Abstract
Viperin (RSAD2) is an interferon-stimulated antiviral protein that belongs to the radical S-adenosylmethionine (SAM) enzyme family. Viperin's iron–sulfur (Fe/S) cluster is critical for its antiviral activity against many different viruses. CIA1 (CIAO1), an essential component of the cytosolic iron–sulfur protein assembly (CIA) machinery, is crucial for Fe/S cluster insertion into viperin and hence for viperin's antiviral activity. In the CIA pathway, CIA1 cooperates with CIA2A, CIA2B, and MMS19 targeting factors to form various complexes that mediate the dedicated maturation of specific Fe/S recipient proteins. To date, however, the mechanisms of how viperin acquires its radical SAM Fe/S cluster to gain antiviral activity are poorly understood. Using co-immunoprecipitation and 55Fe-radiolabeling experiments, we therefore studied the roles of CIA2A, CIA2B, and MMS19 for Fe/S cluster insertion. CIA2B and MMS19 physically interacted with the C terminus of viperin and used CIA1 as the primary viperin-interacting protein. In contrast, CIA2A bound to viperin's N terminus in a CIA1-, CIA2B-, and MMS19-independent fashion. Of note, the observed interaction of both CIA2 isoforms with a single Fe/S target protein is unprecedented in the CIA pathway. 55Fe-radiolabeling experiments with human cells depleted of CIA1, CIA2A, CIA2B, or MMS19 revealed that CIA1, but none of the other CIA factors, is predominantly required for 55Fe/S cluster incorporation into viperin. Collectively, viperin maturation represents a novel CIA pathway with a minimal requirement of the CIA-targeting factors and represents a new paradigm for the insertion of the Fe/S cofactor into a radical SAM protein.
Keywords: interferon, iron, iron–sulfur protein, molecular cell biology, sulfur, Cia targeting complex, biogenesis, metal biology, viperin
Introduction
RSAD2 (radical S-adenosylmethionine domain containing 2) is one of the interferon (IFN)-stimulated genes that is strongly up-regulated by type I IFNs, lipopolysaccharide, polyinosinic/polycytidylic acid, or by different virus infections (1–8). Its gene product viperin (virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible) has received increasing attention because of its ability to interfere with the proliferation of numerous RNA and DNA viruses from different families. Overexpression of viperin has been shown to inhibit budding and release of influenza A virus by disrupting lipid raft micro-domains in the plasma membrane (7). In addition, viperin localizes to lipid droplets, a site of replication for hepatitis C virus (HCV)4 and Dengue virus (DENV) (9), and it targets positive-stranded RNA synthesis of tick-borne encephalitis virus (TBEV) (6) and DENV (10). Because the broad range of viperin-affected viruses use different routes of infection and mechanisms of replication, the antiviral mechanism of viperin is unlikely to be virus-specific, yet the molecular mechanism of viperin function is unknown.
Viperin is highly conserved from fungi to lower vertebrates and mammals, and it is a member of the radical SAM iron–sulfur (Fe/S) protein family (6, 11–14). Radical SAM proteins usually perform chemically difficult reactions such as C–C and C–H bond cleavages, C–S bond formations, or alkylations (15, 16). Human viperin (361 residues, molecular mass of 42 kDa) is composed of three domains (17). The N terminus (residues 1–42) harbors an amphipathic α-helix that is important for viperin's localization to the endoplasmic reticulum (18) and to lipid droplets (9). The domain is also required for antiviral activity against Chikungunya virus (19). The C-terminal domain (residues 218–361) has been shown to mediate protein–protein interactions, e.g. with the HCV proviral factor hVAP-33 (20) and with CIA1 (also known as CIAO1), a component of the cytosolic iron–sulfur protein assembly (CIA) machinery (6). The central domain (residues 71–182) is homologous to the MoaA subfamily of radical SAM enzymes (11). It contains a conserved cysteine-rich motif (CXXXCXXC) responsible for binding a [4Fe-4S] cluster that uses SAM as the fourth ligand (15, 16). Mutations of the conserved cysteine residues abrogate viperin's antiviral action against HCV (2), West Nile virus, DENV (3), and TBEV (6). Despite the central importance of the radical SAM domain for viperin function, little is known about the maturation of its Fe/S cluster and about the assembly of SAM-coordinated [4Fe-4S] clusters in general (6).
Studies in yeast and human cells have revealed the importance of the CIA machinery for maturation of cytosolic and nuclear Fe/S proteins (21–23). This essential biosynthetic system is composed of 11 known CIA proteins that act in three main steps of the assembly reaction. First, a [4Fe-4S] cluster is assembled on the scaffold complex CFD1–NBP35 (24–26). This reaction requires a sulfur source from mitochondria and the electron transfer chain composed of NDOR1 (yeast Tah18) and CIAPIN1 (Dre2) (21, 27, 28). Second, the cluster is released from CFD1–NBP35 and transferred to target apoproteins for insertion into the polypeptide chain. This reaction requires the Fe/S protein IOP1 (yeast Nar1) and the CIA-targeting complex CIA1–CIA2B–MMS19, which, as a whole or in part, is required for maturation of most cytosolic-nuclear Fe/S clients, including DNA polymerases, DNA helicases, and nucleic acid metabolism proteins (29–33). In human cells, the CIA2B-related protein CIA2A specifically assists the maturation of iron regulatory protein 1 (IRP1), a protein involved in cellular iron homeostasis (33). Third, the recently identified CIA factors Yae1–Lto1 function as adapters that specifically recruit the Fe/S protein Rli1 (human ABCE1) to the CIA-targeting complex (34). Previous studies have indicated that targeting and insertion of [4Fe-4S] clusters into different client apoproteins require specific combinations of the CIA-targeting factors of the second step of the CIA pathway (31, 33). Depletion of the dedicated CIA-assembly factors impairs Fe/S cofactor insertion and results in a destabilization and eventually degradation of the respective apoproteins.
We have previously shown that viperin requires the physical interaction with CIA1 to become mature and antivirally active (6), implying a critical function of the CIA machinery for antiviral host defense of mammalian cells. In this study, we aimed to more comprehensively characterize the roles of the various CIA-targeting factors in viperin interaction and assembly of the radical SAM Fe/S cluster. To this end, we performed co-immunoprecipitation and 55Fe-radiolabeling experiments to define the relative importance of the CIA factors in viperin binding and maturation. Overall, our studies suggest the existence of a unique CIA-targeting pathway that uses CIA1 but not the other targeting factors for Fe/S cluster insertion into viperin.
Results
Viperin interacts with CIA1, MMS19, CIA2A, and CIA2B
To define the relative roles of the CIA-targeting factors CIA2A, CIA2B, and MMS19 in the maturation of viperin, we first performed co-immunoprecipitation (co-IP) experiments to analyze their potential interaction. HEK293T cells were transiently co-transfected with plasmids encoding non-tagged wild-type viperin and either CIA1-FLAG, CIA2B-HA, CIA2A-Myc, or MMS19-FLAG. Anti-viperin IP followed by immunoblotting revealed that each of the four CIA-targeting factors interacted with viperin (Fig. 1, A–D). In a reverse approach, an antivirally active FLAG-tagged viperin, expressed in FLP-IN T Rex cells (6), recovered all three endogenous CIA-targeting complex components (CIA1–CIA2B–MMS19) in anti-FLAG-IP (Fig. 1E). Because no antibody against CIA2A is available, we could not confirm the interaction of viperin with endogenous CIA2A. Together, our co-IP experiments reveal that viperin specifically interacts with all four CIA-targeting factors.
Figure 1.
Viperin interacts with the late-acting CIA-targeting proteins. HEK293T cells were transiently transfected with plasmids encoding wild-type viperin, FLAG-tagged CIA1 (A), HA-tagged CIA2B (B), FLAG-tagged MMS19 (C), and Myc-tagged CIA2A (D) as indicated (+). After growth for 1 day, cell extracts were subjected to anti-viperin IP. Whole-cell lysates (input) and immunoprecipitated proteins were analyzed by immunoblotting using antibodies as indicated. E, FLAG–viperin expression in FLP-IN T Rex cells was induced by tetracycline (+ Tet, 1 μg/ml in culture medium) for 24 h, followed by IP with an anti-FLAG antibody and sample analysis by immunoblotting. Tubulin served as loading control. The blots are representative of three independent experiments.
Viperin interacts with the CIA1–CIA2B–MMS19 targeting complex via its C terminus and with CIA2A via its N terminus
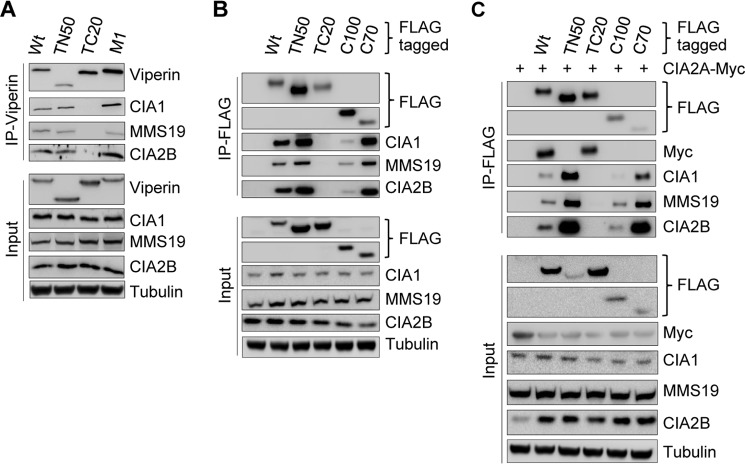
The finding that viperin bound to both CIA2A and CIA2B was surprising because these proteins assist distinct branches of the CIA pathway (33). We therefore sought to identify the viperin segments responsible for interaction with the CIA factors. We used FLP-IN T Rex cells inducibly expressing FLAG-tagged versions of full-length viperin, N- or C-terminally truncated versions (TN50 or TC20 that lack the N-terminal 50 or C-terminal 20 residues, respectively), or the Fe/S cluster-deficient mutant protein M1 in which the three cofactor-coordinating cysteine residues of the radical SAM domain were exchanged for alanine (6). IP of these four viperin versions revealed that the C-terminal region of the protein was important for interaction with CIA1–CIA2B–MMS19, because no co-IP of these CIA-targeting factors was observed with TC20 (Fig. 2A). Notably, the CIA interaction appeared to be strongest for the cluster-deficient viperin mutant M1, suggesting a preferential binding to the apo-form of viperin. The finding that the CIA-targeting complex proteins interact with the C terminus of viperin was further supported by co-IP experiments performed with plasmid-encoded, FLAG-tagged viperin fragments encompassing the C-terminal 100 (C100; residues 261–361) or 70 (C70; 291–361) amino acid residues. The C70 fragment bound to the CIA-targeting complex similarly as wild type and TN50 viperin (Fig. 2B). Although C100 is larger, the binding of C100 to the CIA-targeting complex was weaker compared with C70. One explanation might be improper folding of C100. These data suggest that the viperin C terminus acts as the necessary and sufficient interaction site for the CIA1–CIA2B–MMS19 targeting complex.
Figure 2.
CIA1–CIA2B–MMS19 targeting complex interacts with the C terminus and CIA2A with the N terminus of viperin. A, FLP-IN T Rex cells were treated with tetracycline (1 μg/ml) to induce expression either of wild-type viperin (Wt), viperin versions lacking the TN50 or TC20 residues, or the viperin Fe/S cluster binding mutant M1 (Cys to Ala mutations). After cell growth for 1 day, viperin was immunoprecipitated, and both whole cell lysates (Input) and immunoprecipitates (IP-Viperin) were analyzed by immunoblotting. Tubulin served as loading control. B, HEK293T cells were transiently transfected with plasmids encoding N-terminally FLAG-tagged wild-type viperin, viperin mutants TN50 and TC20, and viperin fragments consisting of the C100 or C70 residues. The FLAG proteins were precipitated by an anti-FLAG antibody, and samples were analyzed by immunoblotting. C, HEK293T cells were transiently transfected as in B and additionally received a plasmid encoding Myc-tagged CIA2A. Anti-FLAG–viperin IP and sample analysis were performed as in B.
A strikingly different result was obtained for the viperin interaction with CIA2A. Co-IP and immunoblotting revealed that viperin interacted with plasmid-expressed CIA2A-Myc through its N terminus rather than its C terminus, because wild-type and TC20 viperin bound CIA2A-Myc efficiently, whereas neither the TN50 mutant protein nor the C100 and C70 fragments interacted (Fig. 2C). In contrast, the latter three viperin variants recovered endogenous CIA1–CIA2B–MMS19, confirming the structural integrity of the C-terminal CIA-targeting complex interaction site. Taken together, our results demonstrate that viperin associates with the two CIA2 isoforms at separate domains.
CIA1 and CIA2B promote the interaction of MMS19 with viperin
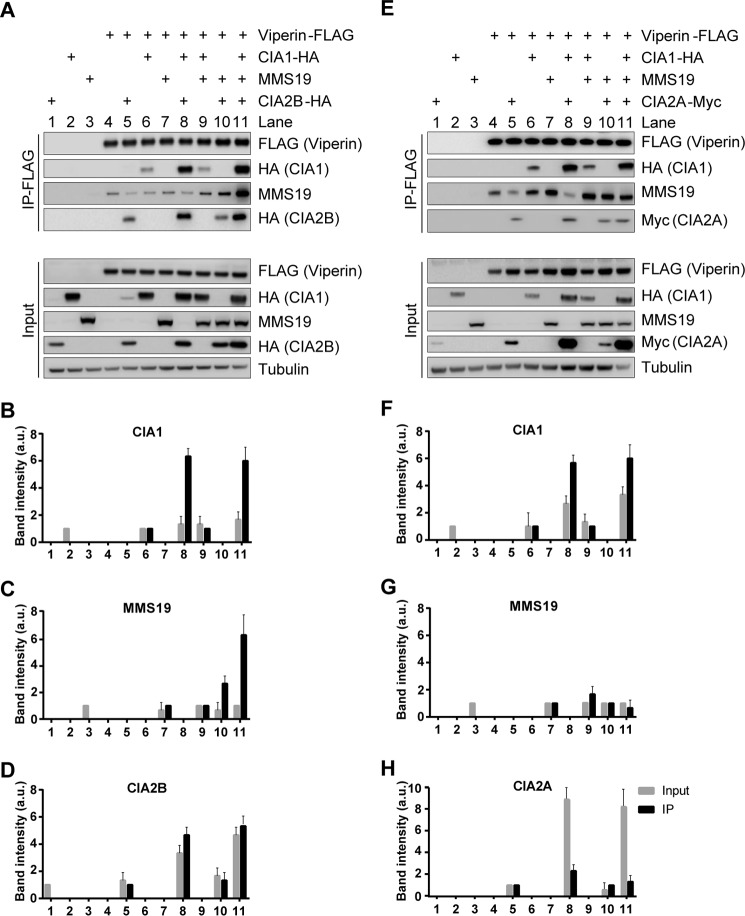
Previous functional analyses of individual CIA-targeting complex components have suggested that unique combinations of these constituents specifically assist the maturation of different apoprotein subsets, a finding mirrored by different target Fe/S protein interaction patterns (33). The co-purification of the CIA-targeting complex constituents CIA1–CIA2B–MMS19 together with viperin raised the question whether all of these maturation factors directly contact the radical SAM enzyme or whether there is a binding hierarchy. First, we determined how the expression of individual CIA-targeting factors might influence viperin binding to the remaining components. HEK293T cells were transiently co-transfected with a plasmid encoding FLAG-tagged wild-type viperin and with varying combinations of plasmids encoding the CIA-targeting factors. Protein–protein interactions were analyzed by anti-FLAG affinity purification and immunoblotting (Fig. 3A). For quantitative analysis, antigen-associated chemiluminescence signals were recorded and then normalized to the amount of immunoprecipitated viperin that was normalized to both the amount of tubulin and viperin in the total cell lysate (Fig. 3, B–D, black bars). For comparison, levels of plasmid-encoded proteins within the input lysates were also normalized to tubulin (Fig. 3, B–D, gray bars). The densitometric analysis revealed that the interaction of viperin with CIA1 was enhanced in the presence of CIA2B (Fig. 3, A and B, lanes 8 and 11) but not by MMS19 (lane 9). Conversely, viperin interaction with CIA2B was enhanced by co-expression of CIA1 (Fig. 3, A and D, lanes 8 and 11) and correlated with elevated CIA2B protein levels present in the input lysate (lanes 8 and 11, gray bars). In contrast, expression of MMS19 altered neither the cellular CIA2B protein level nor the interaction between CIA2B and viperin (Fig. 3, A and D, lane 10). Likewise, co-expression of CIA1 or CIA2B along with FLAG–viperin hardly influenced protein level or recovery of MMS19 (Fig. 3, A and C, lanes 9 and 10). Notably, expression of the entire CIA1–CIA2B–MMS19 targeting complex substantially improved the interaction of each of the complex constituents by FLAG–viperin, including MMS19 (Fig. 3, A–D, lane 11).
Figure 3.
CIA1 and CIA2B mediate the indirect interaction of MMS19 with viperin. A and E, HEK293T cells were transiently transfected with plasmids encoding FLAG-tagged viperin and tagged (HA or Myc) or non-tagged CIA-targeting factors as indicated (+). Anti-FLAG–viperin immunoprecipitation and sample analysis by immunoblotting were performed similar to Fig. 2. Tubulin staining served as reference. The chemiluminescence associated with CIA1-HA (B and F), MMS19 (C and G), CIA2B-HA (D), or CIA2A-Myc (H) was quantified. The signal intensity of input samples was normalized to tubulin (gray bars), and the signals associated with the immunoprecipitated samples were normalized to immunoprecipitated viperin, which was normalized to both the amount of tubulin and viperin in the total cell lysates (black bars, mean values ± S.D.; n = 3).
We next compared the behavior of CIA2A to that of CIA2B. Co-expression of CIA2A and CIA1 together with viperin improved the recovery of CIA1 by FLAG–viperin (Fig. 3, E and F, lane 8), similar to the situation of CIA2B (Fig. 3, A and B, lane 8). In turn, the binding of CIA2A by viperin only slightly increased in the presence of CIA1 (Fig. 3, E and H, lane 8), although co-expression of CIA1 and CIA2A led to elevated cellular protein levels of both factors (Fig. 3, F and H, lane 8). The increased CIA1 levels were likely caused by stabilizing effects of CIA2A and/or CIA2B (33), yet could only partially explain the high recovery of CIA1 by viperin (Fig. 3, B and F, lane 8, black versus gray bars). The results argue for an additional supportive function of the CIA2 proteins in the CIA1–viperin interaction. Similar to CIA1, also MMS19 hardly influenced the interaction of CIA2A with viperin (Fig. 3E, lane 10). Taken together, our co-immunoprecipitation results suggest that the main interactors of viperin are CIA1 and both CIA2 isoforms, whereas MMS19 associates with viperin only indirectly via CIA1 and CIA2B.
Interaction between viperin and the CIA-targeting complex is mediated mainly by CIA1
To better understand which of the CIA-targeting factors are in direct physical contact with viperin, we depleted individual CIA-targeting complex constituents by RNAi technology and then measured the viperin interaction of the residual CIA factors. Knockdown efficiencies of the CIA mRNAs (10–20% of control cells) were estimated by quantitative real-time PCR (supplemental Fig. S1, A–D), and the steady-state protein levels of each CIA factor were assessed by immunoblotting (supplemental Fig. S1E). In line with previous observations (33), individual depletion of CIA1, CIA2B, or MMS19 decreased the steady-state protein levels of the remaining CIA-targeting complex components, despite the fact that the mRNA levels of these proteins were not affected (supplemental Fig. S1). This observation is consistent with the mutual stabilization of the CIA-targeting complex components (cf. Fig. 2) (33). Because the decreased levels of the remaining CIA factors may complicate the interpretation of subsequent interaction studies, we improved the RNAi approach by additional plasmid-based expression of the remaining CIA constituents. This maintained their high abundance as revealed by immunoblotting of cell extracts (Fig. 4A, input). The only exception was the low level of CIA2A in the absence of CIA1 (Fig. 4A, lanes 3 and 7), consistent with the mutual stabilization of these CIA factors.
Figure 4.
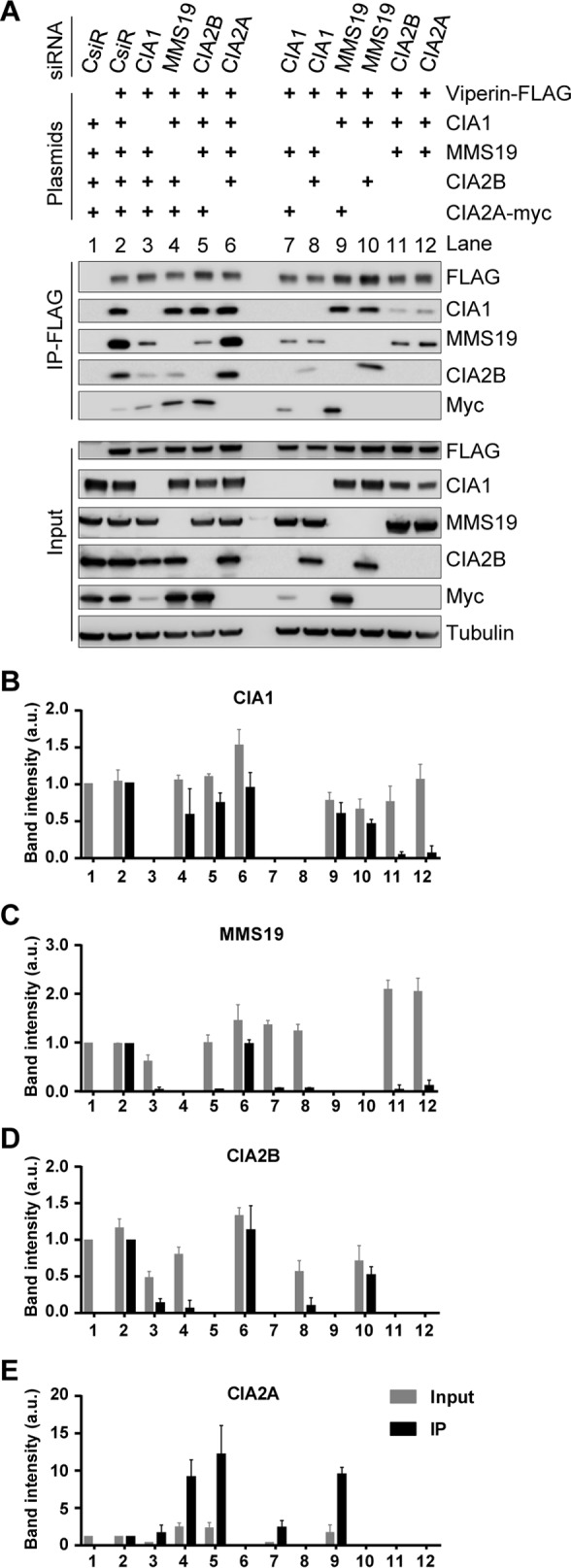
CIA-targeting factors CIA1, CIA2B, and CIA2A directly interact with viperin. A, HEK293T cells were transiently transfected with control siRNAs (CsiR) or siRNAs directed against the indicated CIA-targeting factors. In addition, cells received plasmids coding for the remaining CIA-targeting complex components and FLAG–viperin as indicated (+). Anti-FLAG immunoprecipitation and sample analysis were performed according to Fig. 2. Tubulin and FLAG staining served as reference, and representative blots are shown. The chemiluminescence associated with CIA1 (B), MMS19 (C), CIA2B (D), and CIA2A-myc (E) was quantified as in Fig. 3. Mean values and standard deviation from three independent experiments are shown.
With this improvement of the RNAi depletion approach, we analyzed the interaction of plasmid-encoded FLAG-tagged viperin with the different CIA-targeting factors by immunoprecipitation as in Fig. 3. The interaction was optimal when all three CIA-targeting complex components, i.e. CIA1, CIA2B, and MMS19, were co-expressed (Fig. 4, A–D, lanes 2 and 6). In samples containing low levels of both CIA2 isoforms, i.e. upon depletion of CIA2A with no ectopic expression of CIA2B and vice versa (Fig. 4A, lanes 11 and 12), the association of viperin with CIA1 was severely impaired, suggesting that each CIA2 protein can stabilize the viperin–CIA1 interaction. Inversely, CIA2B bound to viperin only when CIA1 was abundant (Fig. 4, A and D, lanes 2, 6, and 10), whereas CIA2A associated with viperin also in the absence of CIA1 (Fig. 4, A and E, lanes 3 and 7), even though binding was enhanced by co-expression of CIA1 (lanes 4, 5, and 9). We also observed that CIA1 can weakly associate in the absence of the C terminus of viperin, probably via CIA2A, but only in the overexpression system (supplemental Fig. S2). When we analyzed the behavior of MMS19, this protein appeared to indirectly bind to viperin, because their co-purification required overexpression of both CIA1 and CIA2B (Fig. 4, A and C, lanes 2 and 6). The remaining weak interaction between MMS19 and viperin in the absence of CIA1 or CIA2B (Fig. 4A, lanes 3, 5, 7, 8, and 11) is best explained by incomplete RNAi knockdown. This observation indicates that MMS19 associates with viperin by binding to the CIA1–CIA2B complex (see also Fig. 3). Conversely, MMS19 depletion had no effect on the viperin interaction with CIA1, CIA2B, or CIA2A confirming their direct association with viperin (Fig. 4, A–E, lanes 4, 9, and 10). In summary, our results indicate that the major viperin binding partner is CIA1, whose interaction is enhanced by both CIA2A and CIA2B, mainly by exerting a stabilization of CIA1. Although CIA2B binds to viperin in a CIA1-dependent fashion at the C terminus, CIA2A can associate independently with the N terminus of viperin. MMS19 interacts with viperin only via CIA1 and CIA2B as part of the CIA-targeting complex.
Fe/S cluster insertion into viperin is not dependent on CIA2A, CIA2B, or MMS19
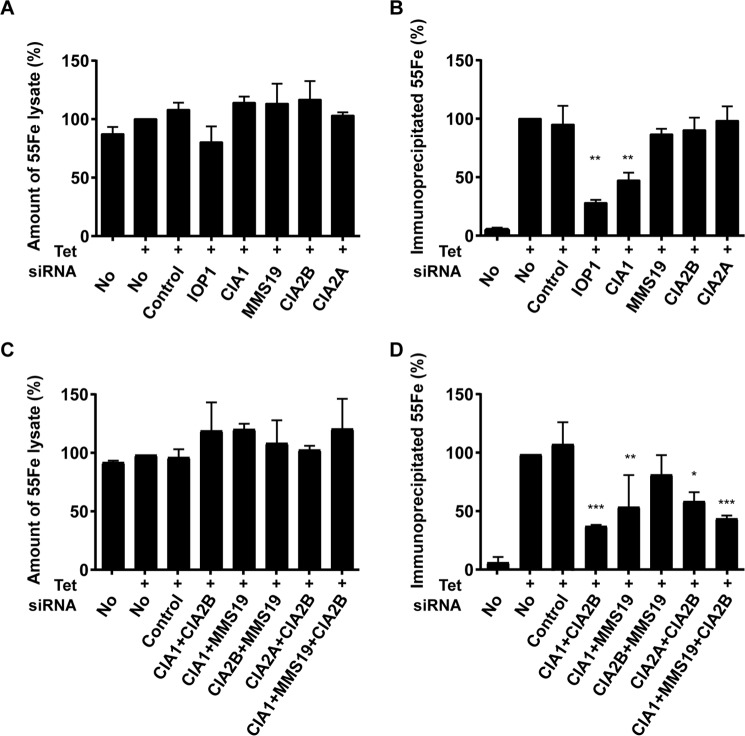
The binding hierarchy observed for viperin interaction with the CIA1–CIA2B–MMS19 complex and CIA2A raised the question as to which individual CIA constituents contribute to Fe/S cluster insertion into viperin. We directly examined the maturation process by following the in vivo incorporation of 55Fe into viperin as a measure of Fe/S cluster assembly at the radical SAM domain (6). To this end, FLP-IN T Rex cells expressing FLAG-tagged viperin were transiently transfected with siRNAs directed against the CIA-targeting complex components and against the general CIA factor IOP1, a protein that is of critical importance for maturation of all cytosolic-nuclear Fe/S proteins and thus is thought to act upstream of the CIA-targeting complex (35). After two successive rounds of siRNA transfection at a 3-day interval, cells were supplemented with 55Fe-loaded transferrin, and viperin expression was induced for 48 h. Cells were harvested, and the 55Fe content of the cell lysate (indicative of total cellular iron uptake; Fig. 5A) and of immunoprecipitated viperin (Fig. 5B) were determined by scintillation counting. The efficient depletion of the individual CIA proteins was verified by immunoblot analysis (supplemental Fig. S3, A and B). IRP1 was used as an indicator for CIA2A knockdown efficiency, as IRP1 stability is dependent on CIA2A (31). The strongest impairment of viperin maturation was observed upon depletion of IOP1 (Fig. 5B), resulting in a 4-fold decrease in 55Fe/S cluster insertion. This result is consistent with the central role of IOP1 in cytosolic-nuclear Fe/S protein assembly (36–38). Among the CIA-targeting complex components, only depletion of CIA1 resulted in a significant effect (55% decrease) on 55Fe incorporation into viperin, whereas iron uptake remained normal (Fig. 5, A and B). In contrast, individual depletion of MMS19, CIA2B, or CIA2A did not result in pronounced effects on cellular 55Fe uptake or viperin 55Fe/S cofactor maturation, even though the expected substantial effects on other cytosolic and nuclear Fe/S proteins such as IRP1 (CIA2A), GPAT (CIA1), and POLD1 (IOP1, CIA1, and MMS19) were observed (supplemental Fig. S3, A–D)(31, 33).
Figure 5.
CIA1 is the only CIA-targeting factor that is crucial for Fe/S cluster insertion into viperin. A and B, HEK FLP-IN T Rex cells (capable of induced FLAG–viperin expression as in Fig. 1E) were depleted for the indicated CIA components by RNAi technology. The cells were grown in the presence of 55Fe-labeled transferrin for 2 days and were then treated with tetracycline to induce FLAG–viperin expression for 1 day. A, subsequent to cell harvest and lysis, the 55Fe content of the total lysate was determined by scintillation counting and expressed as the ratio of 55Fe per total protein relative to mock-transfected, FLAG–viperin-expressing control cells (No, +). B, lysates were subjected to anti-FLAG immunoprecipitation. 55Fe incorporation into FLAG–viperin was determined by scintillation counting and expressed relative to the radioactivity precipitated from mock-transfected control cells (No, +). C and D, HEK FLP-IN T Rex cells were depleted for multiple combinations of CIA components by RNAi as indicated and treated as above. Mean values and standard deviations from three independent experiments are shown. ***, p < 0.001; **, p < 0.01; *, p < 0.05 Student's t test, calculated and compared with the control.
Because depletion of individual CIA-targeting factors apart from CIA1 showed no detectable effect on viperin maturation, we tested whether combined depletion of two or three CIA-targeting factors might impair this process. When CIA1 was among the depleted proteins, we consistently observed a 2–3-fold decrease in 55Fe/S cluster incorporation into viperin, yet no diminution in cellular 55Fe uptake (Fig. 5, C and D, and supplemental Fig. S3E), similar to the single knockdown of CIA1. In contrast, the combined depletion of CIA2B and MMS19 did not affect the maturation of viperin relative to control cells reassuring that only CIA1 of the CIA-targeting complex is essential for viperin maturation. Interestingly, the combined depletion of CIA2A and CIA2B slightly impaired maturation of viperin by 40%. This effect was still less pronounced than depletion of CIA1 alone and likely is best explained by the destabilization of CIA1 in the absence of its two CIA2-binding partners (supplemental Fig. S3C), again suggesting that CIA1 depletion rather than the CIA2 proteins themselves was the cause of this maturation decline. Taken together, these results suggest a direct and important function of CIA1 in the maturation of viperin, whereas CIA2B, MMS19, and CIA2A appear to play dispensable roles despite their association with viperin. Their function therefore might be restricted to the stabilization of CIA1 thus ensuring its optimal binding to viperin.
Discussion
This study identified critical cell biological requirements for [4Fe-4S] cluster assembly of the antiviral radical SAM protein viperin. Because this radical SAM Fe/S cluster has been found to be essential for the antiviral function of viperin, detailed knowledge about its maturation mechanism is crucial for understanding the physiological basis of viperin activation. Previously, we have defined an essential role of the CIA-targeting factor CIA1 in Fe/S cluster insertion into the radical SAM domain of viperin. However, the potential roles of the CIA1-interacting proteins MMS19, CIA2B, and CIA2A have not yet been addressed (31, 33). Likewise, the assembly of a radical SAM-type of Fe/S cluster in general has not been investigated so far. Here, we identified the CIA proteins MMS19, CIA2B, and CIA2A as interaction partners of viperin, in addition to CIA1 (6). Binding of each of the four CIA-targeting factors was independent of the ability of viperin to bind a Fe/S cluster because mutation of the three conserved cysteine ligands (mutant M1) strengthened rather than weakened the interaction. This finding suggests that the CIA-targeting factors interact with the apo-form of viperin, a mechanism expected for bona fide maturation factors and consistent with an earlier report for the assembly of the nuclear Fe/S protein XPD (39). The binding events therefore may be expected to be tightly connected to the Fe/S cluster maturation process (discussed below).
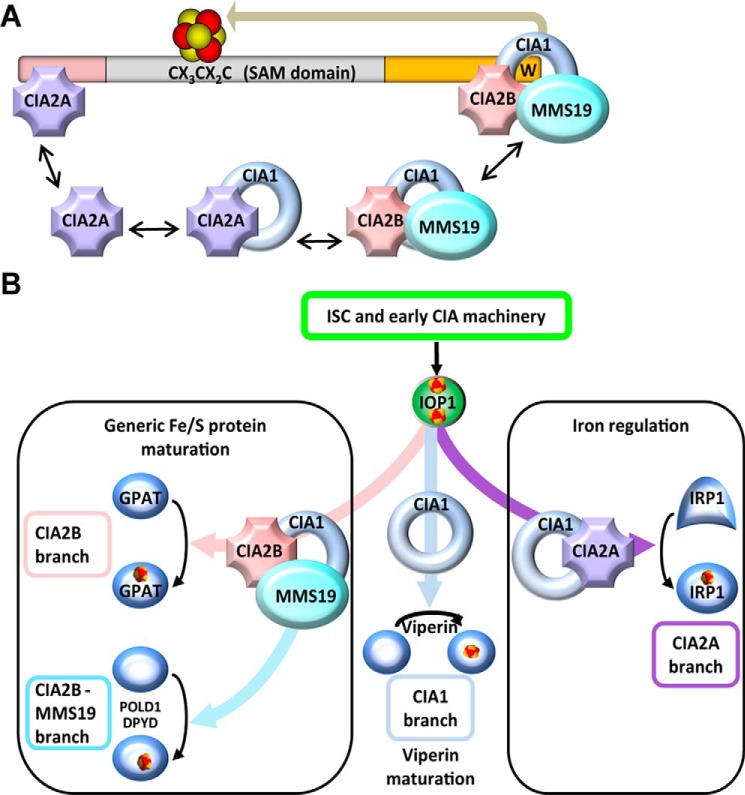
It was surprising to find that both isoforms of human CIA2 bound to viperin, because previously we have reported radically different CIA2A and CIA2B interactomes with hardly any overlap for the two proteins (33). A molecular explanation for viperin binding to both CIA2 isoforms came from the mapping of the precise interaction domains on viperin for the four CIA targeting proteins by performing co-immunoprecipitation experiments with viperin mutants lacking the N or C termini and with C-terminal viperin fragments (Fig. 2). These studies showed that CIA2B (in complex with CIA1 and MMS19) and CIA2A bind to the C and N termini, respectively, of viperin (Fig. 6A). Co-immunoprecipitation studies performed after RNAi depletion of individual CIA factors suggested a hierarchy for binding of CIA1, CIA2B, and MMS19. At the C terminus, the major viperin interaction partner appears to be CIA1, because its depletion substantially decreased the association of both CIA2B and MMS19, indicating that the viperin association of the latter two proteins is mediated by CIA1 (Fig. 4A, left). The sum of our interaction analysis data is best interpreted by the view that viperin, via its C terminus, directly associates with CIA1, which recruits CIA2B, and this complex facilitates binding of MMS19 (Fig. 6A). This interpretation is supported by the finding that MMS19 depletion does not affect the association of CIA1 to viperin. The fact that in this case the CIA2B interaction with viperin is weakened is readily explained by the stabilizing role of MMS19 for CIA1 and CIA2B (31). Overall, the most efficient interaction was formed, when all three CIA-targeting complex proteins were present. This is consistent with their mutual stabilization. Collectively, our data suggest that the CIA1–CIA2B–MMS19 complex tightly interacts with the C terminus of viperin, yet the primary binding partner is CIA1.
Figure 6.
Model for the mode of interaction of CIA-targeting factors with viperin and for the unique maturation pathway. A, multimeric CIA-targeting complexes are present in the cell. As shown here, the CIA1–CIA2B–MMS19 complex interacts with the C terminus of viperin via direct contacts of both CIA1 and CIA2B. The two factors in turn recruit MMS19 to viperin. CIA2A binds at the N terminus of viperin and is stabilized by CIA1. Insertion of the radical SAM Fe/S cluster in the middle domain is mediated by C-terminally bound CIA1 only (see Ref. 6 and this study). B, Fe/S cluster maturation of viperin represents a novel branch of the CIA pathway that is initiated by the mitochondrial ISC machinery, early-acting CIA components, and IOP1 (20, 21). The components of the CIA1–CIA2B–MMS19-targeting complex operate in various combinations to facilitate Fe/S cluster insertion into dedicated client proteins, including GPAT, POLD1, and DPYD. CIA2A is specifically required for the maturation of IRP1 thus affecting cellular iron homeostasis and is stabilized by binding to CIA1. In contrast to all other known client Fe/S proteins, CIA1 appears to mediate Fe/S cluster insertion into the radical SAM protein viperin independently of the other CIA-targeting factors, thus representing a minimal requirement for CIA proteins.
CIA2A binding to the N-terminal region of viperin occurred independently of the other CIA-targeting factors, as their depletion did not affect the CIA2A–viperin binding efficiency. This was particularly true for the depletion of CIA1, but it may not immediately be evident (33). CIA1 knockdown resulted in lower cellular levels of CIA2A because of the missing stabilizing effect of CIA1. Nevertheless, the amount of viperin-bound CIA2A per total cellular CIA2A remained high, clearly indicating that CIA2A can associate with viperin without the other three CIA factors. Conversely, depletion of CIA2A did not affect the extent of viperin binding to the CIA-targeting complex (Fig. 4) suggesting that the binding events at the viperin termini are independent. In fact, CIA2A binding to viperin is even increased under CIA2B or MMS19 depletion conditions. This effect may have been caused indirectly by multiple reasons, including (i) the stabilization of CIA2A by viperin and/or CIA1 binding and (ii) a competition of CIA2A with CIA2B for complex formation with CIA1 (Fig. 6, A and B). Taken together, viperin can interact with CIA2A and the CIA-targeting complex CIA1–CIA2B–MMS19 at opposing termini in independent binding events. The interaction network between the CIA factors and viperin largely contributes to the stability of the CIA factors in the cytosol.
The preferential binding of CIA1, CIA2B, and MMS19 to the C terminus of viperin and the CIA2A interaction with viperin's N terminus raised the interesting question of what the contribution of each of these independent binding events to Fe/S cluster assembly may be. This fundamental prerequisite for viperin activation could be directly addressed by radiolabeling and immunoprecipitation experiments employing the 55Fe isotope to estimate Fe/S cluster assembly in vivo. Depletion of the general CIA component IOP1 strongly deceased the radiolabeling of viperin by a factor of 4, highlighting the essential requirement of the CIA system for Fe/S cluster assembly on viperin (Fig. 5). In contrast, the knockdown of the other CIA-targeting factors, with the exception of CIA1 (6), did not significantly decrease 55Fe incorporation into viperin, even though other cytosolic and nuclear Fe/S proteins are impaired under this condition (31, 33). This result clearly demonstrates that CIA1 not only directly associates with viperin but also performs the predominant if not exclusive role as a targeting factor in Fe/S cluster insertion. Viperin is the first known cytosolic Fe/S protein whose maturation solely depends on the early, general part of the CIA machinery and on CIA1, but not on the other three CIA-targeting factors. The Fe/S cluster maturation mechanism of viperin thus represents a novel branch of the late phase of the CIA pathway (Fig. 6B).
The importance of CIA1 for viperin maturation was also evident from double and triple RNAi depletion approaches for the CIA factors (Fig. 5D). Whenever CIA1 was depleted, 55Fe/S cluster formation was severely impaired. In contrast, double knockdown of CIA2B and MMS19 yielded wild-type efficiencies of Fe/S cluster insertion into viperin. As an exception, depletion of both CIA2A and CIA2B slightly (40%) diminished 55Fe radiolabeling of viperin. At first glimpse, this result may suggest that at least one of the two CIA2 isoforms is required for an efficient maturation process. However, this effect of the simultaneous depletion of the two CIA2 isoforms on viperin Fe/S cluster assembly may be better explained by their strong stabilizing effect on CIA1 (see above). In the absence of the two CIA2 proteins, CIA1 strongly vanishes (supplemental Fig. S3C), and this may then indirectly lead to the observed maturation defect. Together, these findings suggest that the diminution of the viperin Fe/S cluster assembly efficiency in the absence of both CIA2A and CIA2B is caused by the concomitant depletion of the key maturation factor CIA1 which functions without CIA2B and MMS19.
CIA2A binding to viperin raises the question of the functional meaning of this association. A direct role in Fe/S protein maturation seems unlikely from our experiments because (i) CIA2A depletion does not hamper 55Fe/S association with viperin, and (ii) CIA2A binding to the viperin N terminus cannot substitute for the complete abrogation of Fe/S cluster insertion observed upon deletion of the viperin C terminus, including the C-terminal conserved tryptophan as the binding site of the CIA-targeting complex (6). Currently, the physiological meaning of the CIA2A–viperin interaction is not understood, requiring future studies to examine the implications of this complex formation on viperin function in the antiviral response in an infected cell.
Experimental procedures
Plasmids and siRNAs
Eukaryotic expression vector pI.18 was kindly provided by Jim Robertson (National Institute for Biological Standards and Control, Hertfordshire, UK) and was used for the expression of N-terminally FLAG-tagged wild-type viperin, TN50 viperin (residues 51–361), and viperin M1 (Cys to Ala exchange of Fe/S cluster-ligating residues) (6). TC20 viperin (residues 1–341), C100 viperin (residues 261–361), C70 viperin (residues 291–361), as well as CIA1, CIA2B, and MMS19 (without or with C-terminal FLAG tags and HA tags, respectively) were cloned into pI.18 using standard PCR-cloning methods. KOD Hot Start polymerase (Novagen), restriction enzymes, and T4 DNA ligase (Fermentas) were used according to the manufacturers' recommendations. All plasmids were DNA-sequenced to verify correctness, and oligonucleotide primer sequences are available upon request. The following siRNAs were purchased from Ambion: IOP1 (s34746, s34747, and s34748), CIA1 (s17970 and s17971), MMS19 (s34553 and s34552), CIA2A (s38636 and ss38638), CIA2B (s28462 and s28461), and negative control (negative control #1).
Tissue culture, transfection, co-immunoprecipitation, and 55Fe incorporation
HEK293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum (FCS) and passaged in a ratio of 1:5 every 3 or 4 days. HEK293T FLP-IN T Rex cells inducibly expressing N-terminally FLAG-tagged viperin and viperin mutants (2) were propagated in DMEM supplemented with 5% tetracycline-free FCS (PAA) and passaged similar to normal HEK cells. Protein expression was induced by 1 μg/ml tetracycline (Sigma) for 24–48 h. HEK293T cells were seeded a day before plasmid transfection. The concentration of DNA and the ratio of transfection reagent (GeneJuice) were used as suggested by the manufacturer (Merck Millipore).
For siRNA transfection, HEK293T cells were seeded into 6-well plates. One day later, a final concentration of 10 nm siRNA was transfected using the transfection reagent (jetPRIME) according to the manufacturer's (Polyplus) recommendations, followed by medium exchange after 24 h. The next day, transfected cells were reseeded, and the transfection procedure was repeated. In some experiments, protein-encoding plasmids were transfected 24 h after the second round of siRNA application using the jetPRIME transfection reagent. Knockdown efficiency was determined by PCR and immunoblotting at day 5 after siRNA transfection. Co-IP and 55Fe incorporation studies were performed as described previously (6).
Antibodies
Mouse (ms) α-viperin (ab107359, dilution (d) 1:1000), rabbit (rb) α-viperin (ab73864, d 1:5000), rb α-CIA1 (ab123297, d 1:500), rb α-CIA2B (ab103227, d 1:1500), rb α-MMS19 (ab188156, d 1:500), rb α-tubulin (ab6046, d 1:4000), rb α-HA epitope (ab9110, d 1:4000), and rb α-Myc epitope (ab9106, d 1:2500) were distributed by Abcam. The rb α-FLAG epitope (F7425, d 1:5000), ms α-FLAG epitope (200472, d 1:2500), and rb α-POLD1 (15646–1-AP, d 1:1000) were obtained from Sigma, Stratagene, and Proteintech, respectively. The ms α-IRP1 (clone 295B) was a kind gift of R. Eisenstein (Madison, WI) (24), and rb α-GPAT was a kind gift of H. Puccio (Illkirch, France) (24). The rb α-IOP1 was raised against recombinant full-length protein in the laboratory of R. Lill. To this end, an N-terminally His-tagged version had been expressed in Escherichia coli and purified by nickel-nitrilotriacetic acid affinity chromatography. Horseradish peroxidase-conjugated secondary antibody was obtained from Pierce, and antigen detection was performed using the SuperSignal West Pico or Femto kit (Pierce).
RNA extraction and real-time RT-PCR
Total cellular RNA was extracted using the Nucleospin RNA II kit (Macherey-Nagel) according to the manufacturer's recommendations. Aliquots of 600 ng or 1 μg of RNA were used to synthesize cDNA with the Quantitect reverse transcription (RT) kit (Qiagen). All real-time RT-PCRs were performed with 7900HT fast-time PCR system (Applied Biosystems). In the subsequent PCRs, the γ-actin, CIA1, CIA2B, MMS19, and CIA2A mRNAs were detected with QuantiTect primers QT00996415, QT00004158, QT00003724, QT00078330, and QT01027348, respectively, using the QuantiTect SYBR Green RT-PCR kit (Qiagen).
Author contributions
A. S. U., O. S., and C. P. designed and performed the experiments, analyzed, compiled, and interpreted the data, and wrote the manuscript. R. R. performed the experiments. A. K.Ö. and R. L. designed, interpreted, compiled the data, wrote the manuscript, and provided funding. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgment
We thank Karin Edlund at Clinical Microbiology, Virology, Umeå University, for excellent technical assistance and help with cloning.
Note added in proof
In the version of this article that was published as a Paper in Press on June 14, 2017, some immunoblots in Figs. 1 and 2 were inadvertently swapped. Additionally, some panels contained an incorrect immunoblot. These errors have now been corrected and do not affect the results or conclusions of this work.
This work was supported in part by the Laboratory for Molecular Medicine Sweden (MIMS), the Umeå Center for Microbial Research (UCMR), Swedish Research Council Grant 2011-2795, Swedish Foundation for Strategic Research Grants ICA10-0059 and FFL12-0089 (to A. K. Ö.), and EMBO Short Term Fellowship ASTF444-2015) (to A. S. U.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–3.
- HCV
- hepatitis C virus
- DENV
- Dengue virus
- TBEV
- tick-borne encephalitis virus
- IP
- immunoprecipitation
- SAM
- S-adenosylmethionine
- CIA
- cytosolic iron–sulfur protein assembly
- ms
- mouse
- rb
- rabbit
- d
- dilution.
References
- 1. Helbig K. J., Lau D. T., Semendric L., Harley H. A., and Beard M. R. (2005) Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology 42, 702–710 [DOI] [PubMed] [Google Scholar]
- 2. Jiang D., Guo H., Xu C., Chang J., Gu B., Wang L., Block T. M., and Guo J. T. (2008) Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82, 1665–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang D., Weidner J. M., Qing M., Pan X. B., Guo H., Xu C., Zhang X., Birk A., Chang J., Shi P. Y., Block T. M., and Guo J. T. (2010) Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 84, 8332–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nasr N., Maddocks S., Turville S. G., Harman A. N., Woolger N., Helbig K. J., Wilkinson J., Bye C. R., Wright T. K., Rambukwelle D., Donaghy H., Beard M. R., and Cunningham A. L. (2012) HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood 120, 778–788 [DOI] [PubMed] [Google Scholar]
- 5. Szretter K. J., Brien J. D., Thackray L. B., Virgin H. W., Cresswell P., and Diamond M. S. (2011) The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J. Virol. 85, 11557–11566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Upadhyay A. S., Vonderstein K., Pichlmair A., Stehling O., Bennett K. L., Dobler G., Guo J. T., Superti-Furga G., Lill R., Överby A. K., and Weber F. (2014) Viperin is an iron–sulfur protein that inhibits genome synthesis of tick-borne encephalitis virus via radical SAM domain activity. Cell. Microbiol. 16, 834–848 [DOI] [PubMed] [Google Scholar]
- 7. Wang X., Hinson E. R., and Cresswell P. (2007) The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2, 96–105 [DOI] [PubMed] [Google Scholar]
- 8. Zhang Y., Burke C. W., Ryman K. D., and Klimstra W. B. (2007) Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 81, 11246–11255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hinson E. R., and Cresswell P. (2009) The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic α-helix. Proc. Natl. Acad. Sci. U.S.A. 106, 20452–20457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Helbig K. J., Carr J. M., Calvert J. K., Wati S., Clarke J. N., Eyre N. S., Narayana S. K., Fiches G. N., McCartney E. M., and Beard M. R. (2013) Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl. Trop. Dis. 7, e2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shaveta G., Shi J., Chow V. T., and Song J. (2010) Structural characterization reveals that viperin is a radical S-adenosyl-l-methionine (SAM) enzyme. Biochem. Biophys. Res. Commun. 391, 1390–1395 [DOI] [PubMed] [Google Scholar]
- 12. Duschene K. S., and Broderick J. B. (2010) The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 584, 1263–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sofia H. J., Chen G., Hetzler B. G., Reyes-Spindola J. F., and Miller N. E. (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goodwin S. B., M'Barek S. B., Dhillon B., Wittenberg A. H., Crane C. F., Hane J. K., Foster A. J., Van der Lee T. A., Grimwood J., Aerts A., Antoniw J., Bailey A., Bluhm B., Bowler J., Bristow J., et al. (2011) Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 7, e1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J., Woldring R. P., Román-Meléndez G. D., McClain A. M., Alzua B. R., and Marsh E. N. (2014) Recent advances in radical SAM enzymology: new structures and mechanisms. ACS Chem. Biol. 9, 1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Landgraf B. J., McCarthy E. L., and Booker S. J. (2016) Radical S-adenosylmethionine enzymes in human health and disease. Annu. Rev. Biochem. 85, 485–514 [DOI] [PubMed] [Google Scholar]
- 17. Mattijssen S., and Pruijn G. J. (2012) Viperin, a key player in the antiviral response. Microbes Infect. 14, 419–426 [DOI] [PubMed] [Google Scholar]
- 18. Hinson E. R., and Cresswell P. (2009) The N-terminal amphipathic α-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J. Biol. Chem. 284, 4705–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teng T. S., Foo S. S., Simamarta D., Lum F. M., Teo T. H., Lulla A., Yeo N. K., Koh E. G., Chow A., Leo Y. S., Merits A., Chin K. C., and Ng L. F. (2012) Viperin restricts chikungunya virus replication and pathology. J. Clin. Invest. 122, 4447–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang S., Wu X., Pan T., Song W., Wang Y., Zhang F., and Yuan Z. (2012) Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33. J. Gen. Virol. 93, 83–92 [DOI] [PubMed] [Google Scholar]
- 21. Netz D. J., Mascarenhas J., Stehling O., Pierik A. J., and Lill R. (2014) Maturation of cytosolic and nuclear iron–sulfur proteins. Trends Cell Biol. 24, 303–312 [DOI] [PubMed] [Google Scholar]
- 22. Paul V. D., and Lill R. (2015) Biogenesis of cytosolic and nuclear iron–sulfur proteins and their role in genome stability. Biochim. Biophys. Acta 1853, 1528–1539 [DOI] [PubMed] [Google Scholar]
- 23. Sharma A. K., Pallesen L. J., Spang R. J., and Walden W. E. (2010) Cytosolic iron–sulfur cluster assembly (CIA) system: factors, mechanism, and relevance to cellular iron regulation. J. Biol. Chem. 285, 26745–26751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stehling O., Netz D. J., Niggemeyer B., Rösser R., Eisenstein R. S., Puccio H., Pierik A. J., and Lill R. (2008) Human Nbp35 is essential for both cytosolic iron–sulfur protein assembly and iron homeostasis. Mol. Cell. Biol. 28, 5517–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Netz D. J., Pierik A. J., Stümpfig M., Mühlenhoff U., and Lill R. (2007) The Cfd1-Nbp35 complex acts as a scaffold for iron–sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 3, 278–286 [DOI] [PubMed] [Google Scholar]
- 26. Hausmann A., Aguilar Netz D. J., Balk J., Pierik A. J., Mühlenhoff U., and Lill R. (2005) The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron–sulfur protein assembly machinery. Proc. Natl. Acad. Sci. U.S.A. 102, 3266–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Banci L., Bertini I., Calderone V., Ciofi-Baffoni S., Giachetti A., Jaiswal D., Mikolajczyk M., Piccioli M., and Winkelmann J. (2013) Molecular view of an electron transfer process essential for iron–sulfur protein biogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, 7136–7141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Netz D. J., Stümpfig M., Doré C., Mühlenhoff U., Pierik A. J., and Lill R. (2010) Tah18 transfers electrons to Dre2 in cytosolic iron–sulfur protein biogenesis. Nat. Chem. Biol. 6, 758–765 [DOI] [PubMed] [Google Scholar]
- 29. Seki M., Takeda Y., Iwai K., and Tanaka K. (2013) IOP1 protein is an external component of the human cytosolic iron–sulfur cluster assembly (CIA) machinery and functions in the MMS19 protein-dependent CIA pathway. J. Biol. Chem. 288, 16680–16689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Wietmarschen N., Moradian A., Morin G. B., Lansdorp P. M., and Uringa E. J. (2012) The mammalian proteins MMS19, MIP18, and ANT2 are involved in cytoplasmic iron–sulfur cluster protein assembly. J. Biol. Chem. 287, 43351–43358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stehling O., Vashisht A. A., Mascarenhas J., Jonsson Z. O., Sharma T., Netz D. J., Pierik A. J., Wohlschlegel J. A., and Lill R. (2012) MMS19 assembles iron–sulfur proteins required for DNA metabolism and genomic integrity. Science 337, 195–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gari K., León Ortiz A. M., Borel V., Flynn H., Skehel J. M., and Boulton S. J. (2012) MMS19 links cytoplasmic iron–sulfur cluster assembly to DNA metabolism. Science 337, 243–245 [DOI] [PubMed] [Google Scholar]
- 33. Stehling O., Mascarenhas J., Vashisht A. A., Sheftel A. D., Niggemeyer B., Rösser R., Pierik A. J., Wohlschlegel J. A., and Lill R. (2013) Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron–sulfur proteins. Cell Metab. 18, 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paul V. D., Mühlenhoff U., Stümpfig M., Seebacher J., Kugler K. G., Renicke C., Taxis C., Gavin A. C., Pierik A. J., and Lill R. (2015) The deca-GX3 proteins Yae1–Lto1 function as adaptors recruiting the ABC protein Rli1 for iron–sulfur cluster insertion. Elife 4, e08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Song J. Y., Marszalek J., and Craig E. A. (2012) Cysteine desulfurase Nfs1 and Pim1 protease control levels of Isu, the Fe-S cluster biogenesis scaffold. Proc. Natl. Acad. Sci. U.S.A. 109, 10370–10375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Balk J., Pierik A. J., Netz D. J., Mühlenhoff U., and Lill R. (2004) The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23, 2105–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Balk J., Pierik A. J., Aguilar Netz D. J., Mühlenhoff U., and Lill R. (2005) Nar1p, a conserved eukaryotic protein with similarity to Fe-only hydrogenases, functions in cytosolic iron-sulphur protein biogenesis. Biochem. Soc. Trans. 33, 86–89 [DOI] [PubMed] [Google Scholar]
- 38. Song D., and Lee F. S. (2008) A role for IOP1 in mammalian cytosolic iron–sulfur protein biogenesis. J. Biol. Chem. 283, 9231–9238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vashisht A. A., Yu C. C., Sharma T., Ro K., and Wohlschlegel J. A. (2015) The association of the xeroderma pigmentosum group D DNA helicase (XPD) with transcription factor IIH is regulated by the cytosolic iron–sulfur cluster assembly pathway. J. Biol. Chem. 290, 14218–14225 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.