Abstract
Canine piroplasmosis is a significant disease in dogs caused by Babesia and Theileria parasites. The clinical manifestations range from mild illness to serious disease depending on the parasite species and the physical condition of the infected dog. Canine piroplasmosis has been reported to be prevalent in China. However, no molecular evidence of the disease has been reported in pet dogs from Wuhan. In this study, 118 blood samples were randomly collected from pet dogs in veterinary clinics. The blood samples were subjected to both microscopic examination and reverse line blot (RLB) hybridization assays to detect piroplasm infection. Parasites were observed in 10 blood samples via microscopic examination, whereas there were 14 Babesia gibsoni-positive RLB tests. Phylogenetic analysis was performed after the 18S rRNA and ITS gene sequences from the 14 positive samples were cloned and sequenced. The results confirmed the existence of B. gibsoni in this area. This is the first molecular report of canine babesiosis in pet dogs from Wuhan, China. Pet dogs are companion animals, and the prevalence of babesiosis will be of concern in daily life. This study will help veterinarians better understand the prevalence of canine babesiosis and provide a guide for disease control in pet dogs.
Keywords: Babesia gibsoni, babesiosis, reverse line blot, 18S rRNA, pet dog, companion animal
Introduction
Piroplasmosis is a serious disease caused by an intracellular hemoprotozoan with a worldwide distribution. It can infect animals as well as humans (Service, 2001; Solano-Gallego and Baneth, 2011; Schnittger et al., 2012). Canine piroplasmosis is one of the most important tick-borne infectious diseases. It is now regarded as a common and significant disease of dogs because several different species have been identified (Yisaschar-Mekuzas et al., 2013). The clinical manifestations range from mild illness to serious disease, depending on the infecting parasite species and the nutritional status, age, and immune condition of the dog (Muhlnickel et al., 2002; Schoeman, 2009; Schnittger et al., 2012). Typical symptoms include fever, anemia, pallor, jaundice, hemoglobinuria, splenomegaly, and weakness (Beck et al., 2009; Bajer et al., 2014).
There are eight Babesia and Theileria species that infect canines, which are classified as four large and four small species (Kjemtrup et al., 2006; Schoeman, 2009; Yisaschar-Mekuzas et al., 2013). The large species are Babesia canis vogeli, Babesia canis canis, Babesia canis rossi, and Babesia sp. (unnamed) which was identified in dogs in North Carolina (Zahler et al., 1998; Carret et al., 1999; Irwin, 2009). B. gibsoni is a small piroplasm that is distributed worldwide. Three other small piroplasms are Babesia conradae, Babesia vulpes, and Theileria sp. (unnamed; Kjemtrup et al., 2006; Matjila et al., 2008; Baneth et al., 2015). As tick-transmitted parasites, the prevalence of piroplasmosis depends on the distribution of the transmitted tick vectors. However, dog bites, blood transfusions, and transplacental transmission may represent alternative routes of transmission (Fukumoto et al., 2005; Vichova et al., 2014).
According to the records of the Chinese Center for Disease Control and Prevention, there were 130 million dogs in China in 2012. In Wuhan city, the human population is more than 12,000,000, and there are ~1,000,000 dogs. The south and east regions of China are the most endemic regions for these parasites (Wei et al., 2012; Chen et al., 2014). In Shanghai, the seroprevalence of B. gibsoni was determined to be 9.23% via indirect ELISA (Cao et al., 2015). Yao et al. reported that B. gibsoni is the main species responsible for canine babesiosis in Nanjing (Yao et al., 2014). In Jiangxi, the rates of positivity for B. canis vogeli and B. gibsoni are 4.94 and 2.47%, respectively, as tested by species-specific PCR (Zheng et al., 2017). In 2017, Niu et al. first reported the identification of Theileria sinensis in pet dogs from Gansu province of China, providing the first report of T. sinensis in dogs worldwide (Niu et al., 2017).
However, there have been no reports describing canine piroplasmosis in Wuhan, China. Therefore, the aim of this study was to investigate the occurrence of piroplasma infection in pet dogs in Wuhan.
Materials and methods
Sample collection
According to official information from the Wuhan Animal Health Inspection Institute, China, there are 89 veterinary clinics and hospitals in Wuhan, China. In the present study, a total of 118 blood samples from pet dogs were randomly collected from five clinics with the permission of the dogs' owners. All samples were screened via both microscopic examination and reverse line blot (RLB) hybridization assays at the College of Veterinary Medicine of Huazhong Agricultural University.
DNA extraction
Genomic DNA was extracted from 200 μl of EDTA anti-coagulated blood using the TIANamp Genomic DNA Kit (TransGen Biotech, Beijing, China) according to the manufacturer's instructions. The concentrations of the extracted DNA were measured with a NanoDrop 2000 (Thermo Scientific, USA). The isolated DNA samples were used immediately or stored at −20°C.
Reverse line blot hybridization assay
A pair of primers, RLB-F2 (5′-GAC ACA GGG AGG TAG TGA CAA G-3′) and RLB-R2 (5′-biotin-CTA AGA ATT TCA CCT CTG ACA GT-3′; Nijhof et al., 2003, 2005), was used to amplify the V4 variable region of the 18S rRNA gene of both Babesia and Theileria. Touchdown PCR was performed in a total volume of 25 μl, containing 2.5 μl of 10 × PCR buffer, 2 μl of 2.5 mM dNTP Mixture, 0.1 μM each primer, 0.3 μl of 5 U/μl rTaq polymerase (Takara Biotechnology, China), 2.5 μl of extracted genomic DNA, and double distilled water. As positive and negative controls, we used genomic DNA from B. orientalis that was stored in our laboratory and RNase-free water, respectively.
Oligonucleotide probes (Table 1) containing an N-terminal N-(trifluoracetamidohexyl-cyanoethyl, N,N-diisopropyl phosphoramidite [TFA])-C6 amino linker were synthesized by Augct (Beijing, China). Six known canine piroplasms (B. canis, B. vogeli, B. rossi, B. gibsoni, B. conradae, and B. vulpes) and related Babesia and Theileria species were targeted by these probes. An RLB hybridization assay was then conducted as previously described (Gubbels et al., 1999). Briefly, a Biodyne C membrane was activated at room temperature using 16% (wt/wv) 1-ethyl-3-(3-dimethyl-amino-propyl) carbodiimide (EDAC) (Sigma, USA) for 10 min, after which the oligonucleotide probes were covalently linked to the membrane at optimal concentrations (Table 1) in 0.5 M NaHCO3 for 1 min in a miniblotter. The membrane was subsequently inactivated by 100 mM NaOH for 8 min after washing in 2 × SSPE/0.1% SDS at 60°C for 5 min and then either directly used or stored at 4°C in 20 mM EDTA, pH 8.0. For the assays, 10 μl of PCR product was added to 140 μl of 2 × SSPE/0.1% SDS after denaturing at 100°C for 10 min, followed by immediate cooling on ice. The denatured PCR products were then added to the miniblotter, which was a pre-prepared Biodyne C membrane, and hybridized at 42°C for 60 min. The membrane was subsequently washed twice in preheated 2 × SSPE/0.5% SDS at 50°C for 10 min, incubated for 30 min at 42°C in 2 × SSPE/0.5% SDS with 2.5 μl of streptavidin-POD conjugate (Roche Diagnostic, Germany), washed twice in preheated 2 × SSPE/0.5% SDS at 42°C for 10 min, and finally washed twice in 2 × SSPE for 5 min at room temperature. Hybridization detection was performed using chemiluminescence.
Table 1.
Oligonucleotide RLB probes used in the study and their references.
| Probe | Sequence (5′-3′) | Concentration (pmol) | References |
|---|---|---|---|
| Babesia canis vogeli | AGC GTG TTC GAG TTT GCC | 200 | Matjila et al., 2008 |
| Babesia gibsoni | CAT CCC TCT GGT TAA TTT G | 200 | Matjila et al., 2007 |
| Babesia canis canis | TGC GTT GAC CGT TTG AC | 200 | Matjila et al., 2008 |
| Babesia canis rossi | CGG TTT GTT GCC TTT GTG | 100 | Matjila et al., 2008 |
| Babesia conradae | CGT TCC CTT CGG GGC | 200 | Yisaschar-Mekuzas et al., 2013 |
| Babesia orientalis | CCT CTT TTG GCC GTC TCA CT | 400 | He et al., 2012 |
| Babesia occultans | CCT CTT TTG GCC CAT CTC GTC | 400 | He et al., 2012 |
| Babesia bigemina | CGT TTT TTC CCT TTT GTT GG | 100 | Gubbels et al., 1999 |
| Babesia ovis | TGC GCG CGG CCT TTG CGT T | 100 | Schnittger et al., 2004 |
| Babesia bovis | CAG GTT TCG CCT GTA TAA TTG AG | 100 | Gubbels et al., 1999 |
| Babesia sp. (xinjiang) | GCG GGT TTC GTC TAC TTC GCT TTG T | 400 | He et al., 2012 |
| Babesia sp. (sable) | GCG TTG ACT TTG TGT CTT TAGC | 400 | Oosthuizen et al., 2008 |
| Babesia microti | GAC TTG GCA TCT TCT GGA | 400 | Nijhof et al., 2003 |
| Babesia crassa catch-all | GTT GGC TTA TCT TTT TAC TTT | 100 | Schnittger et al., 2004 |
| Babesia genus-specific 1 | ATT AGA GTG TTT CAA GCA GAC | 100 | Bhoora et al., 2009 |
| Babesia genus-specific 2 | ACT AGA GTG TTT CAA ACA GGC | 100 | Bhoora et al., 2009 |
| Babesia/Theileria genus specific | TAA TGG TTA ATA GGA RCR GTT G | 100 | Gubbels et al., 1999 |
| Theileria genus specific | ATT AGA GTG CTC AAA GCA GGC | 200 | He et al., 2012 |
| Theileria lestoquardi | CTT GTG TCC CTC CGG G | 400 | Schnittger et al., 2004 |
| Theileria taurotragi | TCT TGG CAC GTG GCT TTT | 400 | Gubbels et al., 1999 |
| Babesia vulpes | CTT ATC ATT AAT TTC GCT TCC GAA CG | 400 | Yisaschar-Mekuzas et al., 2013 |
| Theileria annulata | CCT CTG GGG TCT GTG CA | 400 | Gubbels et al., 1999 |
| Theileria ovis | TTG CTT TTG CTC CTT TAC GAG | 400 | Schnittger et al., 2004 |
| Theileria mutans | CTT GCG TCT CCG AAT GTT | 400 | Gubbels et al., 1999 |
| Theileria orientalis 1 | GGC TTA TTT CGG ATG ATA CTT GT | 400 | He et al., 2012 |
| Theileria orientalis 2 | GGC TTA TTT CGG ATG ATA CTT GT | 400 | He et al., 2012 |
| Theileria sp. (buffalo) | CAG ACG GAG TTT ACT TTG T | 400 | Oura et al., 2004 |
| Theileria buffeli | GGC TTA TTT CGG WTT GAT TTT | 400 | Gubbels et al., 1999 |
| Theileria sinensis | TCG CAT CTC TTG CTG AGT GC | 400 | He et al., 2012 |
Cloning and sequencing of 18S rRNA genes and its regions
The partial 18S rRNA gene and the ITS region were amplified from 14 samples that tested positive via RLB using the primer pairs P1/P2 and ITSF/ITS2, respectively (Table 2). PCR amplification of the 18S rRNA gene and ITS sequences was performed in a total volume of 50 μl, with 10 μl of 5 × TransStart FastPfu Buffer, 5 μl of a 2.5 mM dNTP Mixture, 0.1 μM of each primer, 1 μl of TransStart FastPfu DNA Polymerase (Takara Biotechnology, Dalian, China), 2.5 μl of genomic DNA, and double distilled water. The conditions for PCR amplification of the 18S rRNA gene were as follows: an initial denaturation step at 95°C for 2 min; 35 cycles of denaturation for 20 s at 95°C, annealing for 20 s at 55°C, and extension for 45 s at 72°C; and a final extension step of 5 min at 72°C. The PCR amplification conditions for the ITS region were almost the same as for the 18S rRNA gene, except that the annealing temperature used was 54°C in this step. The PCR products were purified using the Easypure Quick Gel Extraction Kit (TransGen Biotech, Beijing, China). The purified amplicons were cloned into the pMD19-T vector (Takara Biotechnology, China), which was then transformed into E. coli JM109 cells (TaKaRa Biotechnology, China) according to the manufacturer's instructions. Three positive colonies of each sample were selected for sequencing (ABI PRISM 377 DNA sequencer).
Table 2.
Primers used to amplify the 18S rRNA gene and ITS region.
| Primer | Sequence | Amplicon size (bp) |
|---|---|---|
| P1 | 5′-AACCTGGTTGATCCTGCCAGTAGTCAT-3′ | 1,700 |
| P2 | 5′-GAT CCT TCT GCA GGT TCA CCT AC-3′ | |
| ITS F | 5′-GAGAAGTCGTAACAAGGTTTCCG-3′ | 1,100 |
| ITS 2 | 5′-ACAATTTGCGTTCAATCCCA-3′ |
Phylogenetic analysis
The obtained 18S rRNA and ITS sequences were subjected to BLAST analysis in the GenBank database. Multiple sequence alignment with related genes was conducted using MAFFT (version 7) (Katoh and Frith, 2012), and the alignment was edited with BioEdit (version 7.0.9). Phylogenetic trees based on the 18S rRNA and ITS nucleotide sequences were constructed using MEGA6 software (Tamura et al., 2013). All analyses were performed with 1,000 bootstrap replications.
Ethics statement
This study was approved by the Scientific Ethic Committee of Huazhong Agricultural University (permit number HZAUDO-2014-006). All pet dogs were handled in accordance with the Animal Ethics Procedures and Guidelines of the People's Republic of China. All samples were collected under the permission of the pet dogs' owners.
Results
Microscopy and RLB results
All samples (n = 118) were screened via both microscopy and RLB. Babesia-like parasites were observed in 10 samples through microscopic examination (Figure 1). The clinical records showed that those 10 dogs had fever, anemia, pallor, and even hemoglobinuria. Fourteen samples, including the 10 microscopy-positive samples, tested positive for B. gibsoni by RLB.
Figure 1.
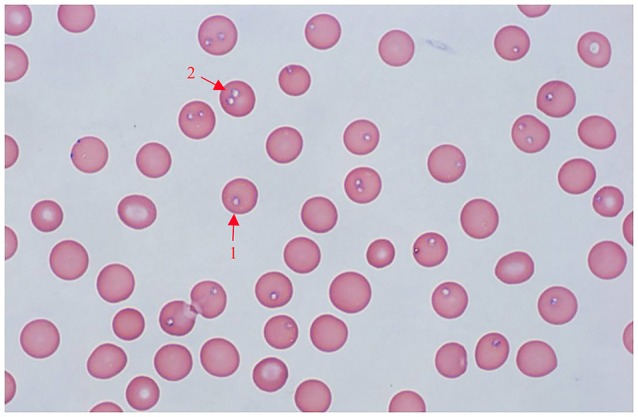
Giemsa-stained thin blood smear of Babesia gibsoni in dog erythrocytes. Final magnification is 1000X, oil. 1, Single pyriform; 2, double pyriform.
18S rRNA and its sequencing
The nucleotide sequences of the 18S rRNA and ITS genes obtained in this study were submitted to GenBank under accession numbers KP666155-KP666168 and KP666141-KP666154, respectively.
Nucleotide sequence analysis
Blast analysis showed that the sequenced 18S rRNA genes shared a high identity of 99.2–99.9% with the 18S rRNA genes of B. gibsoni (DQ184507). The ITS sequences shared high identity with B. gibsoni (EU084673). Nucleotide sequence variations within the 18S rRNA and ITS gene sequences were observed. The identity of the obtained 18S rRNA sequences ranged from 99.4 to 100%, with a 0–10 bp difference. Additionally, the ITS genes obtained in this study showed 98.9–100% identity, with a 0–12 bp nucleotide difference.
Phylogenetic analysis
Phylogenetic analysis was performed to provide a better understanding of the diversity of the sequences. The neighbor-joining tree showed that all obtained 18S rRNA sequences clustered together with the 18S rRNA gene sequences of B. gibsoni and fell into the Babesia clade. Three large canine species (B. vogeli, B. canis, and B. rossi) fell into the same clade, while the rest of the canine species formed different clades (Figure 2). All ITS sequences obtained in this study also fell into the same group with B. gibsoni (Figure 3).
Figure 2.

Neighbor-joining tree based on the full-length 18S rRNA gene sequences obtained in this study (▴) and the related sequences. The 18S rRNA of Plasmodium falciparum was employed as an outgroup. GenBank accession numbers are indicated in parentheses. A bootstrap test of 1,000 replicates was used, and values are given at the nodes.
Figure 3.
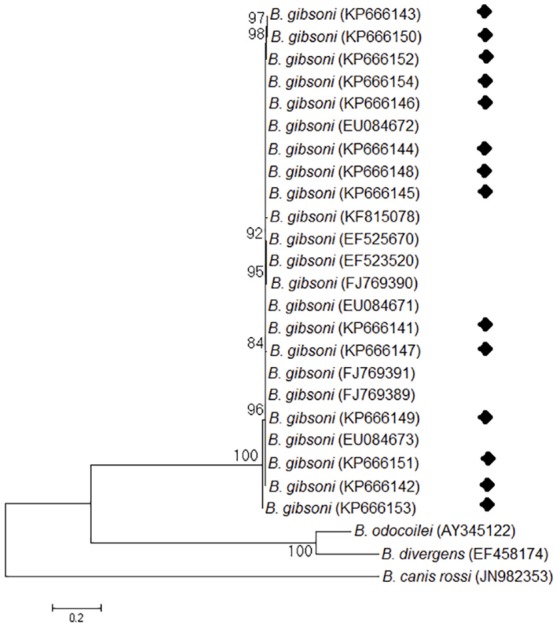
Neighbor-joining tree based on the ITS sequences obtained in this study (♦) and the related piroplasma. The ITS sequences of B. rossi were employed as an outgroup. GenBank accession numbers are indicated in parentheses. A bootstrap test of 1,000 replicates was used, and values are given at the nodes.
Discussion
In this study, microscopic examination and RLB were used to detect canine piroplasmosis. RLB has been widely employed for Babesia and Theileria detection since this technique enables the simultaneous detection and discrimination of infections caused by these pathogens (Gubbels et al., 1999; Georges et al., 2001). This assay is very helpful for identifying new species and novel genotypes (Chaisi et al., 2011; Khan et al., 2013). Fourteen samples out of 118 (11.86%) were positive by RLB, and all positive samples were singly infected with B. gibsoni. The rate of positivity by RLB was higher than that obtained through microscopic examination because RLB is more sensitive.
Regarding canine babesiosis in China, Chen et al. (2014) identified B. gibsoni in two dogs in Henan. B. gibsoni is the most widespread species in China; it has been reported in Shandong, Jiangsu, Anhui, Shanghai, Zhejinag, Nanjing, Jiangxi, and Guangxi. B. canis vogeli is the other species reported in dogs, whose epidemic areas include Jiangxi and Gansu (Wei et al., 2012; Yao et al., 2014; Niu et al., 2017; Zheng et al., 2017). Very recently, Niu et al. reported T. sinensis in pet dogs in Gansu, representing the first report of T. sinensis in dogs (Niu et al., 2017). Previous studies have indicated that canine babesiosis is mainly epidemic in eastern and southern China, while limited information is available on its prevalence in central China. In this study, 118 samples were randomly collected from pet dogs and screened via RLB. Fourteen samples were hybridized with a species-specific probe for B. gibsoni. Sequence analysis of the 18S rRNA gene and the ITS regions showed that the obtained sequences shared high identity with the 18S rRNA and ITS genes of B. gibsoni, respectively. Phylogenetic trees based on the 18S rRNA gene and ITS region were generated. The results confirmed that all positive dogs exhibited single infection with B. gibsoni. We assume that B. gibsoni is the only species infecting dogs in Wuhan. The initial purpose of this study was to detect the occurrence of piroplasm in pet dogs, and only limited samples were collected and analyzed. An epidemiological investigation considering the dog population and characteristics such as age and breed should be performed as this study showed a high rate of positivity, of 11.86%. B. gibsoni was previously thought to be prevalent only in Asian countries (Irwin and Jefferies, 2004; Goo et al., 2008). However, it has spread to South Africa, America, Europe and many other areas around the world with notable speed (Birkenheuer et al., 2005; Matjila et al., 2007). The disease can spread through biting, fighting, and transplacental transmission rather than simply via ticks, which is why B. gibsoni has spread so quickly worldwide (Jefferies et al., 2007; Yeagley et al., 2009). As babesiosis is a tick-borne disease, its distribution normally depends on the prevalence of the ticks responsible for transmission. However, no ticks were found on the dogs sampled in this study. We infer that either the ticks had already dropped off of the dogs when they were taken to the clinic, or B. gibsoni was inherited from the dogs' parents or through activities such as dog fighting.
The clinical manifestations of dogs infected with B. gibsoni vary from subclinical to severe to even fatal, based on the physical condition of the host (Schnittger et al., 2012). Among the 14 positive dogs, four of the samples tested negative by microscopic examination. These four dogs exhibited subclinical infections without significant clinical symptoms. There will be a greater risk when these dogs are subjected to immunosuppressive conditions. On the other hand, regardless of whether significant clinic manifestations are present, infected dogs can become reservoirs and infect other B. gibsoni-free dogs.
In conclusion, this study provides the first molecular record from Wuhan, China using a molecular RLB assay to simultaneously detect canine piroplasmosis in pet dogs. B. gibsoni was the only identified species. The results showed a considerable rate of positivity in pet dogs. As pets are considered companion animals and play increasingly important roles in humans' lives, their health deserves greater attention. It is necessary to pay attention and monitor this disease in dogs.
Author contributions
LH and XM wrote the draft of the manuscript. JZ and LH designed the study and corrected the manuscript. LH, XM, JiH, YH, PH, JuH, LY, NM, and LS collected samples and performed the molecular assays.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Basic Science Research Program (973 program) of China (Grant No. 2015CB150302), the National Key Research and Development program of China (2017YFD0501201), the National Natural Science Foundation of China (31302080) and the Fundamental Research Funds for the Central Universities (2662015PY006).
References
- Bajer A., Mierzejewska E. J., Rodo A., Welc-Faleciak R. (2014). The risk of vector-borne infections in sled dogs associated with existing and new endemic areas in Poland. Part 2: occurrence and control of babesiosis in a sled dog kennel during a 13-year-long period. Vet. Parasitol. 202, 234–240. 10.1016/j.vetpar.2014.02.007 [DOI] [PubMed] [Google Scholar]
- Baneth G., Florin-Christensen M., Cardoso L., Schnittger L. (2015). Reclassification of theileria annae as Babesia vulpes sp nov. Parasit. Vec. 8:207. 10.1186/s13071-015-0830-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R., Vojta L., Mrljak V., Marinculic A., Beck A., Zivicnjak T., et al. (2009). Diversity of Babesia and Theileria species in symptomatic and asymptomatic dogs in Croatia. Int. J. Parasitol. 39, 843–848. 10.1016/j.ijpara.2008.12.005 [DOI] [PubMed] [Google Scholar]
- Bhoora R., Franssen L., Closthuizen M. C., Guthrie A. J., Zweygarth E., Penzhorn B. L., et al. (2009). Sequence heterogeneity in the 18S rRNA gene within Theileria equi and Babesia caballi from horses in South Africa. Vet. Parasitol. 159, 112–120. 10.1016/j.vetpar.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Birkenheuer A. J., Correa M. T., Levy M. G., Breitschwerdt E. B. (2005). Geographic distribution of babesiosis among dogs in the United States and association with dog bites: 150 cases (2000-2003). J. Am. Vet. Med. Assoc. 227, 942–947. 10.2460/javma.2005.227.942 [DOI] [PubMed] [Google Scholar]
- Cao J., Yang Q., Zhang J., Zhou Y., Zhang H., Gong H., et al. (2015). Seroprevalence survey of Babesia gibsoni infection and tick species in dogs in East China. Vet. Parasitol. 214, 12–15. 10.1016/j.vetpar.2015.10.002 [DOI] [PubMed] [Google Scholar]
- Carret C., Walas F., Carcy B., Grande N., Precigout E., Moubri K., et al. (1999). Babesia canis canis, Babesia canis vogeli, Babesia canis rossi: differentiation of the three subspecies by a restriction fragment length polymorphism analysis on amplified small subunit ribosomal RNA genes. J. Eukaryot. Microbiol. 46, 298–303. 10.1111/j.1550-7408.1999.tb05128.x [DOI] [PubMed] [Google Scholar]
- Chaisi M. E., Sibeko K. P., Collins N. E., Potgieter F. T., Oosthuizen M. C. (2011). Identification of Theileria parva and Theileria sp. (buffalo) 18S rRNA gene sequence variants in the African Buffalo (Syncerus caffer) in southern Africa. Vet. Parasitol. 182, 150–162. 10.1016/j.vetpar.2011.05.041 [DOI] [PubMed] [Google Scholar]
- Chen Z., Liu Q., Jiao F. C., Xu B. L., Zhou X. N. (2014). Detection of piroplasms infection in sheep, dogs and hedgehogs in Central China. Infect. Dis. Poverty 3:18. 10.1186/2049-9957-3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto S., Suzuki H., Igarashi I., Xuan X. (2005). Fatal experimental transplacental Babesia gibsoni infections in dogs. Int. J. Parasitol. 35, 1031–1035. 10.1016/j.ijpara.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Georges K., Loria G. R., Riili S., Greco A., Caracappa S., Jongejan F., et al. (2001). Detection of haemoparasites in cattle by reverse line blot hybridisation with a note on the distribution of ticks in Sicily. Vet. Parasitol. 99, 273–286. 10.1016/S0304-4017(01)00488-5 [DOI] [PubMed] [Google Scholar]
- Goo Y. K., Jia H., Aboge G. O., Terkawi M. A., Kuriki K., Nakamura C., et al. (2008). Babesia gibsoni: serodiagnosis of infection in dogs by an enzyme-linked immunosorbent assay with recombinant BgTRAP. Exp. Parasitol. 118, 555–560. 10.1016/j.exppara.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Gubbels J. M., De Vos A. P., Van Der Weide M., Viseras J., Schouls L. M., De Vries E., et al. (1999). Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization. J. Clin. Microbiol. 37, 1782–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Feng H. H., Zhang W. J., Zhang Q. L., Fang R., Wang L. X., et al. (2012). Occurrence of Theileria and Babesia species in water buffalo (Bubalus babalis, Linnaeus, 1758) in the Hubei province, South China. Vet. Parasitol. 186, 490–496. 10.1016/j.vetpar.2011.11.021 [DOI] [PubMed] [Google Scholar]
- Irwin P. J. (2009). Canine babesiosis: from molecular taxonomy to control. Parasit. Vec. 2(Suppl. 1), S4. 10.1186/1756-3305-2-S1-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin P. J., Jefferies R. (2004). Arthropod-transmitted diseases of companion animals in Southeast Asia. Trends Parasitol. 20, 27–34. 10.1016/j.pt.2003.11.004 [DOI] [PubMed] [Google Scholar]
- Jefferies R., Ryan U. M., Jardine J., Broughton D. K., Robertson I. D., Irwin P. J. (2007). Blood, Bull Terriers and Babesiosis: further evidence for direct transmission of Babesia gibsoni in dogs. Aust. Vet. J. 85, 459–463. 10.1111/j.1751-0813.2007.00220.x [DOI] [PubMed] [Google Scholar]
- Katoh K., Frith M. C. (2012). Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics 28, 3144–3146. 10.1093/bioinformatics/bts578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. K., He L., Hussain A., Azam S., Zhang W. J., Wang L. X., et al. (2013). Molecular epidemiology of Theileria annulata and identification of 18S rRNA gene and ITS regions sequences variants in apparently healthy buffaloes and cattle in Pakistan. Infect. Genet. Evol. 13, 124–132. 10.1016/j.meegid.2012.09.007 [DOI] [PubMed] [Google Scholar]
- Kjemtrup A. M., Wainwright K., Miller M., Penzhorn B. L., Carreno R. A. (2006). Babesia conradae, sp. nov., a small canine Babesia identified in California. Vet. Parasitol. 138, 103–111. 10.1016/j.vetpar.2006.01.044 [DOI] [PubMed] [Google Scholar]
- Matjila P. T., Leisewitz A. L., Oosthuizen M. C., Jongejan F., Penzhorn B. L. (2008). Detection of a Theileria species in dogs in South Africa. Vet. Parasitol. 157, 34–40. 10.1016/j.vetpar.2008.06.025 [DOI] [PubMed] [Google Scholar]
- Matjila P. T., Penzhorn B. L., Leisewitz A. L., Bhoora R., Barker R. (2007). Molecular characterisation of Babesia gibsoni infection from a pit-bull terrier pup recently imported into South Africa. J. S. Afr. Vet. Assoc. 78, 2–5. 10.4102/jsava.v78i1.277 [DOI] [PubMed] [Google Scholar]
- Muhlnickel C. J., Jefferies R., Morgan-Ryan U. M., Irwin P. J. (2002). Babesia gibsoni infection in three dogs in Victoria. Aust. Vet. J. 80, 606–610. 10.1111/j.1751-0813.2002.tb10961.x [DOI] [PubMed] [Google Scholar]
- Nijhof A. M., Penzhorn B. L., Lynen G., Mollel J. O., Morkel P., Bekker C. P., et al. (2003). Babesia bicornis sp. nov. and Theileria bicornis sp. nov.: tick-borne parasites associated with mortality in the black rhinoceros (Diceros bicornis). J. Clin. Microbiol. 41, 2249–2254. 10.1128/JCM.41.5.2249-2254.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhof A. M., Pillay V., Steyl J., Prozesky L., Stoltsz W. H., Lawrence J. A., et al. (2005). Molecular characterization of Theileria species associated with mortality in four species of African antelopes. J. Clin. Microbiol. 43, 5907–5911. 10.1128/JCM.43.12.5907-5911.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Q., Yang J., Liu Z., Gao S., Pan Y., Guan G., et al. (2017). First molecular detection of piroplasm infection in pet dogs from Gansu, China. Front. Microbiol. 8:1029. 10.3389/fmicb.2017.01029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosthuizen M. C., Zweygarth E., Collins N. E., Troskie M., Penzhorn B. L. (2008). Identification of a novel Babesia sp from a sable antelope (Hippotragus niger Harris, 1838). J. Clin. Microbiol. 46, 2247–2251. 10.1128/JCM.00167-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oura C. A., Bishop R., Wampande E. M., Lubega G. W., Tait A. (2004). The persistence of component Theileria parva stocks in cattle immunized with the ‘Muguga cocktail’ live vaccine against East Coast fever in Uganda. Parasitology 129, 27–42. 10.1017/S003118200400513X [DOI] [PubMed] [Google Scholar]
- Schnittger L., Rodriguez A. E., Florin-Christensen M., Morrison D. A. (2012). Babesia: a world emerging. Infect. Genet. Evol. 12, 1788–1809. 10.1016/j.meegid.2012.07.004 [DOI] [PubMed] [Google Scholar]
- Schnittger L., Yin H., Qi B., Gubbels M. J., Beyer D., Niemann S., et al. (2004). Simultaneous detection and differentiation of Theileria and Babesia parasites infecting small ruminants by reverse line blotting. Parasitol. Res. 92, 189–196. 10.1007/s00436-003-0980-9 [DOI] [PubMed] [Google Scholar]
- Schoeman J. P. (2009). Canine babesiosis. Onderstepoort J. Vet. Res. 76, 59–66. 10.4102/ojvr.v76i1.66 [DOI] [PubMed] [Google Scholar]
- Service M. W. (2001). The Encyclopedia of Arthropod-Transmitted Infections: Of Man and Domesticated Animals. Liverpool, UK: CABI Publishing. [Google Scholar]
- Solano-Gallego L., Baneth G. (2011). Babesiosis in dogs and cats–expanding parasitological and clinical spectra. Vet. Parasitol. 181, 48–60. 10.1016/j.vetpar.2011.04.023 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichova B., Miterpakova M., Iglodyova A. (2014). Molecular detection of co-infections with Anaplasma phagocytophilum and/or Babesia canis canis in Dirofilaria-positive dogs from Slovakia. Vet. Parasitol. 203, 167–172. 10.1016/j.vetpar.2014.01.022 [DOI] [PubMed] [Google Scholar]
- Wei F. R., Lan Q. X., Zhu D., Ye J. H., Liu Q., Zhang Y. (2012). Investigation on Babesia in ticks infested on police dogs in selected areas of China. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 30, 390–392. Available online at: http://www.jsczz.cn:8080/Jweb_jsczz/CN/abstract/abstract2596.shtml [PubMed] [Google Scholar]
- Yao D. W., Jiang J. Y., Yu Z. Z., Yao D. Q., Yang D. J., Zhao Y. B. (2014). Canine Babesiosis in China caused by Babesia gibsoni: a molecular approach. Iran. J. Parasitol. 9, 163–168. [PMC free article] [PubMed] [Google Scholar]
- Yeagley T. J., Reichard M. V., Hempstead J. E., Allen K. E., Parsons L. M., White M. A., et al. (2009). Detection of Babesia gibsoni and the canine small Babesia ‘Spanish isolate’ in blood samples obtained from dogs confiscated from dogfighting operations. J. Am. Vet. Med. Assoc. 235, 535–539. 10.2460/javma.235.5.535 [DOI] [PubMed] [Google Scholar]
- Yisaschar-Mekuzas Y., Jaffe C. L., Pastor J., Cardoso L., Baneth G. (2013). Identification of Babesia species infecting dogs using reverse line blot hybridization for six canine piroplasms, and evaluation of co-infection by other vector-borne pathogens. Vet. Parasitol. 191, 367–373. 10.1016/j.vetpar.2012.09.002 [DOI] [PubMed] [Google Scholar]
- Zahler M., Schein E., Rinder H., Gothe R. (1998). Characteristic genotypes discriminate between Babesia canis isolates of differing vector specificity and pathogenicity to dogs. Parasitol. Res. 84, 544–548. 10.1007/s004360050445 [DOI] [PubMed] [Google Scholar]
- Zheng W., Liu M. M., Moumouni P. F., Liu X., Efstratiou A., Liu Z., et al. (2017). First molecular detection of tick-borne pathogens in dogs from Jiangxi, China. J. Vet. Med. Sci. 79. 10.1292/jvms.16-0484 [DOI] [PMC free article] [PubMed] [Google Scholar]


