examined the expression of gnd1, which encodes the pentose phosphate enzyme phosphogluconate dehydrogenase, as a control. The RNA blot analysis revealed that zwf1 and gnd1 mRNAs were abundantly expressed in wild-type and loz1Δ cells (Fig. 4C). In contrast, we were unable to detect SPAC3C7.13c using this method. Due to the low expression of SPAC3C7.13c, our further experiments focused on zwf1. Genome-wide deletion studies in S. pombe suggest that zwf1Δ cells are inviable (25). Nevertheless, we were able to delete zwf1 in a diploid background and isolate viable, albeit slow growing, haploid zwf1Δ cells using tetrad dissection analysis. We were also able to isolate viable haploid strains lacking zwf1 and gcd1 from crosses of zwf1Δ to gcd1Δ. When the growth phenotypes of these mutants were compared, cells lacking gcd1 did not display any significant growth defect on YES medium, or in this medium supplemented with zinc or the zinc chelator EDTA (Fig. 5). In contrast, zwf1Δ cells had a strong growth defect under all conditions. Importantly, cells lacking both zwf1 and gcd1 exhibited a more severe growth defect than cells lacking only zwf1. These results suggest redundancy between Zwf1 and Gcd1, and that Gcd1 functions overlap with Zwf1.
Discussion
Enzymes involved in gluconate metabolism are found in many organisms, however, few studies have examined the biological role of these enzymes in vivo. Here we demonstrate that gcd1 encodes a novel NADP+-dependent glucose dehydrogenase that is required for gluconate synthesis, and that the Idn1 gluconate kinase is necessary for gluconate breakdown in vivo. As previous studies have shown that Idn1 specifically phosphorylates gluconate to produce 6-phosphogluconate (5, 18), and Gcd1 functions overlap with Zwf1, we propose that in fission yeast the gluconate shunt provides an alternative route for directing glucose into the pentose phosphate pathway that bypasses hexokinase and Glc-6-P dehydrogenase.
Studies of the gluconate shunt enzymes have so far mostly been limited to organisms that are able to metabolize 6-phosphogluconate by the ED pathway (6–8). Despite the established role for these enzymes, glucose dehydrogenase and gluconate kinase activities have been detected in mammals and S. pombe, which both lack the key enzymes of the ED pathway (5, 13, 26). An important factor that has previously limited studies of gluconate metabolism in these organisms in vivo is that the genetic identity of the glucose dehydrogenase(s) was unclear. The identification of Gcd1 has therefore provided a means of examining gluconate metabolism in organisms without a functional ED pathway.
Gcd1 differs from traditional Glc-6-P dehydrogenases in that it contains nonconsensus amino acids in its putative Glc-6-P binding pocket. In the well characterized Glc-6-P dehydrogenases from humans and Leuconostoc mesenteroides, lysine and tyrosine residues within the Glc-6-P binding pocket are critical for Glc-6-P binding and catalysis (21, 22, 27). In Gcd1 the equivalent tyrosine is replaced by a phenylalanine, whereas the lysine is replaced with a glutamate (Fig. 3B). As Glc-6-P is itself negatively charged, the incorporation of a negatively charged glutamate into the Glc-6-P binding pocket instead of a positively charged lysine could be an important feature of Gcd1 that results in it preferentially using glucose instead of Glc-6-P as a substrate. An enzyme with similarities to Gcd1 is found in flies. Drosophila contains two putative Glc-6-P dehydrogenases, Zw and CG7140. Zw contains consensus amino acids in the Glc-6-P binding pocket, whereas CG7140 contains amino acid substitutions that replace the conserved tyrosine and lysine residues (supplemental Fig. S3). Although it is currently unknown if CG7140 is able to use glucose as a substrate, the similarity of CG7140 with Gcd1 suggests that it also may have a broader substrate specificity. Other organisms may therefore express and utilize Gcd1-like NADP+-dependent glucose dehydrogenases.
Although homologs of Gcd1 appear to be limited to fission yeast and flies, glucose dehydrogenase and gluconate kinase activities have been detected in mammals suggesting that they also may use a related pathway (9, 12, 13, 26, 28). It is therefore interesting to note that cells lacking gcd1 and idn1 accumulate ∼15-fold higher levels of gluconate relative to the wild-type control (Fig. 4F). In addition, the molecular weight of the glucose dehydrogenase that was originally purified from S. pombe was 66.5 kDa (5) and the molecular mass of Gcd1 is 53.5 kDa. Together these observations suggest the presence of a Gcd1-independent mechanism for generating gluconate in S. pombe. Thus, future studies characterizing this alternative pathway for gluconate production in fission yeast may provide further information regarding gluconate metabolism in mammals.
A clue to the function of the gluconate pathway in fission yeast was revealed by our observations that gcd1 is regulated at a transcriptional level by Loz1 and zinc, and that cells lacking zwf1 and gcd1 have a more severe growth defect than cells lacking only zwf1. Loz1 plays a central role in zinc homeostasis by regulating the expression of genes required for zinc uptake and zinc storage (14). The Loz1-dependent derepression of gcd1 in zinc-limited cells therefore suggests that increased flux of glucose through the gluconate pathway is important for zinc homeostasis or cell survival during longer periods of zinc limitation. Studies of zinc homeostasis in S. cerevisiae have found that MET30 expression is increased in zinc-limited cells (23). Met30 is a component of the SCF(Met30) ubiquitin ligase that targets the Met4 transcription factor for degradation. As Met4 typically activates the expression of genes required for the NADPH expending process of sulfate assimilation, it is thought that the zinc-dependent regulation of MET30 and Met4 conserves NADPH for defense against the increased levels of oxidative stress that arise from zinc starvation (23). Because flux of glucose through the gluconate shunt into the pentose phosphate pathway would also yield 2 NADPH molecules per glucose, increased expression of gcd1 could be an alternative mechanism to protect zinc-deficient cells from the oxidative stress associated with this growth condition. However, we did not see any major changes in NADPH regeneration in cells lacking gcd1 and we observed increased levels of NADP+ and NADPH in zwf1Δ and zwf1Δ gcd1Δ cells (supplemental Fig. S4). One potential limitation with the above experiments is that other NADPH-producing pathways could be up-regulated and compensate for the loss of zwf1 and/or gcd1. In support of this hypothesis, in S. cerevisiae disruption of the ZWF1 gene results in a methionine auxotrophy, because high levels of NAPDH are required to sustain methionine biosynthesis (29, 30). However in S. pombe, neither zwf1Δ nor zwf1Δ gcd1Δ are methionine auxotrophs (data not shown). Thus, a better understanding of the net contributions of different routes of NADPH synthesis in S. pombe may shed additional light into the role of Zwf1 and Gcd1 in NADPH regeneration. Additional important experiments include the use of different labeled forms of glucose and gluconate to determine the extent to which glucose in cells is metabolized by the gluconate shunt and pentose phosphate pathway.
In addition to its role in NADPH regeneration, the pentose phosphate shunt is a major anabolic pathway that produces pentose sugars that are biosynthetic precursors of nucleic acids and amino acids. An alternative explanation for increased expression of gcd1 in zinc-limited cells is therefore that it generates needed biosynthetic precursors. In support of this hypothesis, higher levels of ergothioneine, a derivative of the amino acid histidine, were detected in loz1Δ and loz1-1 cells (Table 1). Although it is not yet known if the Loz1-dependent increase in ergothioneine levels are dependent upon the gluconate shunt, ergothioneine has antioxidant functions (24). Increased ergothioneine production in zinc-deficient cells may therefore be a different metabolite route to protect cells from oxidative stress.
Another metabolite that accumulated to higher levels in loz1Δ cells was 2,3-butandiol (Table 1). In S. cerevisiae, deletion of the alcohol dehydrogenase genes ADH1, ADH3, and ADH5 results in a large increase in the synthesize of 2,3-butandiol, potentially as a mechanism to reduce the buildup of the toxic metabolite acetaldehyde (31). Interesting future studies would therefore be to test whether the increase in 2,3-butandiol in loz1Δ cells is a mechanism to help these cells contend with the low expression of adh1 in this strain. Another phenotype of cells expressing the loz1Δ and loz1-1 alleles is that they hyperaccumulate zinc. Although most cells do not naturally hyperaccumulate zinc, an interesting exception is found in the prostate. Prostate epithelial cells are atypical in that they possess high levels of zinc in the cytosol and mitochondria to facilitate citrate secretion (32, 33). The high levels of zinc in the mitochondria inhibit the activity of the Fe-S requiring enzyme aconitase that oxidizes citrate to isocitrate, which in turn allows citrate to build up for secretion. It is therefore noteworthy that citrate and cis-aconitate accumulated in loz1Δ and loz1-1 cells. Thus, future studies to determine why the levels of metabolites such as citrate and ergothioneine are altered in loz1Δ cells may further our knowledge of zinc homeostasis in yeast and other organisms.
Experimental procedures
Strains, media, and plasmids
A full list of strains and genotypes can be found in supplemental Table S1. Strains were either grown to exponential phase at 31 °C in YES (yeast extract + supplements) medium, or in a derivative of Edinburgh minimal medium that lacks zinc (ZL-EMM) (17). Strain ABY1116 (gcd1-13MYC::kanMX6) was generated by homologous recombination using standard procedures. The Pet32a-Gcd1 fusion construct was generated by introducing gcd1 into the XhoI/SacI site of the vector Pet32a. The idn1-lacZ and gcd1-lacZ reporters were generated by amplifying ∼1 kb of the respective promoter regions with PCR primers that contained BamHI/EagI restriction sites. Each PCR fragment was then cloned into similar sites of the vector JK-lacZ (17). All vectors derived from plasmid JK148 were linearized with NruI before integration into the genome.
RNA blot and immunoblot analysis
Total RNA was extracted by using hot acidic phenol and RNA blots run using standard protocols. Probes for RNA blots were generated from purified PCR products with the MAXIscript T7 kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Crude protein extracts for immunoblot analysis were obtained using a trichloroacetic acid precipitation (34). Crude protein extracts were separated on 10% (w/v) SDS-PAGE gels, and immunoblot analysis was performed using standard procedures. Immunoblots were incubated with the primary antibodies anti-c-Myc (Sigma C3956), anti-GFP (Sigma G1544), and anti-Act1 (Abcam ab3280), and secondary antibodies IR-Dye800CW-conjugated anti-mouse IgG (LICOR) and IRDye680-conjugated anti-rabbit IgG (LICOR). Signal intensities were analyzed using the Odyssey Infrared image system (LICOR).
Recombinant protein purification and glucose dehydrogenase assays
BL21(DE3) pLysS cells containing Pet32a-gcd1 or the empty Pet32a vector were pregrown for 8 h in lysogeny broth + 15% glycerol at 31 °C. gcd1 expression was induced by 0.5 mm 1 mm isopropyl β-d-thiogalactopyranoside and cells were grown for a further 12–15 h at 31 °C before lysis by sonication. Cell lysates were collected by centrifugation, and His-tagged proteins were purified using nickel-nitrilotriacetic acid Super flow (Qiagen) columns pre-equilibrated with lysis buffer (phosphate-buffered saline (PBS) + 0.1% Tween + 10 mm imidazole). Following washing, proteins were eluted with containing PBS + 0.05%, Tween 20, and 250 mm imidazole. Protein fractions were stored in 50% (v/v) glycerol at −80 °C. The concentration and purity of proteins was determined using the Bradford assay and SDS-PAGE analysis, respectively. 1 μg of Gcd1, 1 μg of Pet32a HIS tag, or 0.5 units of ScZwf1 (Sigma G7877) were diluted to 50 μl in PBS + 0.1% Tween 20. The protein was added to a solution containing 1 μm MgCl2, 2.5 mm NADP (Calbiochem 481972), and 2.5 mm with one of the following sugars, glucose (Fisher Scientific D18), galactose (Fisher Scientific BP656), or Glc-6-P (Acros Organics 446980010). Reactions were allowed to go to completion and NADPH levels were measured via absorbance at 340 nm using a Synergy H1 plate reader.
Metabolomics and gluconate measurements
For metabolomic profiling, 50 ml of cells were grown to an A600 of 1.0 in YES medium. Cells were then washed in ddH2O and pellets were frozen at −80 °C. Further sample preparation, extraction of metabolites, ultra high performance LC-MS and GC-MS, and metabolite detection were performed by Metabolon®. For further LC-MS/MS analyses to measure gluconate levels cells were grown as described above. Cells were then washed in ddH2O and cell pellets were frozen at −80 °C. Frozen cell pellets were plunged into a boiling water bath. After the addition of 1 ml of 65 °C 50 μm [U-13C]fumarate, samples were boiled for a further 10 min, vortexed for 5 min in the presence of zirconia beads, boiled for a further 5 min, and then vortexed for a final 5 min. Cell extracts were centrifuged for 10 min, 4 °C, 14,000 × g, and the supernatant filtered through 0.2-μm cellulose acetate filters. Filtered supernatants were frozen at −80 °C and lyophilized until further analysis. Dry pellets were resuspended in 1 ml of ddH2O and filtered through a 3-kDa Amicon Ultra 0.5-ml filter. LC was performed with a ultra high-pressure liquid chromatography 1290 from Agilent Technologies, using an IonPac AS11 column (250 × 2 mm, Dionex) and a guard column AG11 (50 × 2 mm, Dionex) at a flow rate of 0.35 ml/min. A gradient of KOH was generated as previously described (35, 36). The MS/MS analysis was performed with a QTRAP 5500, from AB Sciex (Framingham, MA) in negative polarity using multiple reaction monitoring mode. The gluconate was monitored using the parent/daughter transition of 195.2/74.9 and the following parameters: −50 V declustering potential, −26 V collision energy, −45 V cell exit potential, and −10V entrance potential. Under our chromatographic conditions (anion exchange chromatography with basic eluent), the only form of gluconate (gluconic acid, gluconate, and gluconolactone) detected was gluconate. The level of gluconate was quantified using external standard curves of a commercially available gluconate standard, and by using [U-13C4]fumarate as an internal standard to account for any loss of sample during extraction, filtration etc. For gluconate enzymatic assays, cell pellets were resuspended in 1 ml of H2O. Cells suspensions were boiled for 5 min and lysed by bead beating for 5 min. Gluconate levels were then measured using the d-gluconate/d-glucono-δ-lactone assay kit (Megazyme) per the manufacturer's instructions. Briefly in these assays, residual 6-phosphogluconate was removed from boiled cell extracts by incubation with ATP, NADP+, and 6-phosphogluconate dehydrogenase. After the removal of 6-phosphogluconate, the basal level of NADPH was determined by measuring fluorescence excitation: 340 nm and emission 440 nm. IdnK was then added to generate 6-phosphogluconate from gluconate. As 6-phosphogluconate is a substrate for 6-phosphogluconate dehydrogenase further increases in NADPH levels are proportional to the levels of gluconate within cells. Final gluconate levels were determined by comparing the increase in NADPH levels to that obtained with known concentrations of gluconate. All samples were normalized to protein concentration.
β-Galactosidase assays
β-Galactosidase assays were performed as described previously, and activity units were calculated as follows: (ΔA420 × 1000)/(min × ml of culture × culture absorbance at 600 nm). Errors bars represent mean ± S.D. from three independent experiments.
NADP+/NADPH measurements
This procedure was carried out with a modified version of the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide and phenazine ethosulfate cycling assay described by (23, 37). 5-ml cultures were grown to an A600 ∼1.0 in YES medium. 2 × 1.5 ml of cells were then washed with ddH2O, and were resuspended in 250 μl of 0.1 m HCl or 0.1 m NaOH to measure NADP+ and NADPH levels, respectively. Cells were broken open and the endogenous proteins were denatured by freezing at −80 °C for 30 min, boiling for 3 min, and vortexing with glass beads for 5 min. Cell debris was removed by centrifugation and cycling reactions set up in 96-well plates. For each reaction 20 μl of supernatant was added to 100 μl of 0.1 m Tris, pH 7.4. Samples were mixed by low speed vortexing and the reaction initiated by the addition of 100 μl of detection buffer (0.1 m Tris, pH 7.4, 0.1% (v/v) Tween 20, 10 mm MgCl2, 10 mm Glc-6-P (Acros Organics 446980010), 1 mm 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma M5655), 0.2 mm phenazine ethosulfate (Sigma P4544), and 5 units of ScZwf1 (Sigma G7877)). Reactions were kept in the dark and color development was monitored spectrophotometrically at 595 nm. Reactions were stopped by the addition of 0.45 m NaCl and the concentrations of NADP+ and NADPH were determined based of a standard curve of known concentrations of NADPH. Final concentrations were normalized to the initial cell density of the culture.
Author contributions
M. E. C. conducted most of the experiments and analyses. J. C. C. and A. P. A. assisted with the LC-MS/MS analyses to directly measure gluconate. S. W. generated and conducted the experiments with the zwf1 and gcd1 mutants. A. J. B. conducted the RNA blot analyses and wrote the majority of the paper. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank members of the Bird lab and Dr. R. Michael Townsend for critical reading of the manuscript and Dr. Kurt W. Runge for assistance with strain generation.
This work was supported, in whole or in part, by NIGMS, National Institutes of Health Grant R01GM105695 (to A. J. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Table S1 and Figs. S1–S4.
The descriptive names of genes or proteins discussed in the text are as follows: loz1, loss of zinc sensing 1; S. pombe, adh4, alcohol dehydrogenase 4; ght3, homology to Glut- and Hxt-transporter 3; gnd1, 6-phosphogluconate dehydrogenase 1; idn1, l-IDoNate catabolism 1; pgk1, phosphoglycerate kinase 1; zrt1, zinc-regulated transporter 1; zwf1, ZWischenferment 1; S. cerevisiae, MET30, METhionine requiring 30; MET4, METhionine requiring 4; Drosophila melanogaster-Zw, ZWischenferment; H. sapiens Glu-6-PD, glucose-6-phosphate dehydrogenase 1.
- ED
- Entner-Doudoroff
- EV
- empty vector
- GC-MS
- gas chromatography-mass spectrometry
- LC-MS
- liquid chromatography-mass spectrometry
- YES
- yeast extract plus supplements
- Adh1
- alcohol dehydrogenase 1
- ZL-EMM
- zinc-limited minimal medium.
References
- 1. Stanton R. C. (2012) Glucose-6-phosphate dehydrogenase, NADPH, and cell survival. IUBMB Life 64, 362–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stincone A., Prigione A., Cramer T., Wamelink M. M., Campbell K., Cheung E., Olin-Sandoval V., Grüning N. M., Krüger A., Tauqeer Alam M., Keller M. A., Breitenbach M., Brindle K. M., Rabinowitz J. D., and Ralser M. (2015) The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. Camb. Philos. Soc. 90, 927–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lunt S. Y., and Vander Heiden M. G. (2011) Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27, 441–464 [DOI] [PubMed] [Google Scholar]
- 4. Peekhaus N., and Conway T. (1998) What's for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J. Bacteriol. 180, 3495–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tsai C. S., Ye H. G., and Shi J. L. (1995) Carbon-13 NMR studies and purification of gluconate pathway enzymes from Schizosaccharomyces pombe. Arch. Biochem. Biophys. 316, 155–162 [DOI] [PubMed] [Google Scholar]
- 6. Chen X., Schreiber K., Appel J., Makowka A., Fähnrich B., Roettger M., Hajirezaei M. R., Sönnichsen F. D., Schönheit P., Martin W. F., and Gutekunst K. (2016) The Entner-Doudoroff pathway is an overlooked glycolytic route in cyanobacteria and plants. Proc. Natl. Acad. Sci. U.S.A. 113, 5441–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Patra T., Koley H., Ramamurthy T., Ghose A. C., and Nandy R. K. (2012) The Entner-Doudoroff pathway is obligatory for gluconate utilization and contributes to the pathogenicity of Vibrio cholerae. J. Bacteriol. 194, 3377–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fliege R., Tong S., Shibata A., Nickerson K. W., and Conway T. (1992) The Entner-Doudoroff pathway in Escherichia coli is induced for oxidative glucose metabolism via pyrroloquinoline quinone-dependent glucose dehydrogenase. Appl. Environ. Microbiol. 58, 3826–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campbell D. P., Carper W. R., and Thompson R. E. (1982) Bovine liver glucose dehydrogenase: isolation and characterization. Arch. Biochem. Biophys. 215, 289–301 [DOI] [PubMed] [Google Scholar]
- 10. Ferri S., Kojima K., and Sode K. (2011) Review of glucose oxidases and glucose dehydrogenases: a bird's eye view of glucose sensing enzymes. J. Diabetes Sci. Technol. 5, 1068–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cavener D. R., and MacIntyre R. J. (1983) Biphasic expression and function of glucose dehydrogenase in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 80, 6286–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Metzger R. P., Wilcox S. S., and Wick A. N. (1964) Studies with rat liver glucose dehydrogenase. J. Biol. Chem. 239, 1769–1772 [PubMed] [Google Scholar]
- 13. Rohatgi N., Nielsen T. K., Bjørn S. P., Axelsson I., Paglia G., Voldborg B. G., Palsson B. O., and Rolfsson Ó. (2014) Biochemical characterization of human gluconokinase and the proposed metabolic impact of gluconic acid as determined by constraint based metabolic network analysis. PLoS ONE 9, e98760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corkins M. E., May M., Ehrensberger K. M., Hu Y. M., Liu Y. H., Bloor S. D., Jenkins B., Runge K. W., and Bird A. J. (2013) Zinc finger protein Loz1 is required for zinc-responsive regulation of gene expression in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 110, 15371–15376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilson S., and Bird A. J. (2016) Zinc sensing and regulation in yeast model systems. Arch. Biochem. Biophys. 611, 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ehrensberger K. M., Corkins M. E., Choi S., and Bird A. J. (2014) The double zinc finger domain and adjacent accessory domain from the transcription factor loss of zinc sensing 1 (loz1) are necessary for DNA binding and zinc sensing. J. Biol. Chem. 289, 18087–18096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ehrensberger K. M., Mason C., Corkins M. E., Anderson C., Dutrow N., Cairns B. R., Dalley B., Milash B., and Bird A. J. (2013) Zinc-dependent regulation of the Adh1 antisense transcript in fission yeast. J. Biol. Chem. 288, 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsai C. S., Shi J. L., and Ye H. G. (1995) Kinetic studies of gluconate pathway enzymes from Schizosaccharomyces pombe. Arch. Biochem. Biophys. 316, 163–168 [DOI] [PubMed] [Google Scholar]
- 19. Heiland S., Radovanovic N., Höfer M., Winderickx J., and Lichtenberg H. (2000) Multiple hexose transporters of Schizosaccharomyces pombe. J. Bacteriol. 182, 2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dainty S. J., Kennedy C. A., Watt S., Bähler J., and Whitehall S. K. (2008) Response of Schizosaccharomyces pombe to zinc deficiency. Eukaryot. Cell 7, 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cosgrove M. S., Gover S., Naylor C. E., Vandeputte-Rutten L., Adams M. J., and Levy H. R. (2000) An examination of the role of Asp-177 in the His-Asp catalytic dyad of Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase: X-ray structure and pH dependence of kinetic parameters of the D177N mutant enzyme. Biochemistry 39, 15002–15011 [DOI] [PubMed] [Google Scholar]
- 22. Bautista J. M., Mason P. J., and Luzzatto L. (1995) Human glucose-6-phosphate dehydrogenase: lysine 205 is dispensable for substrate binding but essential for catalysis. FEBS Lett. 366, 61–64 [DOI] [PubMed] [Google Scholar]
- 23. Wu C. Y., Roje S., Sandoval F. J., Bird A. J., Winge D. R., and Eide D. J. (2009) Repression of sulfate assimilation is an adaptive response of yeast to the oxidative stress of zinc deficiency. J. Biol. Chem. 284, 27544–27556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheah I. K., and Halliwell B. (2012) Ergothioneine: antioxidant potential, physiological function and role in disease. Biochim. Biophys. Acta 1822, 784–793 [DOI] [PubMed] [Google Scholar]
- 25. Kim D. U., Hayles J., Kim D., Wood V., Park H. O., Won M., Yoo H. S., Duhig T., Nam M., Palmer G., Han S., Jeffery L., Baek S. T., Lee H., Shim Y. S., et al. (2010) Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28, 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harrison D. C. (1931) Glucose dehydrogenase: a new oxidising enzyme from animal tissues. Biochem. J. 25, 1016–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kotaka M., Gover S., Vandeputte-Rutten L., Au S. W., Lam V. M., and Adams M. J. (2005) Structural studies of glucose-6-phosphate and NADP+ binding to human glucose-6-phosphate dehydrogenase. Acta Crystallogr. D Biol. Crystallogr. 61, 495–504 [DOI] [PubMed] [Google Scholar]
- 28. Rolfsson Ó., Paglia G., Magnusdóttir M., Palsson B. Ø., and Thiele I. (2013) Inferring the metabolism of human orphan metabolites from their metabolic network context affirms human gluconokinase activity. Biochem. J. 449, 427–435 [DOI] [PubMed] [Google Scholar]
- 29. Thomas D., Cherest H., and Surdin-Kerjan Y. (1991) Identification of the structural gene for glucose-6-phosphate dehydrogenase in yeast. Inactivation leads to a nutritional requirement for organic sulfur. EMBO J. 10, 547–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Slekar K. H., Kosman D. J., and Culotta V. C. (1996) The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J. Biol. Chem. 271, 28831–28836 [DOI] [PubMed] [Google Scholar]
- 31. Ng C. Y., Jung M. Y., Lee J., and Oh M. K. (2012) Production of 2,3-butanediol in Saccharomyces cerevisiae by in silico aided metabolic engineering. Microb. Cell Fact. 11, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Costello L. C., Feng P., Milon B., Tan M., and Franklin R. B. (2004) Role of zinc in the pathogenesis and treatment of prostate cancer: critical issues to resolve. Prostate Cancer Prostatic Dis. 7, 111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Costello L. C., and Franklin R. B. (2006) The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: connecting the dots. Mol. Cancer 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peter M., Gartner A., Horecka J., Ammerer G., and Herskowitz I. (1993) FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell 73, 747–760 [DOI] [PubMed] [Google Scholar]
- 35. Cocuron J. C., and Alonso A. P. (2014) Liquid chromatography tandem mass spectrometry for measuring 13C-labeling in intermediates of the glycolysis and pentose phosphate pathway. Methods Mol. Biol. 1090, 131–142 [DOI] [PubMed] [Google Scholar]
- 36. Koubaa M., Cocuron J. C., Thomasset B., and Alonso A. P. (2013) Highlighting the tricarboxylic acid cycle: liquid and gas chromatography-mass spectrometry analyses of 13C-labeled organic acids. Anal. Biochem. 436, 151–159 [DOI] [PubMed] [Google Scholar]
- 37. Gibon Y., and Larher F. (1997) Cycling assay for nicotinamide adenine dinucleotides: NaCl precipitation and ethanol solubilization of the reduced tetrazolium. Anal. Biochem. 251, 153–157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.