Abstract
We have developed a user-friendly microfluidic device for the study of gradient-mediated bacterial behaviors, including chemotaxis. This device rapidly establishes linear concentration gradients by exploiting solute diffusion through porous membranes in the absence of convective flows. As such, the gradients are created rapidly and can be sustained for long time periods (e.g., hours), sufficient to evaluate cell phenotype. The device exploits a unique simple bilayer configuration that enables rapid setup and quick reproducible introduction of cells. Its reusability represents an additional advantage in that it need not be limited to settings with microfluidics expertise. We have successfully demonstrated the applicability of this tool in studying the chemotactic response of Escherichia coli to glucose. When coupled with our recent Python program, quantified metrics such as speed, ratio of tumble to run, and effective diffusivity can be obtained from slow frame rate videos. Moreover, we introduce a chemotaxis partition coefficient that conveniently scores swimming behavior on the single-cell level.
I. INTRODUCTION
Concentration gradients of biomolecules are found throughout the human body and play pivotal roles in regulating biological activities, including wound healing, immune responses, and cancer metastasis.1–5 Microorganisms inside the human body can produce, sense, and react to these chemical gradients in multiple ways, some involved in pathogenicity. For instance, biofilm growth of pathogenic bacteria often results in infections. Bacteria within a biofilm exhibit distinct gene expression profiles and metabolic activities in response to concentration gradients of nutrients, signaling compounds, and bacterial wastes within the biofilm.6 This heterogeneity protects cells inside the biofilm from environmental stresses and attacks from the immune system or antimicrobials, thus decreasing therapeutic efficacy.7 While often considered from the perspective of pathogenicity, some gradient-sensing behaviors could be also harnessed for therapeutic purposes. Bacteria can move up or down a chemical gradient in response to a natural attractant or repellent, which is referred to as chemotaxis.8 Leveraging this character, genetically rewired Escherichia coli (E. coli) were attracted to cancer cell lines9,10 and could serve as vectors for drug delivery. Therefore, developing a robust platform for investigation of gradient-mediated bacterial behaviors would be essential.
Over the last several decades, numerous platforms have been devised, such as the Boyden chamber,11 the Dunn slide chamber,12 and the Zigmond chamber,13 which generate chemical gradients mimicking physiological conditions. However, the reproducibility, controllability, and stability of these approaches are limited.2,14–16 To address these issues, researchers have turned to advanced microfluidic systems based on high resolution lithography and soft lithography. Because fabrication methods are programmable, precise, and facile, these approaches have enabled more detailed study of biological phenomena (e.g., morphogen-mediated development,16,17 tumor cell migration,18,19 drug delivery and distribution20). Gradient generators built within microfluidic platforms offer greater resolution and provide well-controlled hydrodynamic and mass transfer conditions.21 Additionally, microfluidics facilitate the integration and manipulation of multiple design spaces, predominately time, chemical species, and chemical concentrations into a single platform.22
Due to the length scales and well-defined stream functions, the majority of microfluidic gradient generators apply laminar flows to establish gradients.23–28 As such, parallel flows of incompressible and miscible fluids mix only by diffusion. By appropriately designing the spatial configurations and the solute concentrations in multiple adjacent streams, gradients can be established across a channel.29 This methodology is particularly suitable for surface-adherent cells as they can be exposed to a steady state gradient with small shear forces. However, these flow-based platforms are unsuitable for many bacterial studies due to two major limitations: (i) bacteria are often non-adherent (planktonic) and thus would experience unsteady concentration gradients as they are transported along the channel by the flow29 and (ii) cells in the device would experience both chemical gradients and shear forces, and because of this, it is challenging to discern the independent effects of each on cell movement.15,30 As a result, flow-free gradient generators have been proposed.8,15,31–38
A common strategy to form chemical gradients within a flow-free device relies on chemical exchange between source and sink reservoirs through flow resistive elements, such as hydrogels15,31–33 or membranes.8,34–38 Submicron-sized pores in these elements generate large hydrodynamic resistances that minimize convective flows and shear forces, leaving diffusive transport unperturbed. Subsequently, concentration gradients can be established without disturbance from flows. Compared to hydrogel-based gradient generators that require in situ fabrication, membrane-based devices are rapidly assembled from uniform components and are easily sealed and integrated with ancillary flow and analytical systems.31,38 We have found that many membrane-based gradient generators facilitate the study of mammalian cells and tissues, with their designs taking advantage of the larger length scales of cell, organoid, and/or tissue morphologies and relatively long time constants. These systems are not ideal, however, for studying motile bacteria and are generally non-reusable.34–38
Previously, Wang et al. developed a bilayer microfluidic gradient generator for bacterial study, which established linear gradients of chemicals by balancing the pressure between source and sink inlets.33 Introduction of bacteria or solutions into their device was based on diffusion, which increased experimental time (>1 h) and washing difficulty. Here, we describe a next generation bilayer membrane-based microfluidic gradient generator (Scheme 1) behavior that can be easily operated in laboratories not specialized in microfluidics. It has the following features: (1) the concentration gradient is formed by diffusion and is steady, sharp, and homogenous for accurate experimental performance; (2) the experimental setup and washing procedures are facile; (3) the device can be coupled to analytical software (e.g., similar to Mohari et al. for non-gradient systems39); and (4) it is reusable. Herein, we describe its design and assembly as well as its performance by illustrating the gradient generation and the tracking of E. coli chemotactic responses.
SCHEME 1.
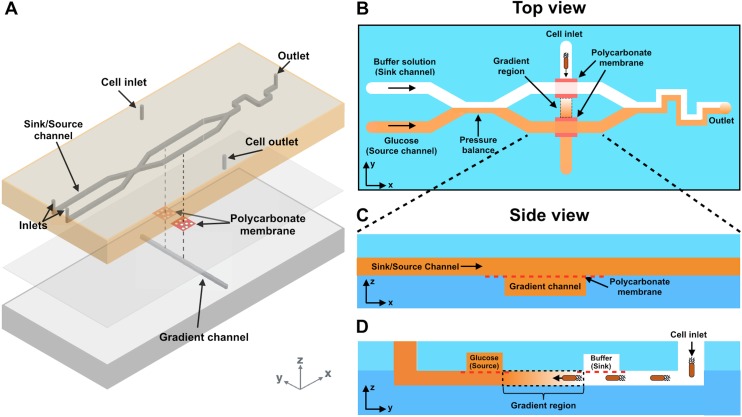
(a) A 3D rendering of a membrane-based bilayer gradient generator. (b)–(d) Schematic top and side representations of the device. Solution of a fixed concentration flows in the source channel while blank buffer flows in the sink channel. The axis that is parallel to the source and sink channels is denoted as the x axis. The convergence of these flows balances the pressure between the two channels. The top and bottom channels are separated by polycarbonate membranes. Small biomolecules and other chemicals diffuse downward through the membrane, forming an orthogonal linear gradient within the bottom channel. The cells are introduced into the cell inlet in the bottom channel. All channels are 50 μm in height and 500 μm in width.
II. MATERIALS AND METHODS
A. Device fabrication
The device consists a top layer and a bottom layer (Scheme 1). The top layer [Scheme 1(a)] contains two parallel channels that deliver either an attractant/repellent (i.e., the source channel) or a buffer solution (i.e., the sink channel). These two channels have a convergence (i.e., the pressure balance) that is structurally similar to a previous work by Irimia et al.40 This convergence dissipates pressure differences between the channels and stabilizes the concentration gradient formed downstream. The length of the convergence and the flow rates of both sink and source channels are optimized in order to avoid chemical diffusion from the source channel to the sink channel when the flow is fully developed. The bottom layer contains a single channel. All channels are 50 μm tall and 500 μm wide. From a top-down view [Scheme 1(b)], the three channels in both layers form two intersections. At each intersection, a polycarbonate membrane is sandwiched between the top and the bottom layers and is large enough to separate channels on both sides [see side views, Schemes 1(c) and 1(d)]. The membrane is porous with each pore being 1 μm in diameter and generates high hydrodynamic resistance (Rh=8 μl/πr4, r = pore radius). Convective flows transmitting through the membrane are thus negligible, and diffusive transport becomes the dominating communication between solutions in the top and the bottom channels.
All layers were made from polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning Co., Midland, MI) cast on an SU-8 master. The SU-8 master was fabricated using standard soft lithography techniques performed in the Maryland Micro and Nano Fabrication Center. Patterns on both layers were designed on AutoCAD (Autodesk, Inc., Mill Valley, CA), converted, exposed, and developed onto a Mylar mask. A 4-in. silicon wafer was cleaned with Piranha solution (concentrated H2SO4 and 30% H2O2, 3:1, v/v) and spin coated with 5 ml SU-8 3050 photoresists (MicroChem, Westborough, MA) at 3000 rpm for 30 s followed by 15 min baking at 95 °C. The pattern was then transferred from the mask to the wafer by exposing the wafer to UV light (405 nm wavelength) at 23.4 mW cm−2 using an EVG 620 mask aligner (Electronic Visions Inc., Phoenix, AZ). The wafer was subsequently developed for 8 min in an SU-8 developer (MicroChem, Westborough, MA). The residual photoresist was removed by rinsing with isopropanol and deionized (DI) water followed by air-drying. The resulting SU-8 master can be reused almost indefinitely.
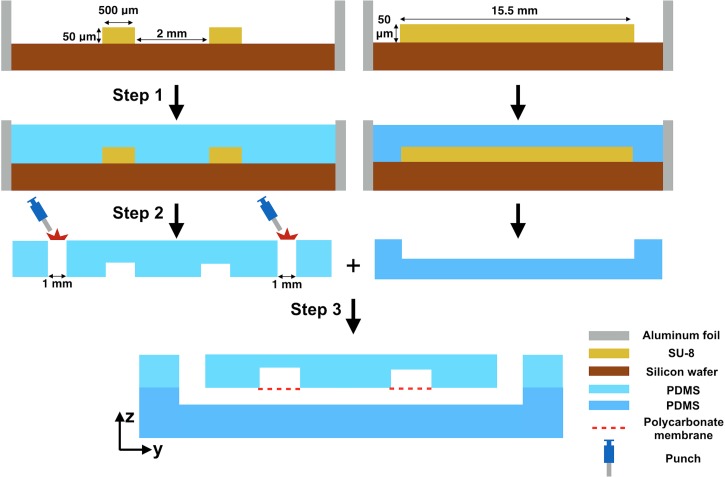
The device fabrication involved three steps (Fig. 1). First, the SU-8 master was surrounded with aluminum foil to prevent leakage. PDMS was prepared by mixing the base and the curing agent (10:1, w/w) and cast over the SU-8 master followed by baking at 60 °C for 2 h. The cured PDMS was peeled from the SU-8 master. Inlets and outlets were perforated using a 1 mm biopsy punch (Miltex, York, PA). The last step was the sandwiching of porous polycarbonate membranes (cut from Corning® transwell) between the top and the bottom PDMS layers. The membranes were first attached to the top PDMS layer at the desired positions. Then, both layers were treated with oxygen plasma (IPC 4000 series plasma system) (Branson, Philadelphia, PA). After the treatment, both layers were bonded with proper alignment.
FIG. 1.
Step-by-step device fabrication. (i) The SU-8 master on a silicon wafer is surrounded by aluminum foil. Freshly mixed PDMS (base and curing agent, 10:1 w/w) is cast onto the SU-8 master. (ii) After baking at 60 °C for 2 h, the PDMS is cooled at room temperature and then peeled from the SU-8 master. (iii) Inlets and outlets are perforated at the positions indicated. The polycarbonate membranes are then sandwiched between the top and bottom PDMS layers.
B. Gradient profile characterization
The device was characterized using fluorescein isothiocyanate (FITC) (Thermo Fisher Scientific, Rockville, MD). Initially, the device was filled with DI water using 1 ml syringes. Outlets of the bottom channel were taped to prevent flow. Flows in the top channels were driven by two independent syringe pumps (Kent Scientific, Genie Plus; Fisher Scientific, Single Syringe Pump), each at a rate of 120 μl h−1. Subsequently, buffer solution in the source channel was replaced with 1.5 × 10−5 M FITC. To avoid FITC diffusion to the sink channel at the convergent site, the pumping rate of FITC was set to a rate of 50 μl h−1. Excess solution at outlet was wiped away. The time was set to be t = 0 when FITC was loaded. A CCD camera, an inverted fluorescence microscope, a FITC filter cube, and a 20X Olympus objective lens were used to image both bright-field and fluorescent images. Images were taken every minute for a total of 100 min. ImageJ (shareware: https://imagej.nih.gov/ij/download.html) was used to analyze fluorescence intensity. The fluorescence intensity was normalized using Eq. (1)34
| (1) |
where I(t) is the intensity of pixels along the gradient at each time point, I0 is the intensity of a pixel across the gradient channel before the introduction of fluorescein, and Imax is the intensity of a pixel across the gradient channel with the highest fluorescein concentration (close to the source channel) at 100 min after starting the gradient generation. Another full characterization of the same device was repeated after 50 times of experiments/washing cycles, demonstrating reusability.
C. Bacterial chemotaxis
E. coli K-12 strain W3110 was used in all chemotaxis experiments. Cells were inoculated and grown overnight in Luria-Bertani (LB) media (Fisher, Pittsburgh, PA) with 50 μg/ml each of ampicillin and kanamycin (Sigma-Aldrich, St. Louis, MO). Cells were reinoculated from the overnight culture and grew to an OD600 of 0.45 at 37 °C, shaking at 250 rpm. Following, samples were spun at 400 g for 5 min and resuspended in freshly made chemotaxis buffer solution (CB) (1× PBS with 0.1 mM EDTA, 0.01 mM L-methionine, and 10 mM sodium DL-lactate).
Prior to bacteria introduction, the bottom channel was primed with Pluronic F-127 (1% w/v, Sigma-Aldrich, St. Louis, MO) for 1 h before replacing with CB. This minimized nonspecific retention of cells to channel walls. Chemotaxis buffer was perfused through both the source and sink channels using 1 mL syringes and syringe pumps. The pumping rate was 120 μl h−1 for both channels. E. coli at an OD600 between 0.7 and 1.0 (in the late exponential growth phase) was introduced into the bottom channel with the same volume for each test. Both ends of the bottom channel were then taped to stop flow. Glucose solution (1 mM D-glucose (Sigma) in CB) was then introduced at a rate of 50 μl h−1 to replace CB in the source channel. The time when glucose was added was set to be t = 0. Bright-field images were taken at 0, 10, 20, and 30 min by a 20× Olympus objective. Cell numbers in each image were counted using ImageJ. The device was washed by manually pumping buffer solution through the system.
D. Single-cell analysis
Twelve-second videos were taken in the center of the bottom channel (∼350 μm wide in the y-axis/∼300 μm wide in the x-axis) at 0, 10, 20, and 30 min by a 20× Olympus objective. Bacteria in each video were detected and tracked using a single-cell, analytic program developed in Python by our group (freely available under request), which was a modified version of the Trackpy code.35 We used Trackpy's core modules for finding and tracking bacteria, whereas the analysis of the measurements was performed using custom-built modules developed specifically to the purposes of this work. The detection of features using Trackpy depended essentially on two parameters: brightness and size. In order to detect as many features as possible in each frame of the dataset, we tested different values and combinations for both parameters and then visually inspected the results until most of the features present in the frame were detected (including artifacts created by the optical system). We found the best results when adopting values of 500 and 15 (in Trackpy's units) for the brightness and size, respectively, and no significant improvement could be achieved by varying these parameters. Based on the displacement of the features between each pair of frames, we were able to filter the results and classify the features into two categories: artifacts (no significant displacement between frames) and bacteria (displacement larger than 1.5 μm between frames). The latter was further divided into two classes: bacteria that were moving and bacteria that were not (stuck in the same place). For the results presented in this paper, we only took into consideration bacteria that were actually moving and tracked for more than 1 s (or about 10 frames with our instrumental setup). The directionality of each bacterium was determined based on the displacement direction along the y-axis comparing the initial and the final time points. The chemotaxis partition coefficient (CPC) was calculated using Eq. (2)
| (2) |
where Ny+ is the total number of cells moving towards the positive y direction (towards glucose), while Ny- is the total number of cells moving towards the negative y direction (away from glucose). In the control groups for all time points, both delivering channels were loaded with CB.
E. Statistical analysis
The goodness of fit of regression models was determined based on the coefficient of determination. The level to accept a model was set at R2 ≥ 0.95. One-way ANOVA was used to determine significant differences between groups. The level of significance was set at α = 0.05.
III. RESULTS AND DISCUSSION
A. Device characterization
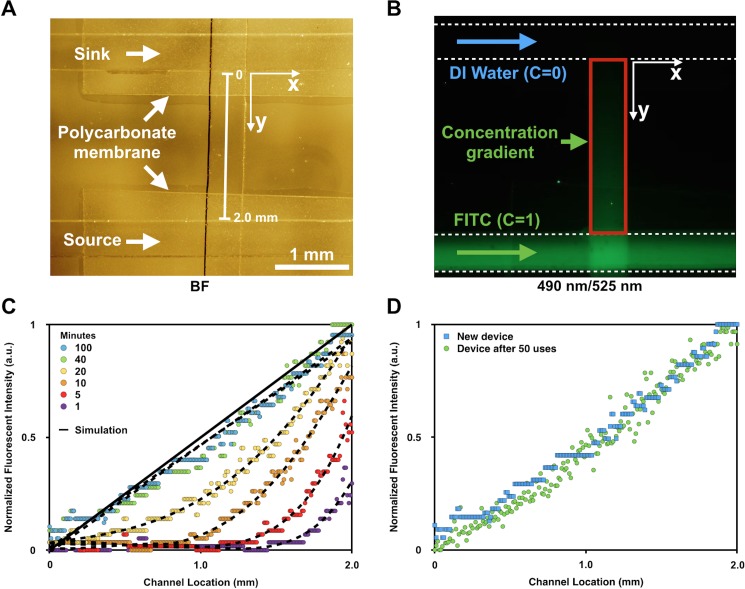
The device was characterized using FITC. This method was based on the observation that the fluorescent light intensity emitted by fluorescein solutions in a microfluidic channel (depth 100 μm) was linearly proportional to the fluorescein concentration. This linearity held when the fluorescein concentration was less than 10−3 M.15 Based on this information, we chose a stock 1.5 × 10−5 M FITC solution for all experiments. Figure 2(a) is a representative bright-field image of both delivery channels in the top layer aligned perpendicularly to the channel in the bottom layer. Two polycarbonate membranes are readily visualized at both intersections. Figure 2(b) depicts a fluorescence image (ex/em: 490/525 nm) in which DI water and FITC were perfused in parallel in the x direction. A FITC gradient is readily seen emanating from the bottom channel, as indicated in the red box.
FIG. 2.
Fluorescence characterization of the concentration gradient within the lower channel. Bright-field (a) and fluorescence (b) images show the bottom channel flanked by parallel sink (c = 0) and source (c = 1) channels. FITC and DI water are flowed into each channel at a constant rate (b). The FITC is detected using an excitation wavelength of 490 nm and an emission filter of 525 nm. The red box (b) marks the region over which the gradient is established. (c) Fluorescence intensity profiles are plotted along the y-axis, the length indicated in (a), for 100 minutes at various increments as noted. A finite element simulated steady state profile is also included. (d) Fluorescence intensity profiles of a freshly made device and an old device (used 50 times) are overlaid. C: Concentration of FITC.
Figure 2(c) depicts the quantified fluorescence intensity of FITC as a function of distance from the sink channel to the source channel (2 mm apart). All data were normalized to the fluorescence intensity at the source (i.e., a scale from 0 to 1). All datasets were least squares fitted by 4th-order polynomial functions. It was not until 40 minutes that the gradient profile could be interpreted as a linear and steady function (R2 = 0.95). We evaluated the long-term stability of the gradient and found that it was maintained at least until 100 min, a period long beyond our need. The steady state gradient profiles displayed no significant differences with results from finite element modeling (Comsol Multiphysics). Due to the linear relationship between fluorescence and fluorescein concentration and the similarity between diffusion coefficients of FITC in water (D = 0.64 × 10−9 m2 s−1)36 and glucose in buffer solution (D = 0.72 × 10−9 m2 s−1),37 we surmised that glucose concentration gradients at different time points could be estimated based on the already measured FITC fluorescent gradient profiles. Also, after 50 repetitions of chemotaxis experiments, the device was re-evaluated and characterized using the same gradient protocols. The profile after 50 uses was not significantly different than that from the freshly made device [Fig. 2(d)].
B. Chemotaxis demonstration
We next demonstrated the chemotactic response of E. coli on both population and single-cell levels. We first examined the bacteria's spatial population distribution under glucose stimuli. Bright-field images of cells in the channel were taken every 10 min at two positions along the bottom channel: position a was close to the sink channel, whereas position b was close to the source channel [Fig. 3(a)]. It should be noticed that the polycarbonate membrane blocked cells beneath from being observed visually. Therefore, positions a and b were not chosen at exactly C = 0 (sink) or C = 1 (source). Instead, they were shifted slightly towards the channel center (≈1.36 mm apart) to ensure that cells were properly viewed. As a result, the glucose concentration at position a was higher than 0 and that at position b was lower than 1 mM at steady state. Cell numbers in a and b were normalized as
| (3) |
| (4) |
where Na and Nb are the total number of cells in a and b, respectively.
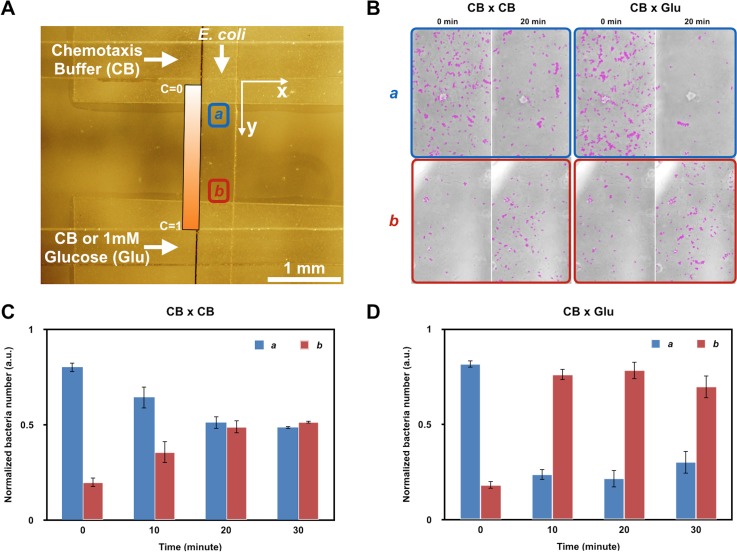
FIG. 3.
Bacterial chemotaxis experiment. (a) Bright-field image indicating the bottom channel (top to bottom) flanked by parallel sink and source channels (horizontally aligned). Imaging positions a and b are selected along the bottom channel but outside the region of the permeable polycarbonate membrane. Position a is closer to the sink channel (CB), while position b is closer to the source channel (Glu). (b) Bright-field images of positions a and b at 0 and 20 min. Cells are marked with purple circles using Imagej TrackMate software. CB × CB: the control group in which chemotaxis buffer (CB) is flowed in both sink and source channels. CB × Glu: the experimental group in which CB in the source channel is replaced with 1 mM glucose in CB. Quantitative results of (b) are shown in (c) and (d). The cell number is normalized to the total cell number in both imaging areas. Data are collected at 0, 10, 20, and 30 min. CB: chemotaxis buffer. Glu: glucose. C: concentration of glucose.
Before glucose addition, cells were unevenly distributed in the channel: the initial cell concentration was much higher at position a than at position b [Fig. 3(b)]. This was due to the loading procedure and importantly can be accommodated in our design. In the control group where both sink and source channels were loaded with chemotaxis buffer, the cell numbers at positions a and b became closer at 10 min and were nearly equal at 20–30 min [Fig. 3(c)]. This result was due to the cell's random dispersion from high cell density areas to low-density areas in the absence of a chemoattractant so that the cell concentration eventually became homogeneous throughout the channel. In contrast, in the experimental group where glucose was introduced at the source, the population distribution changed dramatically, as noticed in the first sample just 10 min into the experiment. The number of bacteria at position b reached nearly three times that of position a. This was remarkable and statistically significant. Interestingly, this population distribution remained the same for the full 30 minutes [Fig. 3(d)]. This result was in accord with the expectation that in the presence of a chemoattractant gradient, cells sensed stimuli and altered the direction of rotation of their flagella to improve their chances of migrating towards the higher concentration of attractant.38 The fact that chemotaxis occurred quicker than the time required for establishing a steady state was due to the minimum glucose concentrations necessary to induce chemotaxis (1 × 10−6 M) being less than the concentrations available to cells in a steady state gradient.39
C. Single-cell analysis
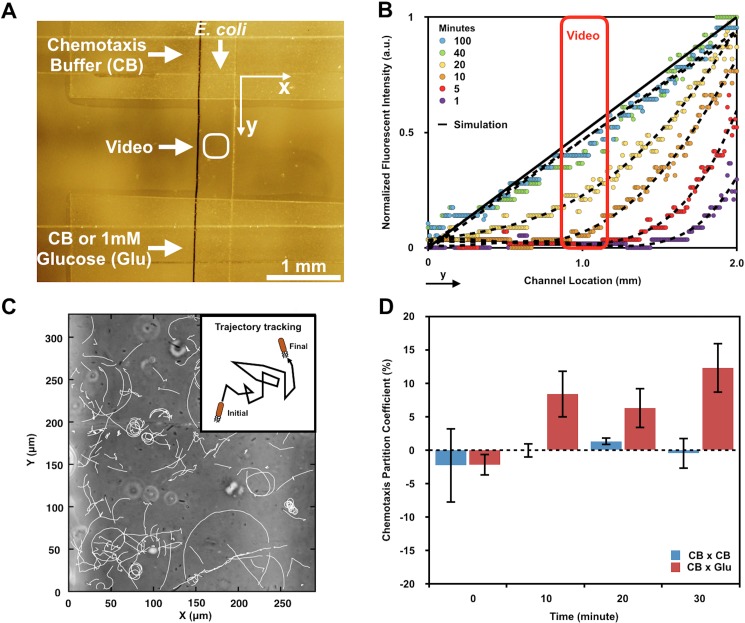
In addition to observing changes in the overall population distribution, we analyzed bacterial chemotactic behavior on the single-cell level by video tracking cells in the center of the bottom channel [Figs. 4(a) and 4(b)]. At every 10 min increment, 12 s videos were recorded and analyzed, as described above. The trajectory of every moving target in a video was marked by a white tail [Fig. 4(c)]. To further quantify the degree of chemotaxis, we calculated a chemotaxis partition coefficient (CPC), a parameter that represented the difference between cell populations that moved towards the chemoattractant and the opposite direction, for each video.24 A CPC of 1 or −1 suggested an absolute net directionality towards or away from the chemoattractant, while a CPC of 0 indicated no net directionality.
FIG. 4.
Single cell analysis. (a) Bright-field image indicating the bottom channel (top to bottom) flanked by parallel sink and source channels (horizontally aligned). Videos are taken at the center of the gradient channel as indicated. Each video scope is about 350 μm wide on the y-axis and 300 μm wide on the x-axis. (b) Fluorescence intensity profiles of FITC are used to predict glucose concentration gradient profiles. The data inside the red box indicate the regional glucose gradient profiles within the video scope. (c) Cells in the video are tracked by a Python single cell analytics program, which generates a trajectory tail after each captured cell in a 11.7 s tracking. (d) The chemotaxis partition coefficient (CPC) of both experimental (CB × Glu) and control (CB × CB) groups is calculated at 0, 10, 20, 30 min. CB: chemotaxis buffer. Glu: glucose.
In both control and experimental groups, bacteria showed small directionalities (CPC < 0.05) at t = 0 [Fig. 4(d)]. After 10 min, however, bacteria in the experimental groups exhibited nearly 10 times stronger directionalities towards glucose than the control groups. This difference remained at this level for 20 min and eventually grew to ∼15× by 30 min. These results were in agreement with the regional concentration gradient profiles exhibiting over these time domains within the video, as depicted in the time resolved gradient profiles in Fig. 4(b). That is, at t = 0, there was no concentration gradient within the experiment during the course of the video. Thus, at early times (e.g., 1 and 5 min) bacteria swam randomly and barely exhibited net directionality. As the glucose concentration gradient gradually became established [concentration profiles transitioned to gold, yellow, and green in Fig. 4(b)], bacteria presumably upregulated chemotaxis machinery and moved towards the source, as demonstrated by significantly increased net directionalities. We observe increased CPC values at 10 minutes when the maximum glucose concentration in the video scope reached ∼1 × 10−4 M (not shown). These results are consistent with a minimum D-glucose concentration needed to trigger a chemotactic response of E. coli (∼1 × 10−6 M).41
IV. CONCLUSIONS
In summary, we have demonstrated that this bilayer membrane-based gradient generator is able to establish stable and sharp concentration gradients in a user-friendly manner. In this device, bacterial introduction and experimental setup can be completed in a simple and quick manner. Furthermore, the device can be reused for at least 50 experiments or as long as the polycarbonate membranes remain intact. Therefore, multiple experiments can be conducted continuously within a short time period. Based on our fluorescence characterization, the gradient evolves to a steady state in less than 40 min and lasts for at least 100 min. Using this device, we have studied the E. coli chemotactic response on both population and single-cell levels. We envision that this gradient generator will be amenable for use in microbiological laboratories with only limited access to microfluidics (just need a microscope and a syringe pump).
We envision this device can also be reconfigured to suit various purposes owing to its unique bilayer structure. For instance, the bottom channel could be replaced by modalities that immobilize planktonic bacteria, thereby resembling biofilms. Such a modification would enable the study of bacterial genetic responses to signal molecules of various concentrations (e.g., autoinducer10) and broaden the range of possible biofilm studies. Also, by including an accessible cell reservoir adjacent to the lower channel, one could subject cells to a steady gradient (e.g., by exposing cells after 30 min). Further, by integrating modalities that enable co-culture of bacteria with mammalian cells in the bottom channel, this device could find utility in investigating inter-kingdom communication under various chemical stimuli. These, in turn, will enable a more detailed study of human disease. Electrochemical and other biosensors may also be integrated into the system to interrogate biochemical information and otherwise monitor cellular metabolic activities in situ and in real-time.42,43 Overall, these contributions advancing both microfluidic technology and in vitro gradient generation will prove beneficial to further investigations of cell and tissue biology.
ACKNOWLEDGMENTS
The authors wish to thank Hana Ueda for helpful discussions. This work was supported with funds from National Science Foundation (CBET #1160005 and DMREF #1435957) and Defense Threat Reduction Agency (HDTRA1-13-0037).
References
- 1. Servant G., Weiner O. D., Herzmark P., Balla T., Sedat J. W., and Bourne H. R., Science (80-) 287, 1037 (2000). 10.1126/science.287.5455.1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lin F., Nguyen C. M. C., Wang S. J., Saadi W., Gross S. P., and Jeon N. L., Ann. Biomed. Eng. 33, 475 (2005). 10.1007/s10439-005-2503-6 [DOI] [PubMed] [Google Scholar]
- 3. Friedl P. and Weigelin B., Nat. Immunol. 9, 960 (2008). 10.1038/ni.f.212 [DOI] [PubMed] [Google Scholar]
- 4. O'Hayre M., Salanga C. L., Handel T. M., and Allen S. J., Biochem. J. 409, 635 (2008). 10.1042/BJ20071493 [DOI] [PubMed] [Google Scholar]
- 5. Yang J. and Weinberg R. A., Dev. Cell 14, 818 (2008). 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 6. Stewart P. S. and Franklin M. J., Nat. Rev. Microbiol. 6, 199 (2008). 10.1038/nrmicro1838 [DOI] [PubMed] [Google Scholar]
- 7. Solano C., Echeverz M., and Lasa I., Curr. Opin. Microbiol. 18, 96 (2014). 10.1016/j.mib.2014.02.008 [DOI] [PubMed] [Google Scholar]
- 8. Diao J., Young L., Kim S., Fogarty E. a., Heilman S. M., Zhou P., Shuler M. L., Wu M., and DeLisa M. P., Lab Chip 6, 381 (2006). 10.1039/B511958H [DOI] [PubMed] [Google Scholar]
- 9. Forbes N. S., Nat. Rev. Cancer 10, 785 (2010). 10.1038/nrc2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu H., Tsao C., Quan D. N., Cheng Y., Servinsky M. D., Carter K. K., Jee K. J., Terrell J. L., Zargar A., Rubloff G. W., Payne G. F., Valdes J. J., and Bentley W. E., Mol. Syst. Biol. 9, 636 (2013). 10.1038/msb.2012.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boyden B. S., J. Exp. Med. 115, 453 (1962). 10.1084/jem.115.3.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zicha D., Dunn G. A., and Brown A. F., J. Cell Sci. 99, 769 (1991). [DOI] [PubMed] [Google Scholar]
- 13. Zigmond S. H., J. Cell Biol. 75, 606 (1977). 10.1083/jcb.75.2.606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Keenan T. M., Hsu C.-H., and Folch A., Appl. Phys. Lett. 89, 114103 (2006). 10.1063/1.2345914 [DOI] [Google Scholar]
- 15. Cheng S.-Y., Heilman S., Wasserman M., Archer S., Shuler M. L., and Wu M., Lab Chip 7, 763 (2007). 10.1039/b618463d [DOI] [PubMed] [Google Scholar]
- 16. Amadi O. C., Steinhauser M. L., Nishi Y., Chung S., Kamm R. D., McMahon A. P., and Lee R. T., Biomed. Microdevices 12, 1027 (2010). 10.1007/s10544-010-9457-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shin Y., Han S., Jeon J. S., Yamamoto K., Zervantonakis I. K., Sudo R., Kamm R. D., and Chung S., Nat. Protocol 7, 1247 (2012). 10.1038/nprot.2012.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang Y., Agrawal B., Sun D., Kuo J. S., and Williams J. C., Biomicrofluidics 5, 013412 (2011). 10.1063/1.3555195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stroka K. M., Jiang H., Chen S. H., Tong Z., Wirtz D., Sun S. X., and Konstantopoulos K., Cell 157, 611 (2014). 10.1016/j.cell.2014.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Z., Samanipour R., Koo K., and Kim K., Sens. Mater. 27, 487 (2015). [Google Scholar]
- 21. Whitesides G. M., Nature 442, 368 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- 22. Toh A. G. G., Wang Z. P., Yang C., and Nguyen N. T., Microfluid. Nanofluid. 16, 1 (2014). 10.1007/s10404-013-1236-3 [DOI] [Google Scholar]
- 23. Jeon N. L., Baskaran H., Dertinger S. K. W., Whitesides G. M., Van L., Water D., and Mehmet T., Nat. Biotechnol. 20, 826 (2002). 10.1038/nbt712 [DOI] [PubMed] [Google Scholar]
- 24. Mao H., Cremer P. S., and Manson M. D., Proc. Natl. Acad. Sci. U. S. A. 100, 5449 (2003). 10.1073/pnas.0931258100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Irimia D. and Toner M., Lab Chip 6, 345 (2006). 10.1039/b515983k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saadi W., Wang S., Francis L., and Jeon N. L., Biomed. Microdevices 8, 109 (2006). 10.1007/s10544-006-7706-6 [DOI] [PubMed] [Google Scholar]
- 27. Englert D. L., Manson M. D., and Jayaraman A., JOVE 38, e1779 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seymour J. R., Marcos, and Stocker R., Am. Nat. 173, E15 (2009). 10.1086/593004 [DOI] [PubMed] [Google Scholar]
- 29. Ahmed T., Shimizu T. S., and Stocker R., Integr. Biol. 2, 604 (2010). 10.1039/c0ib00049c [DOI] [PubMed] [Google Scholar]
- 30. Kim K. P., Kim Y.-G., Choi C.-H., Kim H.-E., Lee S.-H., Chang W.-S., and Lee C.-S., Lab Chip 10, 3296 (2010). 10.1039/c0lc00154f [DOI] [PubMed] [Google Scholar]
- 31. Ahmed T., Shimizu T. S., and Stocker R., Nano Lett. 10, 3379 (2010). 10.1021/nl101204e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang C., Jang S., Amadi O. C., Shimizu K., Lee R. T., and Mitchell R. N., BioMed Res. Int. 2013, 373569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang X., Atencia J., and Ford R. M., Biotechnol. Bioeng. 112, 896 (2015). 10.1002/bit.25497 [DOI] [PubMed] [Google Scholar]
- 34. Abhyankar V. V., Lokuta M. a., Huttenlocher A., and Beebe D. J., Lab Chip 6, 389 (2006). 10.1039/b514133h [DOI] [PubMed] [Google Scholar]
- 35. Kim T., Pinelis M., and Maharbiz M. M., Biomed. Microdevices 11, 65 (2009). 10.1007/s10544-008-9210-7 [DOI] [PubMed] [Google Scholar]
- 36. Mosadegh B., Agarwal M., Tavana H., Bersano-Begey T., Torisawa Y., Morell M., Wyatt M. J., O'Shea K. S., Barald K. F., and Takayama S., Lab Chip 10, 2959 (2010). 10.1039/c0lc00086h [DOI] [PubMed] [Google Scholar]
- 37. Cate D. M., Sip C. G., and Folch A., Biomicrofluidics 4, 44105 (2010). 10.1063/1.3490784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sip C., Nirveek B., and Albert F., Lab Chip 14, 302 (2014). 10.1039/C3LC51052B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mohari B., Licata N. A., Kysela D. T., Merritt P. M., Mukhopadhay S., Brun Y. V., Setayeshgar S., and Fuqua C., MBio 6, e00005 (2015). 10.1128/mBio.00005-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Irimia D., Charras G., Agrawal N., Mitchison T., and Toner M., Lab Chip 7, 1783 (2007). 10.1039/b710524j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adler J., Hazelbauer G. L., and Dahl M. M., J. Bacteriol. 115, 824 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim E., Leverage W. T., Liu Y., White I. M., Bentley W. E., and Payne G. F., Analyst 139, 32 (2014). 10.1039/C3AN01632C [DOI] [PubMed] [Google Scholar]
- 43. Kim E., Liu Y., Bentley W. E., and Payne G. F., Adv. Funct. Mater. 22, 1409 (2012). 10.1002/adfm.201101946 [DOI] [Google Scholar]