Abstract
CHRNA7 is a neurodevelopmental protein involved in differentiation and neurogenesis, which is also named as nicotinic acetylcholine receptors, cholinergic receptor, nicotinic, alpha 7 (neuronal). The protein encoded by this gene forms a homo-oligomeric channel. It is a major component of brain nicotinic receptors displays that are blocked by and sensitive to alpha-bungarotoxin. Studies reports involvement of CHRNA7 protein in different neurological diseases. Non-availability of 3-dimensional (3D) structure leads the study toward structure 3D prediction along with its interaction analysis. The current paper is focused on the structure prediction through homology modeling of CHRNA7 along with binding site prediction using Schrödinger software suite. In continuation of the study, protein-protein interaction analysis is carried out by using string database. Tertiary structure along with binding sites was obtained, and visualized CHRAN7 protein have interaction with CHRNA protein family along with JAK2, AKT1, PICK1 protein that are involved in neurological disease. Structure formation analysis is an important aspect of proteomics studies. Hence, this predicted structure can be used for further advance studies and drug designing. Protein interaction analysis shows that CHRNA7 protein also interact with AKT1 protein which regulate neuronal differentiation and development, that signifies the role of CHRNA7 protein in neurological diseases.
Keywords: CHRNA7, Neuronal development disease, Homology modeling, Schrodinger software, Protein interaction
Introduction
CHRNA7 is an important gene which encodes neuronal acetylcholine receptor subunit alpha-7 protein (uniprot id P36544) responsive for different neuronal expression. It is a member of a superfamily of ligand-gated ion channels which facilitates fast signal transmission at synapses. The uniqueness lies in its structure as its hetero-pentamers are composed of homologous subunits. Conservitivity patterns of these proteins are very significant; till date, the structure of CHRNA7 is not deposited in PDB database.
Since protein structure is related with functionality of protein, hence protein modeling is done to extract protein function from its structure. The proposed structure for each subunit is a conserved N-terminal extracellular domain followed by 3 conserved transmembrane domains, a variable cytoplasmic loop, a fourth conserved transmembrane domain, and a short C-terminal extracellular region.
The protein encoded by this gene forms a homo-oligomeric channel, displays marked permeability to calcium ions, and is a major component of brain nicotinic receptors that are blocked by, and highly sensitive to, alpha-bungarotoxin. Once this receptor binds acetylcholine, it undergoes an extensive change in conformation that affects all subunits and leads to opening of an ion-conducting channel across the plasma membrane [1]. This gene is located in a region identified as a major susceptibility locus for juvenile cyclonic epilepsy and a chromosomal location involved in the genetic transmission of schizophrenia. An evolutionarily recent partial duplication event in this region results in a hybrid containing sequence from this gene and a novel FAM7A gene. After binding to acetylcholine, the AChR responds by an extensive change in conformation that affects all subunits and leads to the opening of an ion-conducting channel across the plasma membrane. The channel is blocked by alpha-bungarotoxin. Gene expression studies shows that CHRNA7 gene is primarily expressed in the posterior amygdalar nucleus and the field CA3 of Ammon's horn in the mouse, and in the mammillary body in humans [2]. Three-dimensional (3D) structure is the most competent requirement for any advance proteomic analysis or drug designing. In extension to this, the current paper deals with detailed structure modeling along with active site predivction of this potential protein that can act as cumulative target for different neurological anomalies.
Homology Modeling
Homology modeling is computational method that predicts 3D structure of protein using known protein structure called as template. Template is most crucial part of homology modeling since protein sequence is modelled on the basis of structure of template protein. Homology modeling is most accurate computational method for protein structure determination and verification of modelled structure is done by Ramachandran plot and statistical analysis [3].
Schrodinger Software
Schrödinger is a scientific leader in computational chemistry, providing software solutions and services for life sciences and materials research. Prime is a powerful and complete tool for generating accurate receptor models for structure-based drug design. It includes comprehensive protein modeling including both rapid and advanced methods for protein homology modeling, chimeric and multimeric models, protein structure quality analysis [4,5].
String Databse (http://string-db.org/)
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) is a biological database and web resource of known and predicted protein-protein interactions. STRING imports protein association knowledge from databases of physical interaction and databases of curated biological pathway knowledge.
Methods
Sequence Retrieval
Neuronal acetylcholine receptor subunit alpha-7, CHRNA7 of Homo sapiens (Human) protein sequence is retrieved from UniProt database UniProt id P36544.
Homology Modeling
Protein structure is modeled by homology modeling method using prime program of Schrödinger software suite. Blast search is performed to identify template protein structure [6]. CHRNA7 protein is modeled using 1bg9_a protein as template sequence. Loops are refined and verification is done by protein refinement program of Schrödinger software [7,8,9]
Binding Site Prediction
Modeled protein structure is used to identify effective binding sites on the basis of structural and physical properties using sitemap program of Schrödinger software. Five binding sites are predicted that can be used for docking and interaction analysis [10,11].
Protein-Protein Interaction Analysis
CHRNA7 protein interacting partners are identified using string databse. And functional characterization of chran7 protein is done on the basis of functionality of interacting protein.
Results
Homology Modeling
Homology modeling of chrna7 using template 2BG9_A. Figure 1 shows the alignment of chrna7 protein sequence with template sequence.
Fig. 1.
Sequence and template alignment.
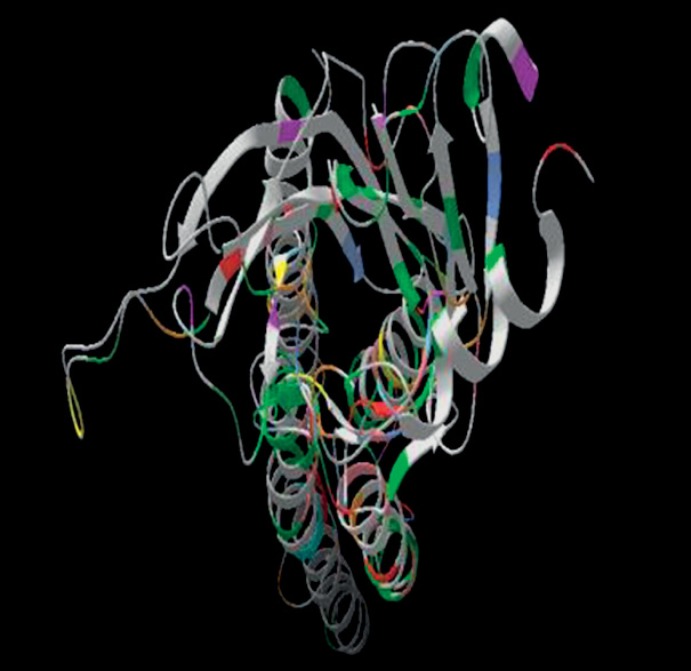
Figure 2 shows the structural alignment of chrna7 protein and template structure colored region represent structurally aligned region. Figure 3 shows the modeled structure.
Fig. 2.
Structural alignment.
Fig. 3.
Model of CHRNA7 protein built on 2BG9_A template sequence.
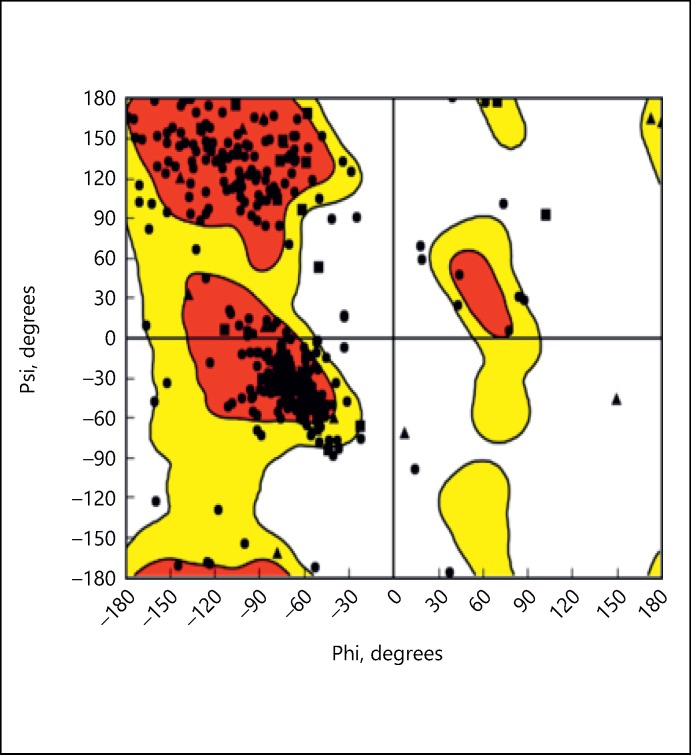
Ramachandran plot is used for protein verification. This shows that 80% of residues lie in favorable region (Fig. 4).
Fig. 4.
Ramachandran plot.
Binding Site Prediction
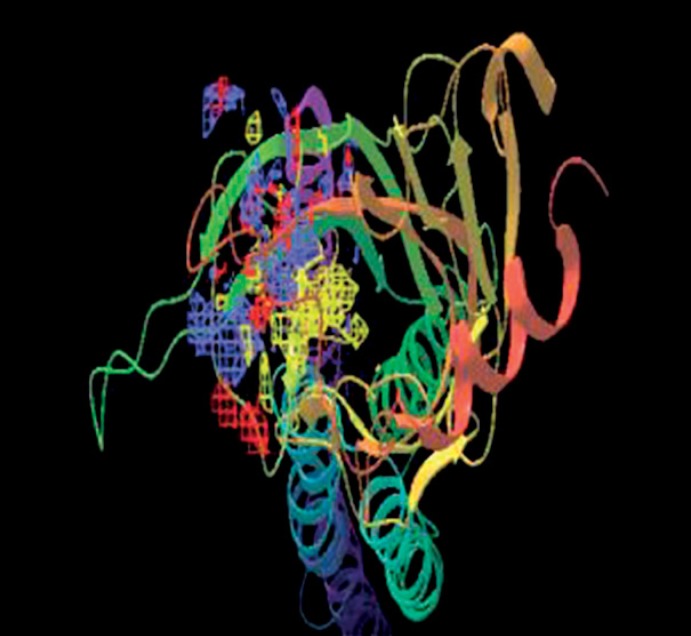
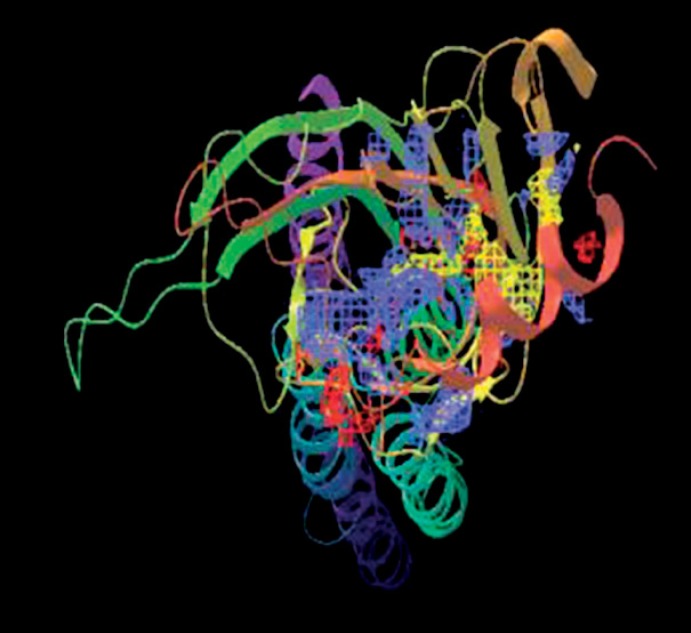
Binding sites prediction is done using sitemap program. Five binding sites were identified and best binding site is identified with a Dscore of 1.228. Figures 5, 6, 7, 8, shows binding sites on modeled chrna7 protein. Table 1 shows 5 binding sites and scores.
Fig. 5.
Sitemap_1_site_1.
Fig. 6.
Sitemap_1_site_2.
Fig. 7.
Sitemap_1_site_3.
Fig. 8.
Sitemap_1_site_4.
Table 1.
Binding sites of CHRNA7 protein
| S. No | Title Site score | Size | D. score | Volume | |
|---|---|---|---|---|---|
| 1 | Sitemap_1_site_1 | 1.160094 | 443 | 1.228 | 985.0 |
| 2 | Sitemap_1_site_2 | 1.131928 | 153 | 1.170 | 233.2 |
| 3 | Sitemap_1_site_3 | 1.130794 | 93 | 1.059 | 218.4 |
| 4 | Sitemap_1_site_4 | 1.002250 | 84 | 1.172 | 136.5 |
| 5 | Sitemap_1_site_5 | 0.702596 | 46 | 0.637 | 126.5 |
Protein-Protein Interaction Analysis
CHRNA7 protein interacting partners is identified using string tool using sequence search tool. Figure 9 shows protein interaction map of chrna7 protein.
Fig. 9.
Protein interaction map using string.
CHRAN7 protein have interaction with CHRNA protein family along with JAK2, AKT1, PICK1 protein that are involved in neurological disease. Table 2 summarizes interacting protein function and involvement of these protein with neurological diseases. This signifies that chrna7 is a protein that is regulated in neurological disease during neuronal development phase of cell.
Table 2.
CHRNA7 interacting protein
| S. No. | Name | Function | Disease | Pathway |
|---|---|---|---|---|
| 1 | CHRNA4 | Neurodevelopment protein involved in differentiation and neurogenesis | Autosomal dominant nocturnal frontal lobe epilepsy | Neuroactive ligand-receptor interaction, Cholinergic synapse and nicotine addiction |
| 2 | CHRNB4, CHRNA3, CHRNA2, CHRNA5, CHRNA6 |
AChR responds by an extensive change in conformation that leads to opening of an ion-conducting channel across the plasma membrane | Frontal lobe epilepsy | Neuroactive ligand-receptor interaction |
| 3 | AKT1 | AKT is responsible of the regulation of glucose uptake by mediating insulin-induced translocation | Neurological diseases | Rap1 signaling pathway, Ras signaling pathway |
| 4 | JAK2 | Janus kinase 2; non-receptor tyrosine kinase involved in various processes such as cell growth, development, differentiation or histone modifications | Polycythemia vera, Budd-Chiari syndrome | Chemokine signaling pathway Jak-STAT signaling pathway |
| 5 | PICK1 | Plays a role in synaptic plasticity by regulating the trafficking and internalization of AMPA receptors | Central nervous system disease and nervous system disease | Transmission across chemical synapses and transmission across chemical synapses |
Conclusion
Chrna7 neurogenesis protein involved in neuronal development and differentiation. Dysfunction of this protein leads to neurological disease such as Alzheimer's, neuroblastoma schizophrenia, and neuroblastoma [12]. Protein interaction analysis shows that CHRNA7 protein also interacts with AKT1 protein, which regulate neuronal differentiation and development, that signifies the role of CHRNA7 protein in neurological diseases [13,14]. Binding sites shows potent binding regions of protein that can be used for docking studies and drug development. Current analysis are apt to conclude the high level of significance in relation to this protein. This designed or predicted 3D structure of high potential protein can be further used for different neurological problems. Our findings may contribute to it as a cumulative target against them which can be fruitful in obtaining better lead molecules through further advance docking studies and can lighten a path in the area of drug designing at preventive or therapeutic aspects. Protein interaction map may give rise a better understanding of this protein with other referential neurologically expressing protein and thus can give a good understanding at system biology level.
Authors Contribution
Ruchi Yadav has done concept generation and network establishments of proteins through bioinformatic tools and MS preparation. Deepshikha has done Insilico experimental bench regarding data/information collection, protein modelling for structure prediction and validations. Dr. Prachi Srivastava has done analysis and interpretations of results along with final assessment and writing of manuscript.
Disclosure Statement
This manuscript complies with ICMJE and authors declared no conflict of interest. This study received no funding or sponsorship of any form.
Acknowledgments
This is not just to follow the custom of writing acknowledgment but to express and record my heartfelt thankfulness to all those who directly or indirectly helped me in this work. Furthermore, we wish to acknowledge the bioinformatics tools used in this study.
References
- 1.Elliott KJ, et al. Comparative structure of human neuronal 2-alpha 7 and beta 2-beta 4 nicotinic acetylcholine receptor subunits and functional expression of the alpha 2, alpha 3, alpha 4, alpha 7, beta 2, and beta 4 subunits. J Mol Neurosci. 1996;7:217–228. doi: 10.1007/BF02736842. [DOI] [PubMed] [Google Scholar]
- 2.Carson R, et al. Genetic variation in the α7 nicotinic acetylcholine receptor is associated with delusional symptoms in Alzheimer's disease. Neuromolecular Med. 2008;10:377–384. doi: 10.1007/s12017-008-8048-8. [DOI] [PubMed] [Google Scholar]
- 3.Schulz-Gasch T, Stahl M. Binding site characteristics in structure-based virtual screening: evaluation of current docking tools. J Mol Model. 2003;9:47–57. doi: 10.1007/s00894-002-0112-y. [DOI] [PubMed] [Google Scholar]
- 4.Release, Schrödinger. ‘1 SiteMap, version 1.7.’. New York: Schrödinger, LLC; 2010. [Google Scholar]
- 5.Schrödinger, Prime. ‘Version 3.5.’. New York: LLC; 2014. [Google Scholar]
- 6.Release, Schrödinger. ‘2 Prime, version 3.’. New York: Schrödinger, LLC; 2013. [Google Scholar]
- 7.Tsutsumi Y. Schrodinger equation. Funkcialaj Ekvacioj. 1987;30:115–125. [Google Scholar]
- 8.Adedeji AO, Singh K, Sarafianos SG. Structural and biochemical basis for the difference in the helicase activity of two different constructs of SARS-CoV helicase. Cell Mole Biol (Noisy-le-Grand) 2012;58:114–121. [PMC free article] [PubMed] [Google Scholar]
- 9.Nayeem A, Sitkoff D, Krystek S., Jr A comparative study of available software for high-accuracy homology modeling: from sequence alignments to structural models. Protein Sci. 2006;15:808–824. doi: 10.1110/ps.051892906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov AA, Barak D, Jacobson KA. Evaluation of homology modeling of G protein-coupled receptors in light of the A(2A) adenosine receptor crystallographic structure. J Med Chem. 2009;52:3284–3292. doi: 10.1021/jm801533x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh KD, et al. Homology modeling, molecular dynamics, e-pharmacophore mapping and docking study of Chikungunya virus nsP2 protease. J Mol Model. 2012;18:39–51. doi: 10.1007/s00894-011-1018-3. [DOI] [PubMed] [Google Scholar]
- 12.Leonard S, et al. Association of promoter variants in the α7 nicotinic acetylcholine receptor subunit gene with an inhibitory deficit found in schizophrenia. Arch Gen Psychiatry. 2002;59:1085–1096. doi: 10.1001/archpsyc.59.12.1085. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, et al. Evidence for linkage disequilibrium between the alpha 7 - nicotinic receptor gene (CHRNA7) locus and schizophrenia in Azorean families. Am J Med Genet. 2001;105:669–674. doi: 10.1002/ajmg.1549. [DOI] [PubMed] [Google Scholar]
- 14.Dempster EL, et al. Episodic memory performance predicted by the 2bp deletion in exon 6 of the “alpha 7-like” nicotinic receptor subunit gene. Am J Psychiatry. 2006;163:1832–1834. doi: 10.1176/ajp.2006.163.10.1832. [DOI] [PubMed] [Google Scholar]