Abstract
We have created 41 clustered charged-to-alanine scanning mutations of the mipA, γ-tubulin, gene of Aspergillus nidulans and have created strains carrying these mutations by two-step gene replacement and by a new procedure, heterokaryon gene replacement. Most mutant alleles confer a wild-type phenotype, but others are lethal or conditionally lethal. The conditionally lethal alleles exhibit a variety of phenotypes under restrictive conditions. Most have robust but highly abnormal mitotic spindles and some have abnormal cytoplasmic microtubule arrays. Two alleles appear to have reduced amounts of γ-tubulin at the spindle pole bodies and nucleation of spindle microtubule assembly may be partially inhibited. One allele inhibits germ tube formation. The cold sensitivity of two alleles is strongly suppressed by the antimicrotubule agents benomyl and nocodazole and a third allele is essentially dependent on these compounds for growth. Together our data indicate that γ-tubulin probably carries out functions essential to mitosis and organization of cytoplasmic microtubules in addition to its well-documented role in microtubule nucleation. We have also placed our mutations on a model of the structure of γ-tubulin and these data give a good initial indication of the functionally important regions of the molecule.
INTRODUCTION
There is now a great deal of evidence that γ-tubulin plays a central role in the nucleation of microtubule assembly from microtubule-organizing centers of fungal and animal cells (reviewed in Wiese and Zheng, 1999; Oakley, 2000). Much remains to be determined about γ-tubulin function, however. For example, the precise mechanism of nucleation remains open to debate. Recent data support the hypothesis that γ-tubulin ring complexes (γ-TuRCs) nucleate microtubule assembly by a template mechanism (Keating and Borisy, 2000; Moritz et al., 2000; Wiese and Zheng, 2000), but alternative explanations are possible (Erickson, 2000). Also, the regions of the γ-tubulin molecule that interact with other proteins are unknown. It is clear that γ-tubulin interacts with two conserved proteins known as Spc97p and Spc98p in Saccharomyces cerevisiae, hGCP2 and hGCP3 (or HsSpc98) in humans, and Dgrip84 and Dgrip91 in Drosophila melanogaster. γ-Tubulin and these proteins form the γ-tubulin small complex, which is a major building block of γ-TuRCs (Oegema et al., 1999) and is an essential component of the S. cerevisiae spindle pole body (SPB) (Geissler et al., 1996; Knop et al., 1997; Knop and Schiebel, 1997; Knop and Schiebel, 1998). The regions of γ-tubulin that interact with these proteins are not known. Similarly, interactions of γ-tubulin with α- and/or β-tubulin are almost certainly essential to the process of microtubule nucleation, but the sites of interaction are not known. Recent in vitro peptide binding data (Llanos et al., 1999) give an interesting initial indication of regions of γ-tubulin that interact physically with α- and β-tubulin, but these regions of interaction have not yet been verified in vivo. In addition, comparison of the sequences of γ-tubulin with α- and β-tubulins in regions corresponding to different polymerization interfaces in the dimer structure shows a conservation of key residues that suggests that γ-tubulin is capable of longitudinal self-assembly (Inclán and Nogales, 2001). Finally, and perhaps most interestingly, there is evidence that γ-tubulin may have important functions in addition to microtubule nucleation. This possibility was proposed on the basis of γ-tubulin localization to the mitotic spindle in mammalian cells (Lajoie-Mazenc et al., 1994), and there is now experimental evidence from Schizosaccharomyces pombe that γ-tubulin has at least one essential role in addition to microtubule nucleation (Paluh et al., 2000). The existence of a second essential function for γ-tubulin was inferred on the basis of a genetic interaction of a γ-tubulin mutation with a deletion of the Pkl1 gene that encodes a kinesin family member, and on the basis of the phenotypes conferred by the mutant γ-tubulin allele. The exact nature of the second essential function remains to be determined but probably involves regulation of microtubule dynamics (Paluh et al., 2000). In addition, C-terminal deletions of the S. cerevisiae γ-tubulin gene cause alterations of microtubule organization that are apparently not due to alterations of microtubule nucleation (Vogel and Snyder, 2000). We reasoned that conditionally lethal mutations might be valuable tools for understanding γ-tubulin interactions with other proteins and for understanding γ-tubulin functions, particularly nonnucleation functions.
Clustered charged-to-alanine scanning mutagenesis has been a highly successful approach for creating conditionally lethal alleles of cytoskeletal proteins (including α- and β-tubulin) and for defining regions essential to the functioning of those proteins (Wertman et al., 1992; Reijo et al., 1994; Richards et al., 2000, and references therein). The principle behind this approach is that charged regions tend to be on the outside of proteins and replacement of charged amino acids with alanines often alters regions of interprotein interactions without causing gross structural changes. Often these replacements are lethal or conditionally lethal and the phenotypes conferred by these mutations can be very helpful in understanding the functions of the protein as well its interactions with other proteins.
We have created 41 mutant alleles of the A. nidulans mipA (γ-tubulin) gene and have replaced the wild-type mipA allele with the mutant alleles by two methods, two-step gene replacement (Dunne and Oakley, 1988) and a novel heterokaryon gene replacement technique that we have developed. Many of the mutant alleles confer a wild-type phenotype, but others are lethal or conditionally lethal. We have determined the microscopic phenotypes of the strongly conditionally lethal alleles and have tested all conditionally lethal alleles for suppression by the antimicrotubule agents benomyl and nocodazole and by the microtubule-stabilizing agent deuterium oxide (D2O). Our results strongly support the notion that γ-tubulin has at least one essential function in addition to the nucleation of microtubule assembly. Finally, we have placed the mutations on a structural model of γ-tubulin. The positions of the mutations give a good initial indication as to the regions of γ-tubulin that are important for function.
MATERIALS AND METHODS
In Vitro Mutagenesis
All mutant alleles were created in plasmid pLO2 (Figure 1) with the use of the Quickchange mutagenesis kit (Stratagene, La Jolla, CA). High-performance liquid chromatography-purified primers used for the mutagenesis were obtained from Operon (Alameda, CA). The mutant alleles were verified by sequencing.
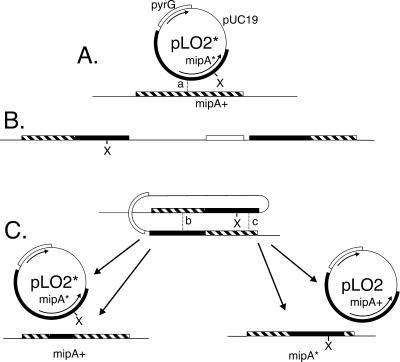
Figure 1.
Two-step gene replacement at the mipA locus. (A) Integration by homologous recombination of pLO2*, a plasmid that carries a mutated copy of mipA (mipA*) as well as the pyrG gene. The mutation is designated with an X. Integration of the transforming plasmid at the wild-type chromosomal mipA locus (mipA+) occurs by crossing over at point a, creating a mutant copy of mipA and a wild-type copy of mipA flanking pUC19 and pyrG sequences (B). Depending on the crossover point, the mutant copy could be at the right or left. (C) Mitotic crossing over leading to eviction of one of the two copies of mipA. If crossing over occurs at point b, a plasmid (pLO2*) carrying the mutant mipA allele will be evicted and the wild-type allele will remain in the chromosome. If crossing over occurs at point c, a plasmid (pLO2) carrying the wild-type mipA allele will be evicted and the mutant mipA allele will remain in the chromosome.
A. nidulans Media, Strains, and Transformation Procedures
YG (5 g of yeast extract and 20 g of d-glucose per liter) was used as a complete liquid medium. YAG (YG with 15 g of agar per liter) and FYG (YG with 25 g of Pretested Burtonite 44c [TIC Gums, Belcamp, MD] per liter) were used as solid complete media. Both were supplemented with 1 ml/l of a trace element solution (Cove, 1966). To support growth of strains carrying pyrG89, YG, YAG, and FYG were supplemented with 10 mM uridine and 10 mM uracil. Liquid minimal medium (6 g/l NaNO3, 0.52 g/l KCl, 0.52 g/l MgSO4 · 7H2O, 1.52 g/l KH2PO4, 10 g/l d-glucose, 1 ml/l of trace element solution, pH adjusted to 6.5 with NaOH before autoclaving) and solid minimal medium (liquid minimal medium with 15 g/l of agar) were derived from those of Pontecorvo et al. (1953). When appropriate, minimal media were supplemented with 10 mM uridine and 10 mM uracil (for strains carrying pyrG89) and/or 10 mM arginine (for strains carrying argB2).
Strain G191 (pyrG89, pabaA1; fwA1, uaY9) was used for two-step gene replacement. The evictants produced by two-step gene replacements carry pyrG89, which confers weak cold sensitivity and would complicate phenotypic analyses of conditionally lethal mutants. We crossed evictants carrying mutant mipA alleles to strain FGSC442 (facB101, riboB2, chaA1, sE15, nirA14) and used pyrG+ segregants for virtually all analyses. These crosses also allowed us to verify that the conditional lethality of the evictants was due to a mipA mutation because the conditional lethality segregated opposite riboB2 in crosses (at least 100 progeny tested for each cross). Strains used for creating heterokaryons are discussed below. Strains R153 (wA3, pyroA4) and LO385 (yA2, biA1; benA33) were used as controls in some experiments.
For transformation, protoplasts were produced with the use of β-d-glucanase (InterSpex, Foster City CA) or a mixture of β-d-glucanase, Driselase (Interspex) and lyticase (Sigma, St. Louis, MO) (Jung et al., 2000). Transformation procedures were as previously described (Oakley et al., 1987b; Jung et al., 2000). For the heterokaryon gene replacement procedure, transformants were selected on minimal medium containing 0.6 M KCl (with no supplements or supplemented with 10 mM arginine as discussed below). Two-step gene replacement was carried out by the method of Dunne and Oakley (1988) except that concentrations of 5-fluoro-orotic acid were varied (depending on temperature and the age of the 5-fluoro-orotic acid) to give optimal conditions for eviction. Evictions of benomyl-dependent, benomyl-suppressed, and putative lethal alleles were carried out with and without benomyl. Because the effects of benomyl are reduced as temperatures increase, we used higher concentrations of benomyl at higher temperatures (0.6 μg/ml at 42°C, 0.4 μg/ml at 30°C, and 0.2 μg/ml at 25°C).
Heterokaryon Gene Replacement
We previously created mipA disruptions in A. nidulans heterokaryons (Oakley et al., 1990; Martin et al., 1997). In these disruptants (Oakley et al., 1990) there are two dysfunctional mipA alleles arranged in tandem. Rearrangements between these two alleles can, in principle, create a wild-type mipA allele and our early experiments indicated that this occurs frequently enough to be a problem. To eliminate this problem, we created a new heterokaryon carrying a γ-tubulin deletion rather than a disruption (Figure 2, A and B) as follows. The coding sequence of mipA in pGEM11zf (Promega, Madison, WI) was replaced by the A. nidulans argB gene. An 1833-bp fragment, including 8 bp upstream of the mipA coding sequence and all but the last four codons of the coding sequence, was removed and replaced by a 1704-bp fragment containing the argB gene. The argB fragment had been amplified from pM006 (Upshall, 1986) by polymerase chain reaction (PCR). The resulting plasmid (pKJ28) contains the argB gene flanked by 1700 bp homologous to the region upstream of the mipA coding sequence and 2300 bp downstream. The length and position of the excised fragment (containing the mipA coding sequence) were chosen to ensure that no other coding sequences upstream or downstream of mipA were disrupted. A linear 5.7-kb fragment amplified from pKJ28 by PCR was used to transform KJ15 to create the deletion heterokaryon. This was verified by Southern blotting, and we call this heterokaryon H26.
Figure 2.
Heterokaryon gene replacement. (A) Deletion of the chromosomal mipA allele. A linear DNA fragment in which the wild-type mipA gene (mipA+) has been replaced by the wild-type argB gene (argB+) is used to transform a strain carrying the wild-type mipA gene. Note the sequences flanking argB+ are identical to those flanking mipA+ in the chromosome. Crossing over occurs in the sequences flanking mipA+ and this produces a replacement of mipA+ by argB+. (B) Creation of a mipA deletion heterokaryon. Protoplasts from a strain carrying pyrG89, argB2, fwA1, and the wild-type mipA allele (mipA+) are transformed with a linear DNA fragment in which mipA has been replaced with argB. Resultant transformant nuclei carry pyrG89, argB2, fwA1, argB+, and the mipA deletion (mipAΔ). Transformant nuclei would not support growth because of the absence of a mipA gene. At some frequency, however, transformant protoplasts fuse with nontransformant protoplasts, producing a heterokaryon. This heterokaryon is viable on medium lacking arginine because transformant nuclei have a functional copy of argB and nontransformant nuclei have a functional copy of mipA. The uninucleate asexual spores (conidia) produced by the heterokaryon are not viable on medium lacking arginine because those carrying the transformant nuclei produce no γ-tubulin and those carrying the nontransformant nuclei do not have a functional copy of argB. If the transformant and nontransformant nuclei fuse, they will produce a diploid nucleus that carries a functional copy of mipA and a functional copy of argB. Conidia carrying such nuclei will be viable on medium lacking arginine. To produce a mipA deletion heterokaryon that allows easy detection of diploids and has other advantages for heterokaryon gene replacements, we have created the heterokaryon shown in C. Protoplasts from the heterokaryon shown in B were fused with protoplasts from a strain carrying pyrG89, argB2, wA3, and mipA+. Several products were produced, but only the desired heterokaryon is shown. As in the heterokaryon shown in B, one class of nuclei is argB+, mipAΔ and the other is argB2, mipA+. Fusion of the two classes of nuclei will produce wA+, fwA+ nuclei that will cause green conidia to be produced. At the right are shown the results of transformation with a plasmid (pLO2*) carrying a mutant mipA allele (see Figure 1 for a diagram of the plasmid).
A limitation with the use of H26 for gene replacements was that the two types of nuclei in the heterokaryon occasionally fused to create a diploid nucleus that carried argB+ and a wild-type mipA allele. Such diploids produced conidia (asexual spores, sometimes called conidiospores) that would confuse our mutant analysis (i.e., would give apparent mipA+ transformants regardless of the phenotype conferred by the transforming mutant mipA allele). To circumvent this problem, we took advantage of the conidial color mutations available in A. nidulans to create a heterokaryon in which such fusions could be detected easily. We prepared protoplasts from germinating conidia of H26 and fused them with protoplasts from strain KJ19, which carries wA3 (white conidia), argB2, and pyrG89 (Figure 2C). Among the products of the fusion were mipA deletant heterokaryons of the type shown in Figure 2C. In these heterokaryons, the nuclei carrying the mipA deletion also carry pyrG89 and a mutation (fwA1) that causes fawn conidial color. The nuclei carrying mipA+ also carry wA3, argB2, and pyrG89. Because conidial color is governed by the single nucleus in each conidium, the heterokaryon produces both fawn and white conidia. If the two types of nuclei fuse, however, the resulting diploid nuclei will be heterozygous for both fwA and wA. Because both alleles are recessive, diploid conidia will be green and colonies that grow from them will also have green conidia. Undesired diploids can, thus, be distinguished easily by observing conidial color. We used two of these heterokaryons (designated H39 and H59) for our studies. These heterokaryons were, in theory, identical and behaved identically in our experiments.
These heterokaryons have several useful features as recipients for mutant mipA alleles. First, because they carry pyrG89, they can be transformed to uridine prototrophy by pLO2 or pLO2* (both of which carry pyrG+). Second, when transformed with pLO2* (pLO2 carrying a mutant mipA allele), all fawn pyrG+ transformants must carry the transforming mutant mipA allele (mipA*) and no wild-type mipA allele (Figure 2C). They, thus, display the phenotype conferred by the mutant allele. Similarly, all white pyrG+ transformants must carry the transforming mutant allele as well as the resident wild-type allele (Figure 2C). Comparing phenotypes of fawn and white transformants thus reveals whether the mutant allele is dominant or recessive. Third, if we obtain white transformants, but no fawn transformants, this indicates that the mutant mipA allele is incapable of supporting growth on its own (i.e., is recessive lethal). If we fail to obtain white transformants, it means that the mutant allele is dominant lethal. Fourth, if we inoculate a portion of our transformation mixtures onto medium lacking arginine and uridine, the white transformants, which carry argB2, cannot grow and only fawn colonies, which carry argB+ and the mutant mipA allele, will grow. Growth of nontransformants and white transformants is sufficiently inhibited that any mutant mipA allele that will support growth (even very slow growth) can be detected.
Immunofluorescence Microscopy
Sterile coverslips were placed in Petri dishes containing liquid complete medium. In most cases coverslips were coated with poly-l-lysine (Ovechkina et al., 1999) before sterilization. Conidia inoculated into the medium adhered to the coverslips and remained attached to the coverslips through our immunofluorescence procedures. Immunofluorescence procedures were based on those of Ovechkina et al. (1999). Cell wall digestion times and concentrations of digestion enzymes were varied to give optimal staining. In particular, high concentrations of β-d-glucanase (32 or 64 mg/ml) were necessary to obtain optimal wall digestion and staining with cultures incubated at 42 or 43°C. We used three anti-γ-tubulin antibodies, an affinity-purified rabbit antibody against A. nidulans γ-tubulin (Oakley et al., 1990); a commercial mouse monoclonal anti-γ-tubulin, GTU-88 (Sigma); and a mouse monoclonal anti-γ-tubulin (G9) generously provided by Dr. Tetsuya Horio (Tokushima University School of Medicine, Tokushima, Japan). Each antibody was tested for specificity by Western blotting. A mouse monoclonal anti-β-tubulin, Tu27B (generously provided by Dr. L. Binder, Northwestern University School of Medicine, Chicago, IL, via Dr. G. Lozano, M. D. Anderson Cancer Center, Houston, TX) and a rat monoclonal anti-α-tubulin, YOL1/34, (Accurate, Westbury, NY) were used to stain microtubules. Tu27B was used in combination with the rabbit polyclonal anti-γ-tubulin and YOL1/34 was used in combination with either of the mouse monoclonal anti-γ-tubulins. Secondary antibodies were from Jackson ImmunoResearch (West Grove, PA) (Cy3-labeled goat anti-rabbit, Cy3-labeled goat anti-mouse, fluorescein isothiocyanate-labeled goat anti-mouse) or Molecular Probes (Eugene, OR) (Alexa Fluor 488-labeled goat anti-rat, Alexa Fluor 488-labeled goat anti-mouse). They were preadsorbed by the manufacturers against serum proteins of relevant species to give minimal cross reactivity in double labeling experiments.
Observations were made on a Zeiss standard microscope, a Nikon Eclipse 800 microscope, or an Intelligent Imaging Innovations Everest wide-field deconvolution system. Images were captured with a Princeton Instruments MicroMax charge-coupled device camera on the Nikon Eclipse 800 controlled by IPLab software or a SensiCam charge-coupled device camera on the Everest controlled by Slidebook (Mac) software. Images were processed with the use of Slidebook, NIH Image, Adobe Photoshop (Mac), or Corel Photopaint (Mac) software. Composite images were assembled with the use of CorelDraw (Mac) software.
RESULTS
Design and Construction of Mutant mipA Alleles
We created all mutant mipA alleles in plasmid pLO2 (Figure 1). In general we followed the algorithm of Wertman et al. (1992) in choosing sites for mutagenesis. We chose regions with at least two charged amino acids (aspartic acid, glutamic acid, lysine, or arginine) in a window of five. We mutated no more than three amino acids to alanine in a single allele. Thus, if four of five amino acids were charged, we created two mutant alleles, each carrying two residues changed to alanine. Although histidines are only very weakly charged at neutral pH (pK of ≈6.5), they are often found in active sites and we mutated histidines in three regions highly conserved in γ-tubulins (H29, H266, H370). A complete list of the mutant alleles created is given in Table 1. For simplicity and brevity, we will refer to each mutant allele by the N-terminal-most amino acid altered. The mutant allele in which the arginine at amino acid 3 and the glutamic acid at amino acid 4 are changed to alanines will, thus, be called mipAR3.
Table 1.
Mutations and phenotypes
| Allele | Amino acids changed to alanine | Phenotype(s) | Allele | Amino acids changed to alanine | Phenotype(s) |
| mipAR3 | R3, E4 | wt | mipAK284 | K284, R287, K288 | cs |
| mipAE28 | E28, H29 | wt | mipAD293 | D293, R296, R297 | wt |
| mipAE38 | E38, E39 | wt | mipAK302 | K302, R304 | wt |
| mipAE43 | E43, D46, R47 | wt | mipAE326 | E326, D328 | wt |
| mipAK48 | K48, D49 | wt | mipAD331 | D331, K334 | wt |
| mipAD56 | D56, D57, R59 | wt | mipAR338 | R338, R340 | cs |
| mipAR63 | R63 | cs, hs | mipAR341 | R341, R342, R343 | rl |
| mipAD68 | D68, E70, R72 | rl | mipAR361 | R361, K362 | wt |
| mipAE116 | E116, E117, D120 | rl | mipAH370 | H370, R371 | weak cs |
| mipAD123 | D123, R124, E125 | weak cs | mipAK388 | K388, R389 | wt |
| mipAD127 | D127, D130 | wt | mipAD395 | D395, R396 | wt |
| mipAE133 | E133 | wt | mipAR398 | R398, K399, R400 | rl |
| mipAE155 | E155, R156 | wt | mipAK408 | K408, K409, E410 | bd |
| mipAD159 | D159, R160 | cs | mipAD415 | D415, D418, E419 | wt |
| mipAK163 | K163, K164 | wt | mipAD421 | D421, E422, R424 | wt |
| mipAR192 | R192, R193 | wt | mipAD429 | D429 | wt |
| mipAR211 | R211 | wt | mipAE433 | E433, E435 | wt |
| mipAR215 | R215, R216 | wt | mipAE438 | E438, R439, E440 | wt |
| mipAR243 | R243 | weak hs | mipAD444 | D444, D446 | wt |
| mipAR264 | R264, H266 | rl | mipAK449 | K449, D450, E451 | wt |
| mipAD278 | D278, D281 | wt | |||
wt, wild type; cs, cold-sensitive; hs, heat-sensitive; rl, recessive lethal; bd, benomyl-dependent.
Gene Replacement Procedures
Two-Step Gene Replacement
One gene replacement strategy that has been used successfully in A. nidulans is the two-step gene replacement (Dunne and Oakley, 1988), which was based on a procedure developed in S. cerevisiae (Botstein and Shortle, 1985), and this is one of the two strategies we have used in this study. Replacement of a chromosomal wild-type mipA allele (mipA+) by a mutant allele (mipA*) carried on plasmid pLO2 is shown in Figure 1. We give the designation pLO2* to pLO2 in which mipA has been mutated. Integration of pLO2* at the mipA locus by homologous recombination (Figure 1A) creates a duplication of the mipA gene, with one wild-type allele and one mutant allele (Figure 1B) separated by pUC19 and pyrG sequences. PyrG is the A. nidulans orotidine-5′-phosphate decarboxylase gene, and it is used as the selectable marker for transformation. After a strain carrying the duplication is established, homologous recombination between the two copies of mipA occurs at a low frequency (Figure 1C). This results in the excision and loss (eviction) of pLO2 or pLO2* and leaves only a single copy of mipA. 5-Fluoro-orotic acid can be used to select for evictants that are pyrG−. Depending on where the crossover leading to excision occurs, the copy of mipA that remains can be wild-type or mutant, and, consequently, some evictants carry the wild-type allele and others carry the mutant allele. If the mutation confers a clear phenotype, evictants carrying the mutant allele can be distinguished from those carrying the wild-type allele. This approach does not, however, allow one to determine easily whether the mutant allele has a wild-type phenotype or is lethal. If the mutant allele has a wild-type phenotype all the evictants (wild-type and mutant) will have a wild-type phenotype. If the mutant allele is lethal, all evictants carrying the mutant allele will die before forming colonies and only evictants carrying the wild-type allele will be seen. In both cases all surviving evictants will be wild type.
Development of a Heterokaryon Gene Replacement Procedure
Because distinguishing between wild-type and lethal alleles is critical for defining functionally important regions of γ-tubulin, we wished to develop a procedure that would allow us to make this distinction. In the yeasts S. cerevisiae and S. pombe, gene replacements are often carried out by simple transformation with a linear fragment carrying the replacing allele and a selectable marker. Such one-step gene replacements are possible in A. nidulans, but problems arise in that the replacements are sometimes incomplete and, in other instances, the transforming DNA inserts in other places in the genome creating insertional mutants (Miller et al., 1985). This creates a very high background of irrelevant mutants, which can be detected by Southern hybridizations but only with significant effort. Demonstrating that an allele is lethal is also difficult with this technique in A. nidulans because the transforming mutant allele may insert randomly in the genome, leaving the wild-type allele intact. If the transforming allele is recessive lethal, correct replacements will be inviable, but transformants in which random insertion occurs may be viable.
A second procedure used in yeasts is the plasmid shuffle technique. In this approach one starts with a strain that has a deletion in the gene of interest and a wild-type allele on a plasmid that contains a marker that allows both positive and negative selections. One transforms the strain with a second plasmid that carries the mutant allele and then evicts the plasmid carrying the wild-type allele. The lack of suitable autonomously replicating vectors has, so far, precluded this approach in A. nidulans.
Because neither of these approaches work easily with A. nidulans, we have developed a novel procedure, heterokaryon gene replacement, that allows us to determine whether alleles are wild-type or lethal (Figure 2). This procedure is based on the facts that A. nidulans is coenocytic and that nuclei carrying a deletion (or disruption) of an essential gene can be maintained indefinitely in a heterokaryon as long as the heterokaryon also carries nuclei with a functional copy of the gene (Osmani et al., 1988; Oakley et al., 1990; Martin et al., 1997). A. nidulans produces uninucleate conidia and many of the conidia produced by the heterokaryon carry the nuclei with the deletion. Although the fact that these nuclei carry a deletion in an essential gene precludes their growth and development into colonies, one can usually germinate the conidia and work with them. We reasoned that we should be able to create a heterokaryon that carries nuclei with a deletion of the mipA gene. We could then transform protoplasts, made from the spores produced by the heterokaryon, with a plasmid carrying a mutant mipA allele. When protoplasts carrying the mipA deletion were transformed, they should carry only the mutant mipA allele and should have the phenotype conferred by the mutant allele. This procedure, which is discussed in more detail in MATERIALS AND METHODS, proved very useful.
In addition to their utility for gene replacement, γ-tubulin deletion heterokaryons are also ideal for observing the phenotypic effects of the γ-tubulin deletion. If conidia from these heterokaryons are incubated in the absence of arginine, the mipA+ conidia, which carry argB2, will not germinate. The γ-tubulin deletant conidia, which are argB+, will germinate and will have the phenotype conferred by the γ-tubulin deletion.
Macroscopic Phenotypes of mipA Alleles
Two-step and heterokaryon gene replacements were carried out for each of the mutant mipA alleles. Results are shown in Table 1. Twenty-eight alleles confer a wild-type phenotype. Five alleles are lethal, five confer cold sensitivity, one confers both cold sensitivity and heat sensitivity, one confers heat sensitivity, and one is dependent on the antimicrotubule agents benomyl or nocodazole for growth. All the lethal and conditionally lethal alleles proved to be recessive.
All conditionally lethal alleles were verified in several ways. The fact that there was a single mipA allele and that it was at the mipA locus was verified by Southern hybridizations. The fact that the conditional lethality was indeed due to the mutation at the mipA locus was verified by crossing to a strain carrying riboB2. RiboB2 is tightly linked to mipA (Weil et al., 1986) and, as expected, the conditional lethality segregated opposite riboB2 in each instance. Finally, the conditionally lethal alleles were recovered by PCR and the mutations were verified by sequencing.
Figure 3 shows the growth rates at various temperatures of strains carrying the conditionally lethal mipA alleles. There is a great deal of variation in the “tightness” of conditional lethality. MipAD123 and mipAH370 are listed as cold-sensitive because their growth is more inhibited relative to wild-type controls at low temperatures than at high temperature. They are only weakly cold-sensitive, however, and cause a slight reduction of growth at all temperatures. MipAD159, on the other hand, is tightly cold-sensitive and mipAR338 is extremely cold-sensitive, barely growing at 37°C or below. MipAR243 is heat-sensitive and the drop off in growth rate at high temperatures is very steep. It grows at nearly wild-type rates at 37°C, is greatly restricted for growth at 42°C, and is much more restricted for growth at 43°C (our unpublished data). The dropoff is sufficiently steep that small differences in temperature within an incubator could make a significant difference in growth rate. We consequently had to be careful in testing growth rates on benomyl, nocodazole, and D2O that test and control plates were immediately adjacent. MipAR63 is tightly heat-sensitive and cold-sensitive but grows reasonably well at 25 and 30°C.
Figure 3.
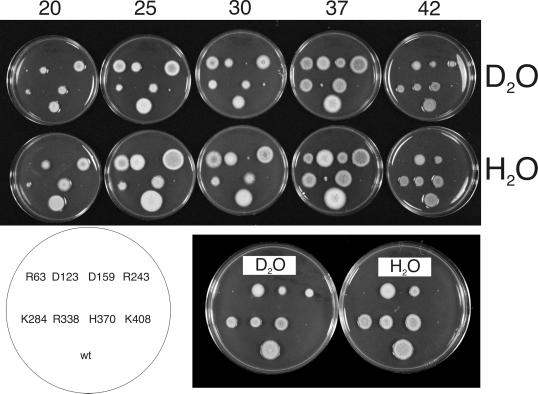
Conditional lethality and benomyl suppression of mipA alleles. The pattern of inoculation was the same on each plate and is shown on the diagram at the bottom (R63 = mipAR63, etc.; wt = wild-type control). The temperature of incubation (in °C) is shown at the left of each row and the concentration of benomyl in micrograms per milliliter is shown at the top of each column. Ethanol was used as a solvent for benomyl and growth on ethanol without benomyl is shown in the second column from the left. YAG is the complete medium to which the ethanol or benomyl was added. Conidia from each strain were stab inoculated and colony size reflects growth rates. All plates at a given temperature were incubated together for the same length of time. Because growth rates for wild-type A. nidulans strains are different at different temperatures, plates at different temperatures were incubated for different times. Plates at 30 and 37°C were incubated for 2 d, 42°C plates for 3 d, 25°C plates for 4 d, and 20°C plates for 7 d. (Note that as shown by the growth of the wild-type control, A. nidulans is somewhat less sensitive to benomyl at high temperatures than at low temperatures.)
Suppression of Conditional Lethality by Benomyl and Nocodazole
A γ-tubulin mutation in S. pombe confers resistance to the antimicrotubule agent thiabendazole (Paluh et al., 2000). Although A. nidulans is somewhat sensitive to thiabendazole, it is much more sensitive to the related antimicrotubule agents benomyl and nocodazole. We consequently tested conditionally lethal alleles for growth at a variety of benomyl concentrations. To our surprise we found that the conditional lethality conferred by several alleles was suppressed by benomyl and nocodazole. Subsequent to our tests, Vogel and Snyder (2000) reported somewhat similar results with cold-sensitive C-terminal deletions of S. cerevisiae γ-tubulin.
Effects of benomyl on the growth of our mutants are shown in Figure 3. Results with nocodazole were similar (our unpublished data). The most dramatic results were with mipAK408. The strain carrying mipAK408 barely grew at any temperature in the absence of benomyl or nocodazole, but at 25°C on 0.2 μg/ml benomyl it grew reasonably well, more than half the growth rate of the wild type. It also grew reasonably well at 30 and 37°C on benomyl. MipAR338 confers extreme cold sensitivity, but the cold sensitivity is dramatically suppressed by benomyl (Figure 3). The cold sensitivity conferred by mipAK284 is also suppressed by benomyl. Other alleles show smaller benomyl effects. The growth of mipAR63 at 37°C and the growth of mipAR243 at 42°C appear to be enhanced by 0.2 μg/ml benomyl. Evictants carrying mipAK284, mipAR338, and mipAK408 were not initially detected by two-step gene replacement. We presume that this was because they grew so slowly relative to evictants carrying the wild-type allele. Colonies carrying these alleles were detected by heterokaryon gene replacement, however, because of the low background. When we became aware of the benomyl rescue of these alleles, we repeated the two-step gene replacement evictions on media with benomyl and we recovered evictants carrying each of these alleles. We also repeated evictions on benomyl for all putative lethal alleles. The alleles listed as lethal in Table 1 were not recoverable by the heterokaryon gene replacement procedure nor by the two-step gene replacement procedure with or without benomyl.
Deuterium Oxide Effects on Growth of γ-Tubulin Mutants
In view of the effects of antimicrotubule agents on γ-tubulin mutants, we wished to examine the effects of a microtubule-stabilizing agent. The most widely used microtubule stabilizer, paclitaxel (taxol), does not affect the growth of A. nidulans but D2O has significant effects (Oakley et al., 1987a). D2O promotes microtubule assembly and suppresses dynamic instability in vitro and in vivo (Panda et al., 2000, and references therein). In A. nidulans, 40% D2O specifically increases the heat sensitivity of the benomyl-resistant, microtubule-hyperstabilizing β-tubulin mutation benA33 and suppresses the cold sensitivity of the benomyl supersensitive, microtubule-destabilizing α-tubulin mutation tubA4 (Oakley et al., 1987a). These results indicate that the effects of D2O on microtubules in A. nidulans are similar to those observed in vitro and in other organisms.
We tested the growth rates of strains carrying all of our conditionally lethal alleles on medium containing 40% D2O at various temperatures (Figure 4). The growth of most strains was affected little by D2O. Several appeared to be slightly more inhibited on D2O than was the wild type. The heat sensitivity of mipAR63 was slightly suppressed by D2O at 37°C, however, and the heat sensitivity of mipAR243 was more significantly suppressed at 42°C.
Figure 4.
Effects of deuterium oxide on the growth of mipA alleles. (Top) Growth at various temperatures in complete medium with 40% D2O (top row) or medium with H2O only (bottom row). Growth temperatures (in °C) are shown above each pair of plates. Each pair of plates at a given temperature was incubated together for the same length of time. Incubation time was 2 d for 30, 37, and 42°C, 4 d for 25°C, and 7 d for 20°C. A diagram showing the inoculation pattern for the strains is at the lower left (R63 = mipAR63, etc.; wt = wild-type control). The 42°C plates are shown at a higher magnification at the lower right. D2O partially suppresses the heat sensitivity conferred by mipAR243 at 42°C and slightly suppresses the heat sensitivity conferred by mipAR63 at 37°C.
Placing mipA Mutations on a Three-Dimensional Model of γ-Tubulin
Given the high level of sequence similarity of γ-tubulin with α- and β-tubulin, the three-dimensional structure of γ-tubulin can be predicted with confidence from the electron crystallographic structures of α- and β-tubulin (Paluh et al., 2000). As a first step in correlating the structure and functions of γ-tubulin we have determined the positions of our mipA alleles on a structural model of γ-tubulin (Figure 5, see Quicktime video, gammut.mov, with online version of this article). In particular, the location of each mutated residue has been analyzed in terms of its proximity to surfaces of tubulin–tubulin interactions as identified for the tubulin dimer (Nogales et al., 1999). A considerable number of our mutants had a wild-type phenotype. Interestingly, all of the mutations located in loops on the surface that, in α- and β-tubulin, faces the lumen of the microtubule were wild type, as well as those in the C-terminal helix H12. In contrast, mutations in this helix confer a slow growth phenotype in yeast α-tubulin and are lethal in yeast β-tubulin (Richards et al., 2000), most likely due to their participation in the binding of motors. Recessive lethal mutations in γ-tubulin include residues in the T2 loop, which is involved in nucleotide binding (D68, E70, R72), residues in helix H3, which is involved in lateral contacts between tubulin dimers (E116, E117, D120), as well as residues between helices H11 and H12 on the longitudinal interface between dimers.
Figure 5.
Positions of mipA mutations on a structural model of γ-tubulin and microtubule bands in swollen conidia of mipAR243. (A) Stereo pair showing the positions of mutants of various phenotypes. (B) Positions of conditionally lethal mutants on the model. (C and D) mipAR243 ungerminated conidia showing bands of microtubules (green). Nuclei are shown in blue. C and D are projections of Z series obtained with an Everest wide-field deconvolution series. Images were captured, noise was reduced by near-neighbor deconvolution, and the Z series was projected into two-dimensional images. Microtubules in these swollen conidia form bands around the periphery of the cytoplasm. In C a few microtubules are separate from the band. In D, all the microtubules form a tight band. C and D are the same magnification (bar in D, 5 μm).
Microscopic Phenotypes of mipA Mutants
Given recent evidence that γ-tubulin has at least one essential function in addition to microtubule nucleation (Paluh et al., 2000; Vogel and Snyder, 2000), we were interested in determining whether our conditionally lethal alleles inhibited microtubule nucleation. We germinated our conditionally lethal alleles (omitting mipAD123 and mipAH370 because they are very weakly conditionally lethal) under restrictive conditions and examined them by immunofluorescence microscopy. We then determined whether cytoplasmic and mitotic spindle microtubules were present and, if so, whether they were normal in appearance. We also determined whether γ-tubulin was present at the SPBs at restrictive temperatures. Finally, we took the opportunity to examine microtubules in mipA deletant germlings from untransformed heterokaryons. For reference, control mipA+ interphase and mitotic germlings are shown in Figure 6.
Figure 6.
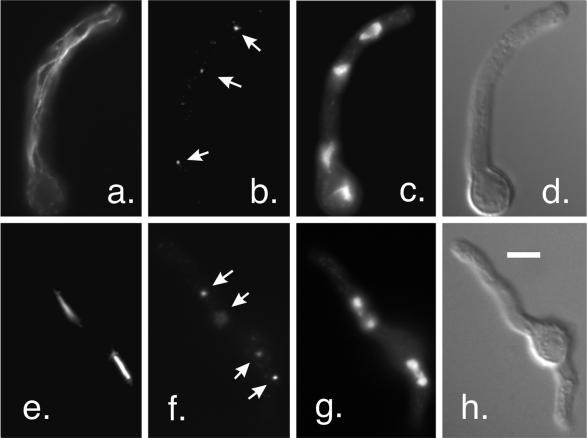
Interphase microtubules and mitotic spindles in a strain with wild-type γ-tubulin. The strain shown is G191, the strain used for two-step gene replacement. (a–d) An interphase germling. Staining with an anti-β-tubulin antibody reveals a normal interphase microtubule array (a). Staining of SPBs (arrows) with an anti-γ-tubulin antibody is shown in b. Nuclei are shown by DAPI staining in c, and the shape of the germling is shown by differential interference contrast in d. (e–h) A germling in anaphase. Two anaphase mitotic spindles are shown in e. γ-Tubulin at the SPBs (arrows) is shown in f. One of the SPBs for the upper spindle is slightly out of the plane of focus. The chromosomes are shown in g. All images are the same magnification (bar in h, 5 μm).
MipA Deletant
MipA deletant germlings from heterokaryons were similar to the germlings carrying a disruption of the mipA gene that had been examined previously (Oakley et al., 1990; Martin et al., 1997). As expected, γ-tubulin was absent from SPBs. Spindles were essentially absent although, very rarely, individual microtubules or very thin bundles of microtubules were seen in the vicinity of chromatin and it is possible that these may have been incompletely formed spindles. As discussed for a mipA disruption (Martin et al., 1997), these could reflect a very small amount of γ-tubulin carried over from the parental heterokaryon. Cytoplasmic microtubules were absent from some germlings but were present in others. When present they differed greatly from normal cytoplasmic microtubule arrays. They often formed long bundles and in other cases were abnormally curved (Figure 7).
Figure 7.
Abnormal cytoplasmic microtubules in mipA deletant germlings. (a and b) Adjacent optical sections showing microtubules stained with an anti-β-tubulin antibody. The arrows indicate a microtubule that curves through >180°. (c and d) DAPI and differential interference contrast images of the same field. (e) Microtubule bundle. The chomosomes appear condensed in this germling (DAPI staining in f), but focusing through the germling revealed that the single microtubule bundle curved around the region containing the chromatin. All images are the same magnification (bar in g, 5 μm).
MipAR63
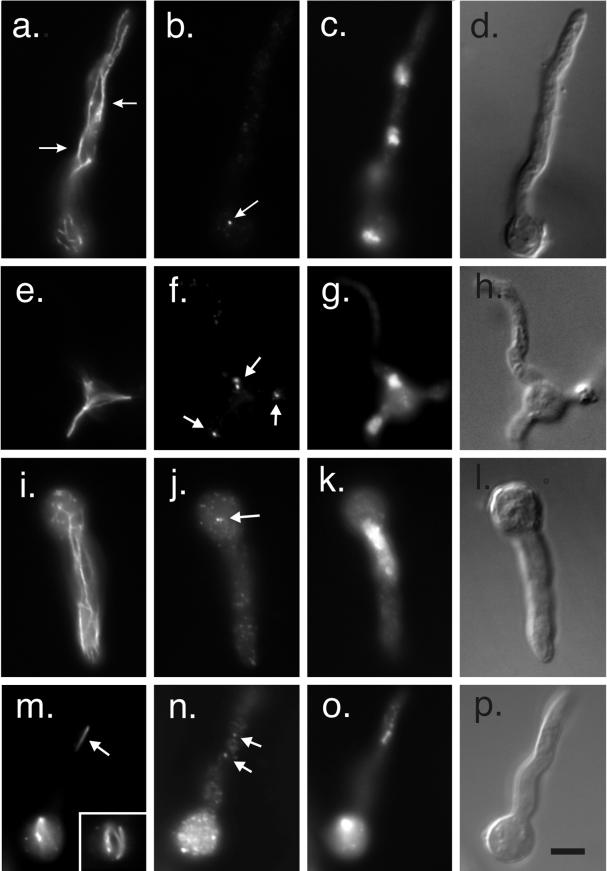
MipAR63 prevents growth at high and low temperatures and partially restricts growth at intermediate temperatures. R63 is a totally conserved residue across the different tubulin families. In yeast the mutation of K59 and R63 to alanines is lethal in α-tubulin (Richards et al., 2000), whereas it is cold-sensitive in β-tubulin (Reijo et al., 1994). This residue could be important for tubulin folding. At the best permissive temperature (30°C) mipAR63 germlings exhibited a more or less wild-type phenotype. We observed occasional slight abnormalities such as clustering of nuclei. At a low restrictive temperature of 20°C, cytoplasmic microtubules were present but were often abnormally curved and in some cases they formed bundles (Figure 8). Spindles were also present but exhibited a variety of structural abnormalities. Some were longer than normal, some were split, and some were multipolar (Figure 8). Others were not rod-shaped as in the wild type, but were spindle-shaped, wide in the middle, and tapered to a point at each pole with microtubules absent from the center. Nuclei were mostly small and the chromatin appeared partially condensed. γ-Tubulin staining was present at spindle pole bodies.
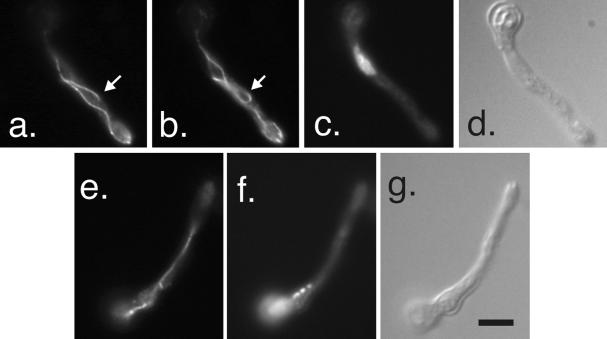
Figure 8.
MipAR63 at low and high restrictive temperatures. (a–d and e–h) Two germlings at a low restrictive temperature (20°C). (a) Interphase germling in which cytoplasmic microtubules have been stained with an anti-β-tubulin antibody. The microtubules in a portion of the germling have formed bundles, two of which are indicated by arrows. (b) An SPB stained with an anti-γ-tubulin antibody (arrow). SPBs on the other nuclei are out of the plane of focus. (c and d) DAPI and differential interference contrast images of the same field. (e–h) A germling with an abnormal mitotic spindle. (e) Spindle stained with an anti-β-tubulin antibody. (f) Four spindle pole bodies (arrows) stained with an anti-γ-tubulin antibody. (g and h) DAPI and differential interference contrast images of the same field. (i–l and m–p) Germlings incubated at a high restrictive temperature of 42°C. (i–l) Interphase germling. (i) More or less normal cytoplasmic microtubule array although there appears to be a little bundling of microtubules. Anti-γ-tubulin staining is shown in j. Dots of γ-tubulin are present through the cytoplasm. A probable SPB pair is shown with an arrow. (k and l) show DAPI and differential interference contrast images of the same field. The nucleus is abnormally large. A mitotic germling is shown in m–p. Two mitotic nuclei are present. One spindle is abnormally thin (arrow in m), whereas the other mitotic nucleus has a more robust curved and split spindle (insert in m which is a different focal plane of the bottom nucleus). Dots of γ-tubulin are distributed through the cytoplasm (n) but two fainter than normal SPBs are associated with the top spindle (arrows in n). All images are the same magnification (bar in p, 5 μm).
At the high restrictive temperature of 42°C, most nuclei were abnormally large and obviously very polyploid. Chromatin was often partially condensed. Cytoplasmic microtubules appeared more or less normal, but in some germlings appeared to be abnormally curved. Mitotic spindles were present but were all highly abnormal. There were a variety of abnormalities but one frequent phenotype was that instead of being rod-shaped, the spindles were spindle-shaped. In other cases the microtubules in the spindle were split into two or more bundles rather than forming a single bundle as in the wild type (Figure 8). In many cases spindles appeared to have fewer than normal microtubules (Figure 8). γ-Tubulin staining at poles was generally less intense than in control cells and there appeared to be more γ-tubulin dots in the cytoplasm than in controls. In fact, SPBs were often not identifiable in interphase cells because their staining was no brighter than that of the cytoplasmic dots. This did not appear to be a staining artifact because bright SPB staining was present in two control strains (a wild-type strain and a strain carrying the heat-sensitive β-tubulin mutation benA33) grown, fixed, and stained in parallel.
Although young germlings were relatively normal in appearance, germlings maintained at 42°C for extended periods of time were often morphologically abnormal. In particular we noted abnormal thickening of portions of the cell walls of many germlings (our unpublished data).
MipAD159
D159, R160 are located on helix H4. Under permissive conditions (37°C) germlings carrying mipAD159 had normal cytoplasmic microtubules and spindles. At a restrictive temperature of 20°C, however, some spindles appeared normal but others exhibited a variety of abnormalities. Some were split or bent (Figure 9). Many germlings had a single large, obviously polyploid nucleus. Cytoplasmic microtubules appeared normal in most germlings but sometimes were bundled or exhibited abnormal curvature (Figure 9).
Figure 9.
MipAD159 at a restrictive temperature (20°C). (a–c) Interphase germling. Microtubules in this germling are abnormally curved (arrowhead in a) or bundled (arrow). (b and c) DAPI and differential interference contrast images of the same field. (d–g) Mitotic germling. (d) Abnormal (bifurcated) spindle stained with an anti-β-tubulin antibody. (e) SPB (arrow) stained with an anti-γ-tubulin antibody. DAPI staining (f) reveals that the nucleus contains a lot of condensed chromatin and is probably polyploid. All images are the same size (bar in g, 5 μm).
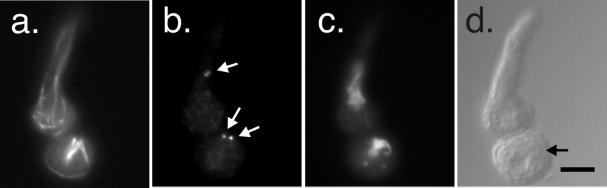
MipAR243
R243 is located in the catalytic T7 loop. This loop, which is on the opposite end of the molecule from the region expected to interact with α-tubulin at the microtubule minus end, has been predicted to be involved in tubulin–tubulin contacts within a γ-tubulin protofilament or dimer (Inclán and Nogales, 2001). Under permissive conditions (25°C), germlings carrying mipAR243 were indistinguishable from wild-type controls. Under restrictive conditions (43°C), however, the phenotype of this allele was very different from the wild type and from our other mutant alleles. One very obvious effect of mipAR243 was that it inhibited germ tube formation. Normally during germination, conidia swell and a germ tube extends. In cultures of mipAR243 at 43°C, nearly all conidia were swollen (indicating that they were metabolically active), but they were inhibited in the formation of germ tubes (Figure 10). Because one encounters occasional swollen conidia without germ tubes in mutant and wild-type cultures, we wished to quantify inhibition of germ tube formation by mipAR243 relative to other strains. We tested strains carrying mipAR243, mipAR63, the wild-type strain R153, and a strain carrying the heat-sensitive β-tubulin mutation benA33. We scored 400 conidia in each of three separate experiments for each strain. Conidia were incubated for 12 h at 43°C in YG medium before fixation. In the strain carrying mipAR243, only 23.3 ± 13.3% of the conidia had germ tubes, whereas in R153 98.2 ± 1.5% had germ tubes, in mipAR63 88.3 ± 4.2% had germ tubes, and in benA33 94.4 ± 0.3% had germ tubes. There is, thus, a slight inhibition of germ tube formation in strains carrying mipAR63 and benA33 relative to the wild type, but a very strong inhibition in the strain carrying mipAR243.
Figure 10.
MipAR243 at a restrictive temperature (43°C). (a–d) Three swollen but ungerminated conidia. (a) Microtubule staining. Microtubules are visible in optical cross section as dots at the peripheries of the swollen conida. SPBs are out of the plane of focus so only cytoplasmic dots of γ-tubulin are visible (b). (c and d) DAPI and differential interference images of the same field. Two nuclei are visible in c but the third is almost out of focus. (e–h) Interphase germling with more or less normal cytoplasmic microtubules (e). There are dots of γ-tubulin in the cytoplasm (f). The nucleus appears partially condensed (g). (i–l) Mitotic germling with an abnormal spindle. The spindle is small and curved (i) and is much shorter than the region occupied by the chromosomes (k). Staining with an anti-γ-tubulin antibody (j) reveals that there are many γ-tubulin dots in the cytoplasm, and it is difficult to distinguish SPBs. All images are the same magnification (bar in l, 5 μm).
In cases in which germ tubes formed, cytoplasmic microtubules were present and apparently normal (Figure 10), but mitotic spindles were rare. The spindles that were present were generally very small and thin (Figure 10), apparently consisting of only a few or perhaps only one microtubule. Chromatin was generally abnormal. Many nuclei were much larger than wild-type nuclei and showed partial or complete chromatin condensation. This was true of both swollen conidia and germlings, and indicates that nuclei were probably going through multiple rounds of DNA replication without nuclear division. γ-Tubulin staining of SPBs was not obvious for most nuclei. Dots of γ-tubulin were present in the cytoplasm (Figure 10) and some of these dots were near the nuclear envelope so it is possible that SPBs were stained faintly. In instances in which spindles could be identified unequivocally, there was γ-tubulin staining of SPBs, but the staining was fainter than in wild-type and other mutant germlings. The lack of spindles and of γ-tubulin staining at the SPBs was apparently not due to poor preparation for immunofluorescence because spindles and SPBs were brightly stained in the wild-type strain R153 and in the heat-sensitive β-tubulin mutant benA33 that was also used as a control (our unpublished data). We obtained the same microtubule staining results with a β-tubulin antibody and in separate experiments with an α-tubulin antibody and the same γ-tubulin staining results with two different antibodies, in three separate experiments.
The fact that spindles seemed rarer than nuclei with condensed chromosomes suggested that many mitotic nuclei (i.e., with condensed chromosomes) might lack spindles completely. To quantify this, we identified germlings and swollen conidia with condensed chromosomes by 4′,6-diamidino-2-phenylindole (DAPI) staining and determined whether mitotic spindles were present. If spindles were present we determined whether they were normal in appearance. Although many cells had partially condensed chromosomes, we scored only cells in which the chromosomes were unequivocally condensed. We scored 200 mitotic cells in two experiments and 67% of the nuclei completely lacked spindles. The remainder had abnormal spindles and no normal spindles were seen. Most of the “spindles” that were seen were so thin that it was not clear that they were, in fact, spindles rather than cytoplasmic microtubules in the vicinity of the condensed chromosomes.
Although cytoplasmic microtubules were apparently organized normally in germlings, we found a very interesting abnormality in many swollen conidia at the restrictive temperature. In approximately one-third of the swollen conidia, microtubules formed a band encircling the cell (Figure 5, C and D). This band was reminiscent of the preprophase band seen in plant cells. The tightness of packing of the microtubules varied, but the microtubules appeared more or less parallel to each other. To our knowledge nothing like this has been reported in A. nidulans.
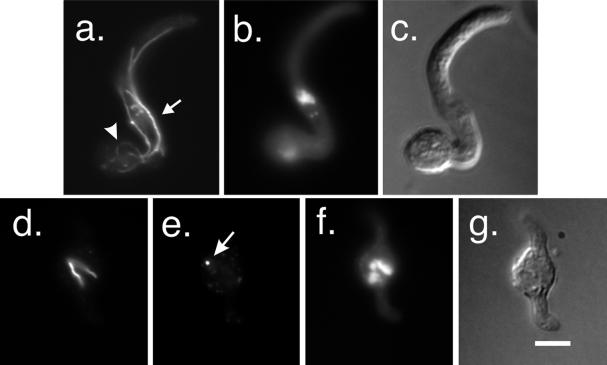
MipAK284
K284, R287, and K288 are in the M-loop that in the α/β-tubulin dimer is essential for lateral contacts between protofilaments. Under permissive conditions (0.2 μg/ml benomyl, 37°C), the vast majority of mitotic spindles in mipAK284 were normal. All mitotic stages were present so it appeared that nuclei were progressing through mitosis normally under these conditions. A few spindles were slightly split and greater abnormalities were seen very rarely. γ-Tubulin staining of SPBs was normal. Cytoplasmic microtubules appeared more or less normal although they may have been slightly more abundant than normal. Nuclei were clustered in some germlings. Under restrictive conditions (20°C without benomyl) mitotic spindles in germlings carrying mipAK284 appeared relatively normal (Figure 11), but a small percentage of mitotic spindles was abnormal with microtubules looped out from the spindle. γ-Tubulin staining of SPBs appeared normal (Figure 11). Cytoplasmic microtubules were less normal in mipAK284 germlings. Many germlings had abnormally curved microtubules (Figure 11) and, unusually for A. nidulans, in some cases short microtubules ran perpendicular to the axis of growth of the germling.
Figure 11.
MipAK284 and mipAR338 under restrictive conditions. (a–h) mipAK284 incubated at 20°C. A mitotic germling is shown in a–d. An apparently normal spindle is present (a) and SPBs are stained by an anti-γ-tubulin antibody (arrows in b). (c) DAPI staining. (d) Differential interference contrast image of the same field. (e–h) Interphase germling with abnormally curved microtubules (arrow in e). SPBs are stained with an anti-γ-tubulin antibody (arrows in f). (g) Interphase nuclei stained with DAPI. (h) Differential interference contrast image of the same field. (i–p) mipAR338 incubated at 37°C without benomyl. (i–l) Mitotic germling. A tripolar spindle is present (i). Two SPBs are stained with an anti-γ-tubulin antibody (arrows in j). At least one additional SPB was present outside of the plane of focus. DAPI staining is shown in k and a differential interference contrast image of the same field is shown in l. An interphase germling is shown in m–p. Cytoplasmic microtubules (m) are not bundled but may be more abundant than normal. Anti-γ-tubulin staining reveals one SPB (arrow in n). Others were visible at other focal planes. All images are the same magnification (bar in p, 5 μm).
MipAR338
R338 and R340 are located in helix H10 and are involved in both longitudinal and lateral interactions between α/β-tubulin dimers. Under permissive conditions (0.2 μg/ml benomyl, 37°C), cytoplasmic microtubules appeared normal. Nearly all spindles were normal and all mitotic stages were present. γ-Tubulin staining of SPBs was normal. Under restrictive conditions (37°C without benomyl) germlings had more or less normal cytoplasmic microtubules (Figure 11). Although it is difficult to quantify cytoplasmic microtubules in A. nidulans, they appeared to be, if anything, more abundant in mipAR338 germlings than in the wild type. Spindles were mostly abnormal. Multipolar spindles were common (Figure 11), and there were a variety of other abnormalities. γ-Tubulin localized to the SPBs at the poles of the spindles. Some germlings had a single large nucleus, whereas others had multiple nuclei. Sometimes there were both abnormally large and abnormally small nuclei in the same germling. These data suggest that mipAR338 allows spindle formation under restrictive conditions, but chromosomal segregation is abnormal and this results in polyploid or aneuploid nuclei with abnormal numbers of SPBs.
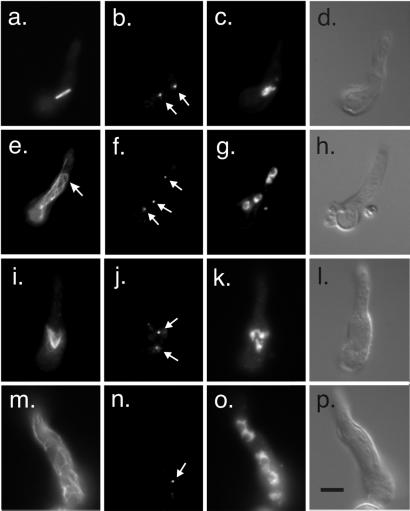
MipAK408
K408, K409, and E410 are residues between the H11 and H12 helices involved in longitudinal contacts of α/β-tubulin dimers. They are on the surface of γ-tubulin expected to interact with α-tubulin in the template model. MipAK408 causes cells to be dependent on benomyl or nocodazole for growth. Under permissive conditions (0.2 μg/ml benomyl, 25°C), germlings carrying mipAK408 had cytoplasmic microtubules that looked normal. Normal-appearing mitotic spindles were present and were in all stages from early prophase to late telophase. Some morphologically abnormal spindles were present but were very rare. γ-Tubulin localization was normal. Under restrictive conditions (without benomyl at 37°C), mipAK408 germlings possessed more or less normal arrays of cytoplasmic microtubules, although cytoplasmic microtubules appeared somewhat more abundant than normal in some germlings (Figure 12). Mitotic spindles were nearly all abnormal in appearance (Figure 12). Multipolar spindles were particularly common. In some spindles, microtubules were splayed out rather than forming a rod as in the wild type. γ-Tubulin localized to the poles of the spindles and, in keeping with the fact that many spindles were multipolar, many nuclei had multiple γ-tubulin-staining spots, undoubtedly corresponding to SPBs. Most germlings had a single abnormally large nucleus, often with two or more nucleoli. These data indicate that spindles form but do not function properly. Nuclear division is inhibited and polyploid nuclei with multiple SPBs result. When these nuclei subsequently enter mitosis, multipolar spindles assemble from the multiple SPBs.
Figure 12.
MipAK408 under restrictive conditions (37°C). A germling is shown as well as a swollen conidium that has not yet extended a germ tube (arrow in d). (a) Microtubules stained with an anti-β-tubulin antibody. The germling is in interphase and the cytoplasmic microtubules are similar to those seen in wild-type cells. The swollen conidium is in mitosis and the spindle is multipolar. (b) Staining with an anti-γ-tubulin antibody. Spindle pole bodies (arrows) are stained in both the germling and conidium. Two additional spindle pole bodies were present in the conidium in a different focal plane. (c) Same field showing chromatin stained with DAPI. (d) Differential interference contrast of the same field. All fields are the same magnification. (bar in d, 5 μm).
DISCUSSION
Alanine-scanning mutagenesis of A. nidulans γ-tubulin has yielded mutations that confer a surprising variety of phenotypes. Our immunofluorescence data show clearly that nucleation of mitotic spindle microtubules is almost completely eliminated in germlings carrying a γ-tubulin deletion. Assembly of spindle microtubules also appears to be inhibited by mipAR243 at restrictive temperatures and may be somewhat inhibited by mipAR63 at high restrictive temperatures. In each case, failure of spindle microtubule assembly seems to correlate with reduced or eliminated localization of γ-tubulin to the SPB. In contrast, our other conditionally lethal mutants have robust, if abnormal, spindles under restrictive conditions as well as robust γ-tubulin staining of SPBs.
Under restrictive conditions, mipAD159, mipAR338, and mipAK408 have extremely abnormal mitotic spindles and large, obviously polyploid, nuclei. These data indicate that, in these mutants, spindles form but do not function properly and nuclei reenter interphase before mitosis is completed. In mipAR338, there may be some partial but unequal segregation of chromatin, which results in the formation of both abnormally large and abnormally small nuclei. Although it is possible that the mutations have additional, less obvious, growth inhibitory effects, the mitotic failures in these strains would certainly be sufficient to inhibit growth. MipAR63 and mipAK284 have abnormal cytoplasmic microtubules at low restrictive temperatures and these may be responsible for, or contribute to, growth inhibition. As mentioned, mipAR243 probably inhibits mitosis by inhibiting the nucleation of mitotic spindle microtubules, and inhibits formation of germ tubes as well.
Immunofluorescence microscopy offers no indication that nucleation of spindle microtubule assembly is inhibited in the majority of our mutants. In principle, however, spindle microtubule assembly might be inhibited enough to render the mitotic spindle dysfunctional but not enough to be obvious by immunofluorescence microscopy. This might lead to failed mitoses and repeated mitotic failures might, in turn, cause many of the mitotic abnormalities we have observed. Our results with benomyl, nocodazole, and D2O are helpful in evaluating this possibility.
The cold sensitivity of two alleles (mipAK284 and mipAR338) was dramatically suppressed by benomyl and nocodazole, and a third allele (mipAK408) was dependent on these compounds for viability. These results indicate that the essential functions inhibited in these mutants under restrictive conditions are partially or completely restored by these compounds. Antimicrotubule agents such as benomyl and nocodazole bind to tubulin dimers and poison them. This shifts the tubulin/microtubule equilibrium away from the tubulin polymer. Benomyl, at 2.4 μg/ml, causes a rapid and essentially complete depolymerization of microtubules in A. nidulans (Ovechkina et al., 1999), and it is possible that there is a partial shift of the equilibrium away from microtubule polymer at the low concentrations of benomyl that restore growth to our mutants. Consistent with this possibility, a subinhibitory concentration of nococazole reduces the average number of cytoplasmic microtubules in wild-type and tub4 mutant strains of S. cerevisiae (Vogel and Snyder, 2000). If such a mechanism were operating in A. nidulans, however, these agents should exacerbate the conditional lethality of any microtubule nucleation defective mutants, not facilitate their growth.
Another activity of antimicrotubule agents at low concentrations is inhibition of microtubule dynamics (Wilson and Jordan, 1994). In animal cells and in vitro, nocodazole reduces microtubule elongation and shortening rates while catastrophe and rescue frequencies may increase or decrease depending on the cell type or experimental system (Vasquez et al., 1997; Mikhailov and Gundersen, 1998). The net result in each system, however, is lower microtubule turnover rates. Data on the effects of benomyl on microtubule dynamics have not, to our knowledge, been published, but benomyl is similar in structure to nocodazole and would be predicted to have similar effects. With respect to our mutants, benomyl and nocodazole might tend to reduce the dynamics of assembled microtubules, but there is no reason to believe that they would facilitate nucleation. Decreased catastrophe frequencies might stabilize assembled microtubules and counteract a nucleation failure, but this would be offset by reduced elongation rates and it is difficult to see how a significant net increase in microtubule number or polymer mass would result.
Our D2O results are also informative in this regard. D2O promotes microtubule assembly and reduces rates of catastrophe (Panda et al., 2000, and references therein). In A. nidulans it exacerbates the heat sensitivity conferred by a microtubule-hyperstabilizing β-tubulin mutation and suppresses the cold sensitivity of a microtubule-destabilizing α-tubulin mutation (Oakley et al., 1987a) so it has effects in A. nidulans in vivo that are similar to those observed in vitro and in vivo in other organisms. If our γ-tubulin mutants were microtubule nucleation-defective, one might expect that they would be suppressed by D2O. This was, indeed, the case for mipAR243 and, to a lesser extent, for mipAR63 at high temperatures, but it was not the case for our other alleles. Extending this logic, if benomyl or nocodazole were suppressing the conditional lethality of these alleles by reducing catastrophe frequencies, D2O should be at least as effective in reducing catastrophe frequencies and suppressing the conditional lethality of these alleles. This may well be the case for mipAR243 because there is very slight improvement in the growth of this strain on benomyl at 42°C and a much more significant improvement of growth on D2O at this temperature. Likewise, growth of mipAR63 was very slightly improved by benomyl and D2O at 37°C. D2O did not suppress the conditional lethality of the strongly benomyl-suppressed alleles, however, and may have slightly worsened their growth (Figure 4).
In summary, the following data indicate that mipAK284, mipAR338, and mipAK408 are not defective in the nucleation of microtubule assembly. First, cytoplasmic microtubules are abundant under restrictive conditions and spindles are robust. This allows us to rule out the possibility that there are serious nucleation defects in these mutants and gives no support for even minor nucleation defects. Second, the cold sensitivity of these alleles is strongly suppressed by benomyl and nocodazole. Most of the known effects of these compounds would exacerbate the growth defects of nucleation-deficient mutants, and the only known activity of these compounds that might suppress a nucleation defect is inhibition of catastrophe. If this were the mechanism of suppression, D2O should suppress the cold sensitivity of these alleles better than benomyl and nocodazole, but it does not suppress at all. MipAR63 at high temperatures and mipAR243 are useful controls in this regard because they appear on the basis of microscopy to be nucleation-defective and the conditional lethality of these alleles is very weakly suppressed by benomyl and nocodazole and, as expected, more strongly suppressed by D2O. We are less certain of whether mipAD159 and mipAR63 at low temperatures are nucleation-defective. They are not significantly suppressed by benomyl or nocodazole and are thus clearly different from mipAK284, mipAR338, and mipAK408, but their microscopic phenotypes and lack of D2O suppression suggest that inhibition of spindle microtubule nucleation is probably not the cause of their conditional lethality. Our data, along with the data of Paluh et al. (2000) and Vogel and Snyder (2000), make a persuasive case that γ-tubulin has one or more essential functions in addition to microtubule nucleation.
Inhibition of Germ Tube Formation by mipAR243
The inhibition of germ tube formation by mipAR243 under restrictive conditions was particularly surprising because the microtubule cytoskeleton of A. nidulans has been subjected to a remarkable variety of genetic and chemical insults over the years and none of these insults has prevented germ tube formation. Benomyl, for example, depolymerizes the microtubule cytoskeleton (Ovechkina et al., 1999) but does not block germ tube formation (Oakley and Morris, 1980). Numerous α- and β-tubulin mutations have been identified in A. nidulans that alter microtubule functions in a variety of ways (Oakley and Morris, 1981; Oakley et al., 1985, 1987a; Oakley and Rinehart, 1985), but germ tubes form in each case. It should be noted that mipAR63 and benA33 caused a slight inhibition of germ tube formation in our experiments but much less than mipAR243.
If germ tubes can form in the absence of microtubules, how is mipAR243 inhibiting this process? Two general possibilities occur to us. One is that γ-tubulin has a direct role in germination. This would be surprising, but certainly cannot be ruled out. The second is that mipAR243 alters the microtubule cytoskeleton such that germination is inhibited. This possibility would imply that microtubules normally have a role in germination (e.g., in establishing the site of germination or in transporting materials required for wall growth to the site of germination). If microtubules are absent, a second system involving microfilaments would be able to carry out the functions required for germination perfectly well. Indeed, we can deduce that microtubules play, at most, a secondary role in this process because germination is inhibited by cytochalasins (Oakley, unpublished) and by mutation in the myoA gene, which encodes an essential myosin I (McGoldrick et al., 1995). This means that inhibiting the microfilament-based system inhibits germination even if functional microtubules are present. Microtubules could assist in the process, however, and if the microtubule cytoskeleton in the conidium were altered (e.g., microtubule polarities were randomized but microtubules were not eliminated), it is possible that components required for germination might be deposited randomly around the cell. This could lead to the swollen conidia we see. In this regard, it is worth noting that although mipAR243 appears to inhibit spindle formation, it is not simply a null mutant. The phenotype is different from that of the γ-tubulin deletant.
γ-Tubulin and Organization of Cytoplasmic Microtubules
The phenotype of our γ-tubulin deletion reinforces and validates previous conclusions made with a γ-tubulin disruption (Oakley et al., 1990; Martin et al., 1997). Mitotic spindles are virtually absent in the deletant and the organization of cytoplasmic microtubules is highly abnormal. These data indicate that γ-tubulin is required for spindle formation but is probably not required for the assembly of microtubules in the cytoplasm.
The abnormal cytoplasmic microtubules are particularly interesting in view of the phenotype of the PL301 γ-tubulin mutation in S. pombe (Paluh et al., 2000). This mutation caused bundling of cytoplasmic microtubules among other things. Microtubule bundles are often seen in the γ-tubulin deletant (Figure 7), in mipAR63 at low temperatures (Figure 8), and at a lower frequency in mipAD159. In addition, cytoplasmic microtubules in mipAR243 often form a circular band just inside the periphery of swollen, ungerminated conidia (Figure 5). Thus, microtubule bundling appears to occur if γ-tubulin is absent and in certain mutants in which its function is altered. In addition, abnormally curved microtubules were present in the deletant, in mipAR63 at low temperatures, and in mipAK284 under restrictive conditions.
What accounts for the abnormal organization of cytoplasmic microtubules in these γ-tubulin mutants? One possibility, suggested by Paluh et al. (2000), is that the bundling is due to suppression of microtubule dynamics in these mutants. In this model, microtubules are assumed to have a tendency to bundle (either by being cross-linked by other proteins or by an intrinsic tendency to bundle). γ-Tubulin would normally promote microtubule dynamics in some way and elimination or mutation of γ-tubulin would lead to reduction in microtubule dynamics and, thus, to bundling. Regardless of what mechanism is responsible, our data and the data of Paluh et al. (2000) indicate that γ-tubulin has a role in controlling the organization of cytoplasmic microtubule arrays in addition to its nucleation function.
γ-Tubulin Structure and Function
The mutations we have created define regions important for γ-tubulin function. A large number of the mutations are on the “inside” face (the side away from the viewer in Figure 5, A and B. See also the Quicktime video, gammut.mov) and the “outside” face of γ-tubulin (the side facing the viewer in Figure 5, A and B). All of these are wild type. This result suggests that both the inside and the outside faces of γ-tubulin are not particularly important for function. On the other hand, mutations on the 4 “sides” of the molecule (the plus and minus faces [top and bottom, respectively, in Figure 5], as well as the helix H3 region [left side in Figure 5] and the M-loop region [right side in Figure 5]) confer lethal or conditionally lethal phenotypes and are clearly important for function. The corresponding regions in α- and β-tubulins are known to be involved in polymerization contacts (Nogales et al., 1999). There are relatively few charged regions on the plus face of the molecule so alanine-scanning mutagenesis produces relatively few mutations in this region. However, of the four mutations we created on this face, all confer significant phenotypes.
Nearly all of the cold-sensitive alleles are on the left and right faces of the molecule, which correspond to interfaces expected to be involved in lateral contacts between tubulin subunits. One cold-sensitive mutation and one lethal mutation are located in helix H10, which, in α- and β-tubulins, is involved both in longitudinal and lateral interactions. There is one lethal mutation on the minus face, but other mutations in this region confer no phenotype. The cold- and heat-sensitive allele (mipAR63) is internal and the heat-sensitive allele (mipAR243) is near the “inside” face.
Do the positions and phenotypes of the mutations tell us anything about how γ-tubulin functions and, in particular, how it functions in microtubule nucleation? Two general mechanisms have been proposed for nucleation, the template mechanism (Oakley 1992; Zheng et al., 1995; Keating and Borisy, 2000; Moritz et al., 2000; Wiese and Zheng, 2000) and the protofilament model of lateral interaction of tubulin dimers with a γ-tubulin protofilament (Erickson and Stoffler, 1996). It is important to remember that a large part of the mass of γ-tubulin complexes (γ-tubulin small complexes and γ-TuRCs) consists of proteins other than γ-tubulin. Two of these proteins (the SPC97 and SPC98 homologues) bind directly to γ-tubulin and are essential for viability (Geissler et al., 1996; Knop et al., 1997; Knop and Schiebel, 1997, 1998; Oegema et al., 1999). It follows that mutations that alter interactions of γ-tubulin with these proteins should have significant phenotypes. Although these proteins have not generally been drawn in diagrams of the protofilament nucleation model, it appears that the only regions of γ-tubulin available for binding of these proteins are the inside and outside of the molecule, as all the other regions are proposed to interact with other γ-tubulin molecules or with the tubulin dimer. Our analysis shows that mutations in these regions do not confer strong phenotypes. In the template nucleation model the SPC97–98 homologues could bind to the minus face, where some mutations confer noticeable phenotypes.
It is also worth noting that although the mutations we have created on the lateral faces of the molecule confer significant phenotypes, they do not appear to inhibit microtubule assembly. This indicates that these regions are important, but does not indicate that they are important for microtubule nucleation.
It is interesting to compare our results to those of Llanos et al. (1999). Llanos et al. (1999) prepared overlapping peptides corresponding to the entire amino acid sequence of human γ-tubulin and assayed binding of these peptides to the tubulin dimer. They identified six peptides that bound specifically and tightly to the tubulin dimer (dissociation constants in the nanomolar range). Although the tightness and specificity of the binding argued that the binding was likely to be of biological importance, the Llanos et al. (1999) data were all obtained in vitro and there was no demonstrated in vivo relevance. We were consequently interested in whether our in vivo data correlated in any way with the in vitro data of Llanos et al. (1999). Although the data of Llanos et al. (1999) were based on human γ-tubulin and ours were obtained with A. nidulans γ-tubulin, the similarity of human and A. nidulans sequences makes comparisons straightforward. Interestingly, all but one of our cold-sensitive alleles fell within regions corresponding to the tubulin-binding peptides of Llanos et al. (1999). MipAD159 is in the γ1 peptide of Llanos et al. (1999), mipAK284 is in γ3, mipAR338 is in γ4, and mipAH370 is in γ5. In addition, the benomyl-dependent allele mipAK408 is in γ6 and the lethal alleles mipA341 and mipAR398 are in γ4 and γ6, respectively. These data suggest that the binding data of Llanos et al. (1999) reflect biologically significant interactions. They also imply that the lethality or conditional lethality of these γ-tubulin alleles may be due to altered interactions of γ-tubulin with tubulin dimers.
New Gene Replacement Procedure for Aspergillus nidulans
The heterokaryon gene replacement procedure we have developed is a valuable addition to the repertoire of molecular genetic techniques available for A. nidulans. It has several advantages over previous procedures. First, it allows one to determine easily whether mutant alleles are lethal or wild type. Second, it allows one to detect mutant alleles that only support very slow growth. With the one-step and two-step gene replacement procedures, slow growing mutant colonies may be rapidly out-paced by colonies carrying the wild-type allele and the mutant colonies may not be detected. Indeed, we found that this was the case for three of our mutant alleles. A third advantage of the procedure is that once the correct heterokaryon has been created, the procedure can be carried out for many alleles with a single set of transformations.
The greatest disadvantage of the heterokaryon gene replacement (aside from the time required to create the correct heterokaryon) is that, under our conditions, we found that the plasmid carrying the mutant allele often integrated nonhomologously or in multiple copies. It should be possible, however, to optimize transformation conditions (e.g., by varying numbers of protoplasts and amounts of transforming DNA) to reduce multicopy integrants. The problem of nonhomologous integration should also be reduced by the use of a plasmid with longer homologous regions flanking the mutant allele.
CONCLUSION
The mutations we have created provide some of the first insights into the regions of γ-tubulin that are important to its functions. Our conditionally lethal alleles confer a surprising variety of phenotypes under restrictive conditions. These phenotypes in combination with our benomyl, nocodazole, and D2O results indicate that γ-tubulin has one or more essential functions in addition to microtubule nucleation. The mutant alleles we have created, moreover, are powerful tools that should be of great value in elucidating these functions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ryan Foley for valuable assistance in preparing several mutant alleles for immunofluorescence and Yuki Inclán for preparing the Quicktime video. The wide field deconvolution microscope facility is supported by a grant from the National Science Foundation. This study was supported by Grant GM-31837 from the National Institutes of Health to B.R.O.
Footnotes
Online version of this article contains video material for some figures. Online version is available at www.molbiolcell.org.
REFERENCES
- Botstein D, Shortle D. Strategies and applications of in vitro mutagenesis. Science. 1985;229:1193–1201. doi: 10.1126/science.2994214. [DOI] [PubMed] [Google Scholar]
- Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- Dunne PW, Oakley BR. Mitotic gene conversion, reciprocal recombination and gene replacement at the benA, β-tubulin locus of Aspergillus nidulans. Mol Gen Genet. 1988;213:339–345. doi: 10.1007/BF00339600. [DOI] [PubMed] [Google Scholar]
- Erickson HP. γ-Tubulin nucleation: template or protofilament? Nat Cell Biol. 2000;2:E93–E96. doi: 10.1038/35014084. [DOI] [PubMed] [Google Scholar]
- Erickson HP, Stoffler D. Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to α/β and γ tubulin. J Cell Biol. 1996;135:5–8. doi: 10.1083/jcb.135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the γ-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Inclán YF, Nogales E. Structural models for the self-assembly and microtubule interaction of γ- δ- and ε-tubulin. J Cell Sci. 2001;114:413–422. doi: 10.1242/jcs.114.2.413. [DOI] [PubMed] [Google Scholar]
- Jung MK, Ovechkina Y, Prigozhina N, Oakley CE, Oakley BR. The use of beta-D-glucanase as a substitute for Novozym 234 in immunofluorescence and protoplasting. Fungal Genetics Newsletter. 2000;47:65–66. [Google Scholar]
- Keating TJ, Borisy GG. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol. 2000;2:352–357. doi: 10.1038/35014045. [DOI] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the γ-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Receptors determine the cellular localization of a γ-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie-Mazenc I, Tollon Y, Detraves C, Julian M, Moisand A, Gueth-Hallonet C, Debec A, Salles-Passador I, Puget A, Mazarguil H, Raynaud-Messina B, Wright M. Recruitment of antigenic γ-tubulin during mitosis in animal cells: presence of γ-tubulin in the mitotic spindle. J Cell Sci. 1994;107:2825–2837. doi: 10.1242/jcs.107.10.2825. [DOI] [PubMed] [Google Scholar]
- Llanos R, Chevrier V, Ronjat M, Meurer-Grob P, Martinez P, Frank R, Bornens M, Wade RH, Wehland J, Job D. Tubulin binding sites on γ-tubulin: identification and molecular characterization. Biochemistry. 1999;38:15712–15720. doi: 10.1021/bi990895w. [DOI] [PubMed] [Google Scholar]
- Martin MA, Osmani SA, Oakley BR. The role of γ-tubulin in mitotic spindle formation and cell cycle progression in Aspergillus nidulans. J Cell Sci. 1997;110:623–633. doi: 10.1242/jcs.110.5.623. [DOI] [PubMed] [Google Scholar]
- McGoldrick CA, Gruver C, May GS. myoA of Aspergillus nidulans encodes an essential myosin I required for secretion and polarized growth. J Cell Biol. 1995;128:577–587. doi: 10.1083/jcb.128.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov A, Gundersen GG. Relationship between microtubule dynamics and lamellipodium formation revealed by direct imaging of microtubules in cells treated with nocodazole or taxol. Cell Motil Cytoskeleton. 1998;41:325–340. doi: 10.1002/(SICI)1097-0169(1998)41:4<325::AID-CM5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Miller BL, Miller KY, Timberlake WE. Direct and indirect gene replacements in Aspergillus nidulans. Mol Cell Biol. 1985;5:1714–1721. doi: 10.1128/mcb.5.7.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. Structure of the γ-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Oakley BR. γ-Tubulin: the microtubule organizer? Trends Cell Biol. 1992;2:1–5. doi: 10.1016/0962-8924(92)90125-7. [DOI] [PubMed] [Google Scholar]
- Oakley BR. γ-Tubulin. Curr Top Dev Biol. 2000;49:27–54. doi: 10.1016/s0070-2153(99)49003-9. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Morris NR. Nuclear movement is β-tubulin-dependent in Aspergillus nidulans. Cell. 1980;19:255–262. doi: 10.1016/0092-8674(80)90407-9. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Morris NR. A β-tubulin mutation in Aspergillus nidulans that blocks microtubule function without blocking assembly. Cell. 1981;24:837–845. doi: 10.1016/0092-8674(81)90109-4. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Kniepkamp KS, Rinehart JE. Isolation and characterization of cold-sensitive mutations at the benA, β-tubulin, locus of Aspergillus nidulans. Mol Gen Genet. 1985;201:56–64. doi: 10.1007/BF00397987. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Rinehart JE. Conditionally lethal tubA, α-tubulin, mutations in Aspergillus nidulans. Mol Gen Genet. 1987a;208:135–144. doi: 10.1007/BF00330434. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. γ Tubulin is a component of the spindle-pole-body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Rinehart JE. Mitochondria and nuclei move by different mechanisms in Aspergillus nidulans. J Cell Biol. 1985;101:2392–2397. doi: 10.1083/jcb.101.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Rinehart JE, Mitchell BL, Oakley CE, Carmona C, Gray GL, May GS. Cloning, mapping and molecular analysis of the pyrG (orotidine-5′-phosphate decarboxylase) gene of Aspergillus nidulans. Gene. 1987b;61:385–399. doi: 10.1016/0378-1119(87)90201-0. [DOI] [PubMed] [Google Scholar]
- Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, Mitchison TJ, Zheng Y. Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani SA, Engle DB, Doonan JH, Morris NR. Spindle formation and chromatin condensation in cells blocked at interphase by mutation of a negative cell cycle control gene. Cell. 1988;52:241–251. doi: 10.1016/0092-8674(88)90513-2. [DOI] [PubMed] [Google Scholar]
- Ovechkina YY, Pettit RK, Cichacz ZA, Pettit GR, Oakley BR. Unusual antimicrotubule activity of the antifungal agent spongistatin 1. Antimicrob Agents Chemother. 1999;43:1993–1999. doi: 10.1128/aac.43.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluh JL, Nogales E, Oakley BR, McDonald K, Pidoux A, Cande WZ. A mutation in γ-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein Pkl1p. Mol Biol Cell. 2000;11:1225–1239. doi: 10.1091/mbc.11.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda D, Chakrabarti G, Hudson J, Pigg K, Miller HP, Wilson L, Himes RH. Suppression of microtubule dynamic instability and treadmilling by deuterium oxide. Biochemistry. 2000;39:5075–5081. doi: 10.1021/bi992217f. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons DW, Macdonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Reijo RA, Cooper EM, Beagle GJ, Huffaker TC. Systematic mutational analysis of the yeast β-tubulin gene. Mol Biol Cell. 1994;5:29–43. doi: 10.1091/mbc.5.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KL, Anders KR, Nogales E, Schwartz K, Downing KH, Botstein D. Structure-function relationships in yeast tubulins. Mol Biol Cell. 2000;11:1887–1903. doi: 10.1091/mbc.11.5.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upshall A. Genetic and molecular characterization of argB+ transformants of Aspergillus nidulans. Curr Genet. 1986;10:593–599. doi: 10.1007/BF00418126. [DOI] [PubMed] [Google Scholar]
- Vasquez RJ, Howell B, Yvon A-MC, Wadsworth P, Cassimeris L. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol Biol Cell. 1997;8:973–985. doi: 10.1091/mbc.8.6.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Snyder M. The carboxy terminus of Tub4p is required for γ-tubulin function in budding yeast. J Cell Sci. 2000;113:3871–3882. doi: 10.1242/jcs.113.21.3871. [DOI] [PubMed] [Google Scholar]
- Weil CF, Oakley CE, Oakley BR. Isolation of mip (microtubule-interacting protein) mutations of Aspergillus nidulans. Mol Cell Biol. 1986;6:2963–2968. doi: 10.1128/mcb.6.8.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertman KF, Drubin DG, Botstein D. Systematic mutational analysis of the yeast ACT 1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. γ-Tubulin complexes and their interaction with microtubule-organizing centers. Curr Opin Struct Biol. 1999;9:250–259. doi: 10.1016/S0959-440X(99)80035-9. [DOI] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- Wilson L, Jordan MA. Pharmacological probes of microtubule function. In: Hyams J S, Lloyd C W, editors. Microtubules. New York: Wiley-Liss; 1994. pp. 59–83. [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.