Abstract
Diffuse intrinsic pontine glioma (DIPG) is a rare but uniformly fatal cancer of the brain, with peak incidence in children of 5–7 years of age. In contrast to most types of human cancer, there has been no significant improvement in treatment outcomes for patients with DIPG. Since DIPG occurs in the brainstem, a vital region of the brain, there are no surgical options for providing relief to patients, and chemotherapy as well as radiation therapy provide palliative relief at best. To date, more than 250 clinical trials evaluating radiotherapy along with conventional cytotoxic chemotherapy, as well as newer biologic agents, have failed to improve the dismal outcome when compared with palliative radiation alone. The recent discovery of somatic oncogenic histone gene mutations affecting chromatin regulation in DIPG has dramatically improved our understanding of the disease pathogenesis in DIPG, and these findings have stimulated the development of novel therapeutic approaches targeting epigenetic regulators for disease treatment. This review will discuss about the role of histone modification in chromatin machinery and epigenetic therapeutic strategies for the treatment of DIPG.
Keywords: histone, methylation, demethylation, pediatric brain tumor, DIPG
Introduction
Brain tumors are the main cause of cancer-related morbidity and mortality in children, representing the second-most common cancer and the most common solid tumor in childhood. Among brain tumors, Diffuse Intrinsic Pontine Gliomas (DIPGs) are the most aggressive primary brain tumors in children.1–3) DIPG originates in the pons and is seen almost exclusively in children, with a median age at diagnosis of 6–7 years.4–7) DIPGs represent approximately 10% of all brain tumors in children, and are typically fatal within two years after diagnosis.8,9)
DIPG remains one of the most severe challenges within pediatric neuro-oncology. The mainstay of treatment for DIPG remains radiation (54–60Gy) therapy, fractionated over a six-week period, which often provides transient symptom relief in approximately 70–80% of patients. No survival benefit has been achieved with alternative radiation strategies or a combination with radiation sensitizers.10,11) Unfortunately, cancer recurrence and patient demise is a certainty, with the median overall survival for patients with DIPG of approximately 10 months. Median progression-free survival is between 7 and 8 months, with survival after relapse measured in weeks. To date, more than 250 clinical trials, including standard and novel chemotherapeutics using a variety of regimens, along with radiation therapy, have been conducted, but none has demonstrated significant improvement in treatment outcomes for patients with DIPG.12)
Major obstacles for the development of effective treatments for DIPGs include extensive tumor cell infiltration at the time of diagnosis, an eloquent anatomic location that is inoperable, and poor response to radiotherapy. In the past 20 years, there has been minimal improvement in DIPG outcomes.13,14) A fundamental limitation in the development of biologically targeted therapies for DIPGs is that the diagnosis is usually confirmed by imaging studies, and surgical biopsy is rarely performed due to associated risk.15) The lack of tissue samples, together with a limited understanding of DIPG tumor biology, has significantly hindered progress in identifying underlying oncogenic steps that contribute to the development of DIPG.
However, recent advances in stereotactic neurosurgery have enabled surgeons to obtain reliable biopsy tissue for histological and genomic analysis, and as well for the establishment of primary tumor cells and patient-derived xenograft (PDX) models, with a low morbidity of less than 4% of patients.16–19) Given the relative safety of stereotactic biopsy, sufficient numbers of DIPGs were eventually collected to enable the identification of recurrent gene alterations, the most common of which results in the substitution of methionine for lysine at position 27 of histone H3 variants (to be referred to as K27M).20,21) Despite being just one of 32 genes that encode histone H3 peptides in a diploid cell, this mutation causes a remarkable hypomethylation of a significant fraction of all wild-type histone H3K27 in cells bearing the K27M mutation.22–26) K27M mutant DIPG is associated with a more aggressive clinical course with poor overall response to therapy.27) The molecular as well as tumor biological consequences of, and therapeutic options for treating K27M DIPG are subjects of intense interest, and the current research described here is intended to advance our understanding of these aspects of this intriguing gene alteration.
Histone modification and chromatin machinery
Identification of the role of histone modification and the functional involvement of chromatin machinery is necessary to understand cancer biology, and to develop effective therapeutic strategies in human cancer. Histones are the major protein component of eukaryotic chromatin and are deposited during DNA synthesis. DNA is packaged by eight histone proteins, and this octamer regulates transcription mediated by selective and reversible modifications of nucleosomal DNA28,29) and histone tails.
Histone modifications are known to control the transition in conformation between genes that are active transcriptionally and those that are inactive, and function as molecular switches, suppressing or activating gene expression or altering the expression levels of genes.30–32) Histone N-terminal tails are subject to post-translational modification, including lysine and arginine methylation, lysine acetylation, serine and threonine phosphorylation, and lysine ubiquitination or sumoylation.32–34) These modifications are catalyzed by enzymes that are called “writers,” which include histone methyltransferases, or “erasers,” including histone demethylase and histone deacetylases (HDACs). There are also proteins called “readers” that are associated with chromatin and are involved in recruiting additional proteins with chromatin-modifying functions.35–37)
Histone acetylation
Acetylation of histones increases a negative charge to the protein, loosening the interaction of negatively charged DNA with the histone, leading to active and open chromatin. Histone acetyltransferases (HATs) as writers and HDACs as erasers antagonistically control histone acetylation. Acetylation is considered a histone mark for transcriptional activation, so HATs are associated with active genes, while HDACs are associated with inactive genes. A recent genome-wide mapping of HAT and HDAC binding on chromatin revealed that both are found at active genes with acetylated histones38) and are targeted to transcribed regions of active genes by phosphorylated RNA Pol II. The majority of HDACs in the human genome function to reset chromatin by removing acetylation at active genes.38)
Acetylated histones recruit specific chromatin-associated bromodomain and extraterminal (BET) proteins.35,39) The BET protein bromodomain-containing 4 (BRD 4) is a lysine acetylation “reader”, and plays a role in initial recognition of acetylated histones. The next step is “mediator” recruitment, which is a transcription initiation cofactor, to promoters, which leads to RNA Pol II phosphorylation. Recently, BRD4 was identified as playing a possibly important role in brain cancer development.40–43)
Histone methylation
In contrast to histone acetylation, methylation of histones does not change the charge of histones. Rather, a docking site is created for chromatin-associated proteins containing specific methyl histone-binding domains. Lysine residues of histones can be mono-, di-, or tri-methylated, and each of these modifications has a specific biologic effect. Transcriptionally active euchromatin, for example, can be found associated with methylation of H3K4, H3K36, and H3K79. Alternatively, methylation of H3K9, H3K27, and H4K20 typically can be found in association with transcriptionally repressed heterochromatin. Of the well characterized modifications, H3K4me3 is associated primarily with active promoters,44,45) while H3K27me3 correlates with silencing by the polycomb repressive complex 2 (PRC2). PRC2 is composed of the SET domain-containing histone methyltransferase EZH2 (enhancer of zeste homolog 2) or its functional homologue EZH1, and histone core accessory proteins (EED, SUZ12, and RbAp48), and the PRC2-associated factors JARID2 and ASXL146–48) (Fig. 1). The H3K27me3 mark is recognized by the PRC1 complex, which represses transcription by several mechanisms, including ubiquitination of histone H2A on lysine K119 and chromatin compaction.
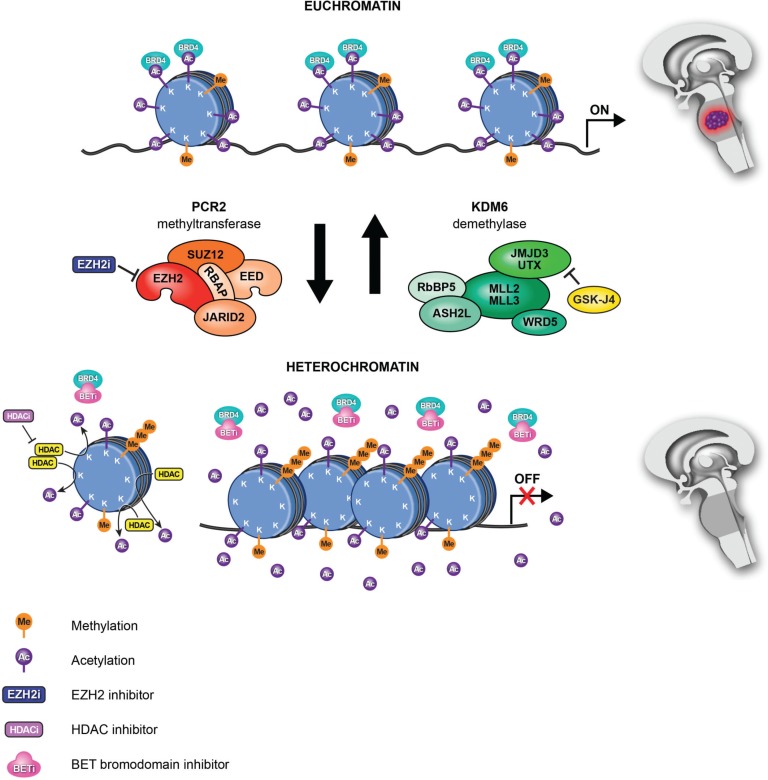
Fig. 1.
New epigenetic therapies for DIPGs. In DIPG, H3K27M mutations (histone H3.1 or H3.3) lead to hypomethylation of H3K27, which promotes a more accessible chromatin state characterized by H3K27 acetylation and aberrant gene expression (upper: Euchromatin). H3K27M mutations inhibits the major H3K27 methylase PRC2. Treatment of DIPG with K27 demethylase inhibitor GSKJ4 results in increased K27me2 and K27me3 and, reduced tumor growth (lower: Heterochromatin). Moreover, treatment with the non-specific HDAC inhibitor panobinostat demonstrated an increase in global H3 acetylation increasing/restoring H3K27me3 levels. In addition, competitive binding with BET bromodomain inhibitors prevents the interaction of BRD4 with acetylated histone, leading to the repression of BRD4 transcriptional targets and also limiting tumor growth. Reprinted by permission from Taylor & Francis Group, LLC: Epigenetics [January 6, Epub ahead of print], copyright 2017.
Histone lysine demethylases (KDMs), which remove methyl group(s) from lysine side chains (Fig. 1), also provide dynamic control of histone methylation. Methyltransferase and demethylases, with their opposing functions, strive to maintain balanced levels of histone methylation. H3K27 methylation is regulated by demethylase, including that of ubiquitously transcribed tetratricopeptide repeat, X chromosome (UTX)/KDM6A and jumonji domain containing-3 (JMJD3)/KDM6B.49–56) UTX and JMJD3 both function as components of a transcriptional activator complex with the MLL2/MLL3 (mixed-lineage leukemia protein 2/3) H3K4 methyltransferases, suggesting dual roles of these enzymes in both removing H3K27 methyl marks and adding methyl groups to H3K4.48)
The genes encoding KDM5A (JARID1A); KDM5C (JARID1C) have been found to have mutations, affecting H3K4 methylation and as has KDM6A (UTX), which affects H3K27 methylation.57–60) The activity of enzymes controlling H3K27 methylation has been found to be both increased and decreased in other cancers,48,61) suggesting that a methylation balance is critical for normal cell growth and for a context-dependent role of polycomb proteins and KDMs in malignancy.
DNA methylation
There have been detailed investigations of DNA methylations in adult GBM, which aided the development of clinical trials, as these relate to tumor recurrence and radiotherapy resistance. By applying genome-wide DNA methylation profiling in pediatric and adult patient cohorts, recurrent age-specific mutations in H3F3A were observed, while tumors with frequent PDGFRA alterations were found in patients wider age range.62) The same approach helped in subgrouping DIPG patients on the basis of CpG island methylation, and identified a subgroup with high-level amplification of MYCN and high-grade histology.63) Observations of the DNA methylation pattern were associated with changes in a specific histone 3 variant mark.19) Recent observation of inactivation of SET-domain-containing histone methyltransferases by K27M variants,23) which may explain the specific DNA methylation pattern in H3.3K27M non-brainstem tumors.61) Since DNA methylation profiles are associated with the K27M mutation irrespective of tumor location, this suggests that the K27M mutation has a role in driving the epigenetic phenotype. Moreover, gain-of-function mutations in ACVR1, FGFR1, and PDGFRA also were found to be associated with H3K27M variants.19)
Histone gene mutations in DIPG
Recent exome sequencing studies of pediatric high-grade gliomas have identified gain-of-function mutations in H3 histone genes: histone 3A (H3F3A) and histone H3b (HIST1H3B), encoding histone H3 variants H3.3 and H3.1 respectively. There are two specific residues that are mutated in histone H3.3, lysine at position 27 for methionine (K27M) or glycine at position 3456) for valine or arginine (G34V/R). K27M is observed in approximately 80% of DIPGs,20,21) mostly involving H3F3A, with the rest involving HIST1H3B.21,27,62–65) G34V/R mutations, however, are observed only in H3F3A.
Since it occurs frequently and confers a poor prognosis, the role of the histone H3K27M mutation has been studied extensively in tumor initiation, maintenance and progression.66,67) Lewis and others have identified a gain-of-function mechanism for this mutation: the K27M mutant protein sequesters PRC2, which normally represses gene expression through histone methylation, and functionally inactivates it.22–25) Suppression of PRC2 by K27M leads to a dramatic reduction of methylation of wild-type H3K27, greatly in excess of the proportion of K27M-to-wild type H3, leading to extensive transcriptional reprogramming of tumor cells.22,24) The effect of K27M on PRC2 function and wild type K27 methylation results in both increased and decreased gene expression. While there is a large-scale decrease in total chromatin H3 di-methylation (K27me2) and tri-methylation (K27me3), and the associated activation of transcription as a resulted of decreased K27 methylation, specific genes are repressed with gain of H3K27 methylation in DIPG. K27M is associated, therefore, with both activation and repression of gene expression. The identification of the specific genes activated and repressed remains are a subject of intensive investigation, with some findings being recently made. Using ChIP-seq and whole-genome sequencing in high-grade gliomas in children, Bender et al. found that DNA hypomethylation and reduced H3K27me3 levels work together to induce gene expression in high-grade gliomas bearing K27M mutations.24) However, Chang et al. found in H3K27M mutant tumors that the genes with increased H3K27me3 were associated with cancer development pathways.22)
It is of interest, and potential significance, that recent study from Drosophila melanogaster constitutively expressing K27M demonstrated that H3K27 acetylation (H3K27ac) levels and associated bromodomain-containing proteins (BRD 1 and 4) are increased in K27M–containing nucleosomes.38) The Drosophila K27M mutant animal models resemble PRC2 loss-of-function phenotypes, causing reduction of H3K27 methylation and derepression of PRC2 target genes, that may indicate similar molecular pathogenesis of K27M pediatric glioma models. Other histone lysine-to-methionine mutations (i.e., H3K9M) have been tested in Drosophila, consistent with H3K27M mutation, showing reduced methylation levels and a possible role in heterochromatic silencing.40)
In a most recent study of patient-derived K27M DIPG models, Piunti et al. found that K27M mutant associates with increased H3K27ac and the heterotypic H3K27M-K27ac nucleosomes colocalize with bromodomain proteins at the loci of actively transcribed genes, whereas PRC2 is excluded from these regions, suggesting that H3K27M does not sequester PRC2 activity.43)
To further understand the role of the H3.3K27M mutation in DIPG tumorigenesis, Funato et al. created a mouse model of DIPG by transducing the gene encoding H3.3K27M into neural progenitors derived from human embryonic stem (ES) cells.68) The oncogenic transformation in neural progenitors was promoted by a synergy between genetic modifications of H3.3K27M, p53 loss, and PDGFRA activation in these particular cells. Importantly, the K27M mutation only transformed neural progenitors derived from ES cells, and not astrocytes derived from these cells or the ES cells themselves. These findings suggest the specificity of K27M-associated transformation for a specific cellular context. H3.3K27M expression also upregulated stem cell–associated genes such as LIN28B, PLAG1, and PLAGL1, while suppression of the same genes reduced growth of these tumor cells.68)
In another study regarding the oncogenic function of H3.3K27M, immortalized human astrocytes were transfected with N-terminally flag-tagged H3.3 wild type (WT) and H3.3K27M, and alterations in both expression and methylation profiles of the H3.3K27M vs. WT expressing cells were observed.63) Molecular and cellular functions, particularly increased cell-to-cell signaling and decreased cell cycle progression and proliferation, were the top pathways altered by H3.3K27M expression. H3.3K27M-expressing cells showed global reduction of H3K27me3 compared with H3.3WT cells. Likewise, reduced H3K27me3 levels relative to WT tumors were observed by immunohistochemical staining of patient-derived K27M DIPGs. However, expression of H3K4me3 and H3K9ac did not change with these mutations.63) These investigations, taken together, reveal the impact of K27M mutations on methylation patterns, gene expression, and transformation.
Histone gene mutations in DIPG are also associated with further genetic alterations.65,69,70) Mutations in activin receptor type 1 (ACVR1, also known as ALK2 (activin receptor-like kinase-2)) are observed frequently (80%) in DIPG with histone H3.1 K27M mutation, but do not occur in other gliomas.17,19,65,71) ACVR1 mutations constitutively activate BMP-dependent transforming growth factor (TGF)-β signaling, with consequent SMAD phosphorylation, leading to increased SMAD target gene transcription. Histone mutations are also associated with oligodendrocyte lineage transcription factors 1 and 2 (OLIG1 and OLIG2) expression, as well as with a forebrain marker, forkhead box G1 transcription factor (FOXG1). High OLIG1 and OLIG2 and low FOXG1 are observed in H3.3 K27M tumors.
Epigenetic targeted therapy for DIPG
Altered epigenetics, either alone or in combination with gene mutations, can play an important role in tumor initiation and progression. The possibility to revert epigenetic changes has recently proven valuable to developing an epigenetic targeted therapy for pediatric brain tumors.26,27,35–37,72–77) Drugs targeting epigenetic modifiers, including the inhibitors of histone methyltransferases, demethylases, HDACs and BET proteins, have emerged recently in clinical trials for these tumors (Table 1).
Table 1.
Epigenetic inhibitors for brain cancer therapy
| Enzyme/protein | Inhibitor | Clinical trials (ongoing or recently completed or terminated) |
|---|---|---|
| UTX /JMJD3 | GSK-J1/J4 | |
| HDAC | Panobinostat | Panobinostat and Stereotactic Radiation Therapy in Treating Patients With Brain Tumors (NCT01324635) |
| Panobinostat in Treating Younger Patients With Progressive Diffuse Intrinsic Pontine Glioma (NCT02899715) | ||
| Trial of Panobinostat in Children With Diffuse Intrinsic Pontine Glioma (NCT02717455) | ||
| Study of LBH589 (Panobinostat) to Treat Malignant Brain Tumors (NCT00848523) | ||
| HDAC | Vorinostat | Study of the Combination of Vorinostat and Radiation Therapy for the Treatment of Patients With Brain Metastases (NCT00838929) |
| Study of Suberoylanilide Hydroxamic Acid (SAHA) With Temsirolimus in Children With Diffuse Intrinsic Pontine Glioma (DIPG) (NCT02420613) | ||
| Vorinostat and Temozolomide in Treating Patients With Malignant Gliomas (NCT00268385) | ||
| N2007-03: Vorinostat and 131-I MIBG in Treating Patients With Resistant or Relapsed Neuroblastoma (NCT01019850) | ||
| Vorinostat and Radiation Therapy Followed by Maintenance Therapy With Vorinostat in Treating Younger Patients With Newly Diagnosed Diffuse Intrinsic Pontine Glioma (NCT01189266) | ||
| Vorinostat, Temozolomide, and Radiation Therapy in Treating Patients With Newly Diagnosed Glioblastoma Multiforme (NCT00731731) | ||
| Vorinostat, Temozolomide, or Bevacizumab in Combination With Radiation Therapy Followed by Bevacizumab and Temozolomide in Young Patients With Newly Diagnosed High-Grade Glioma (NCT01236560) | ||
| Vorinostat and Isotretinoin in Treating Patients With High-Risk Refractory or Recurrent Neuroblastoma (NCT01208454) | ||
| Vorinostat, Isotretinoin and Temozolomide in Adults With Recurrent Glioblastoma Multiforme (GBM) (NCT00555399) | ||
| Magnetic Resonance Spectroscopy Imaging in Predicting Response to Vorinostat and Temozolomide in Patients With Recurrent or Progressive Glioblastoma (NCT01342757) | ||
| Phase I/II Adaptive Randomized Trial of Bevacizumab Versus Bevacizumab Plus Vorinostat in Adults With Recurrent Glioblastoma (NCT01266031) | ||
| High-Dose Vorinostat and Fractionated Stereotactic Body Radiation Therapy in Treating Patients With Recurrent Glioma (NCT01378481) | ||
| Vorinostat and Bortezomib in Treating Young Patients With Refractory or Recurrent Solid Tumors, Including Central Nervous System Tumors and Lymphoma (NCT00994500) | ||
| HDAC | Belinostat | MRSI to Predict Response to RT/TMZ ± Belinostat in GBM (NCT02137759) |
| HDAC | Romidepsin | FR901228 in Treating Patients With Recurrent High-Grade Gliomas (NCT00085540) |
| HDAC | Valproate | Valproate and Etoposide for Patients With Neuronal Tumors and Brain Metastases (NCT00513162) |
| Stereotactic Radiosurgery With Nivolumab and Valproate in Patients With Recurrent Glioblastoma (NCT02648633) | ||
| Sorafenib Tosylate, Valproic Acid, and Sildenafil Citrate in Treating Patients With Recurrent High-Grade Glioma (NCT01817751) | ||
| An International Clinical Program for the Diagnosis and Treatment of Children With Ependymoma (NCT02265770) | ||
| Phase I Study of Temozolomide, Valproic Acid and Radiation Therapy in Patients With Brain Metastases (NCT00437957) | ||
| Valproic Acid and Radiation Followed by Maintenance Valproic Acid and Bevacizumab in Children With High Grade Gliomas or Diffuse Intrinsic Pontine Glioma (NCT00879437) | ||
| Valproic Acid in Treating Young Patients With Recurrent or Refractory Solid Tumors or CNS Tumors (NCT00107458) | ||
| BET | OTX015 | A Trial With Dose Optimization of OTX015 in Recurrent Glioblastoma Multiforme (GBM) Patients (NCT02296476) |
| BET | INCB057643 | A Phase 1/2, Open-Label Safety and Tolerability Study of INCB057643 in Subjects With Advanced Malignancies (NCT02711137) |
| BET | INCB054329 | An Open-Label, Dose-Escalation Study of INCB054329 in Patients With Advanced Malignancies (solid tumors of all types, including brain tumors) (NCT02431260) |
| BET | JQ1 | JQ1 is not being tested in clinical trials due to its short half life. |
| BET | I-BET151 |
Targeting histone methylation
Because global reduction of H3K27 methylation is a key epigenetic event in K27M DIPG, pharmacologic restoration of H3K27 methylation is a rational therapeutic strategy for this lethal pediatric malignancy (Fig. 1). In theory, there are two pharmacological paths to restoring global H3K27 methylation in DIPG: either enhancing PRC2 methyltransferase activity or inhibiting demethylase activity for the lysine 27 residue. Histone H3K27 is methylated by EZH2, a component of PRC2, and is demethylated by the K27 demethylases JMJD3 and UTX.48–56) Recently, a nanomolar inhibitor against JMJD3, GSKJ4, was shown to increase cellular H3K27 methylation in association with its potential use to treat immune disorders78) (Fig. 1). GSKJ4 is considered a prodrug, and its ethyl ester derivative, GSKJ1, is the active histone demethylase inhibitor, and upon hydrolysis GSKJ4 is converted to GSKJ1 in vivo. GSKJ4 has recently been used successfully to treat brainstem gliomas in vitro and in vivo26,71,79–81) (Fig. 1). GSKJ4 treatment promotes increased K27me2 and K27me3 in K27M-mutant DIPG cells, as well as in human astrocytes modified to express K27M.26) GSKJ4 treatment-associated increases in K27me2 and K27me3 are, in turn, associated with a marked dose- and time-dependent inhibition of K27M mutant DIPG cell growth. In contrast, GSKJ4 has little effect against pediatric glioma cells with wild type and G34V mutant H3F3A. In addition to its anti-proliferative activity, GSKJ4 increased the apoptosis of K27M mutant cells, while having no significant effect on apoptosis for cells expressing wild-type or the G34V mutant. Importantly, pharmacologic inhibition of JMJD3 in DIPG orthotopic xenografts reduced tumor growth and significantly extended animal survival but did not achieve the same effect in animals with xenografts containing wild-type H3.3. These results suggested that GSKJ4 anti-tumor activity is specific to K27M mutant tumors, and this K27M anti-tumor activity is associated with increased K27me2 and K27me3. The penetration of GSKJ4 into the brain, including the brainstem where DIPG tumors develop, was assessed by high-performance liquid chromatography following systemic administration of the drug, and suggesting that GSKJ4 be explored as a potential targeted therapy for patients with DIPG.26)
Of interest, GSKJ4 showed a potential therapeutic activity for T-cell acute lymphoblastic leukemia (T-ALL), a hematologic malignancy driven by NOTCH1 signaling, loss-of-function mutations and deletion of the genes EZH2 and SUZ12; JMJD3 is expressed at higher levels in T-ALL relative to other leukemias.58,82,83) GSKJ4 reduced cell proliferation and increased K27M methylation in T-ALL.58) The main mechanism of GSKJ4 in T-ALL appears to be through JMJD3 inhibition.58)
For both T-ALL and DIPG, inhibition of UTX, the other known H3K27 demethylase, did not reduce proliferation of tumor cells. Furthermore, in T-ALL, the genes suppressed by GSKJ4 had significant overlap with genes upregulated by UTX knocking down. Taken together, these results point to distinct roles in chromatin modification for UTX and JMJD3, and suggest that JMJD3 is a likely therapeutic epigenetic target for treating pediatric cancers.
Besides increasing H3K27 methylation by GSKJ4-mediated inhibition of the JMJD3 demethylase, a recent study of patient–derived DIPG models demonstrated that inhibition of residual EZH2 activity decrease H3K27 methylation at the promoter of tumor-suppressor protein p16INK4a (encoded by CDKN2A (cyclin-dependent kinase inhibitor 2A)) and reduces cell proliferation, implicating that EZH2 inhibition presents an epigenetic therapeutic strategy for DIPG.84)
Targeting histone acetylation
The elevated level of H3K27 acetylation in DIPG has also been a potential epigenetic target by treating with HDAC inhibitors (Table 1). HDAC inhibitors increase histone acetylation by inhibiting histone deacetylation, leading to an open chromatin structure and gene activation (Fig. 1). A non-selective FDA-approved potent HDAC inhibitor, panobinostat (LBH589, Farydak), has been developed for the treatment of various cancers85) (Table 1). A recent chemical screening in patient-derived DIPG cells identified panobinostat as having potent anti-tumor activity against DIPG in vitro.86) In fact, panobinostat induced a dose-dependent increase in global H3 acetylation as well as H3K27 methylation, and reduced the expression of oncogenes (e.g., MK167, CCND1). Polyacetylation of the H3 N-terminal tail by panobinostat can ‘detoxify’ K27M-induced inhibition of PRC2 and rescue the H3K27 hypomethylation phenotype. Synergy between panobinostat and GSKJ4 was observed in DIPG cells,86) highlighting the importance of investigating the anti-tumor activities resulting from simultaneous inhibition of multiple histone modifier.
In a recent study in genetic and orthotopic DIPG models, Hennika et al. found, consistent with previous findings,86) that panobinostat demonstrated significant anti-tumor activity in vitro and in short-term in vivo efficacy studies.87) However, the efficacy of panosbinostat was unrelated to H3 status in this study. Moreover, panobinostat treatment at its well-tolerated dose did not increase overall survival in the K27M DIPG models, suggesting that substantial toxicity would arise in using a panobinostat concentration and treatment duration sufficient to provide an overall survival benefit. Given the important functions of histone acetylation in normal physiology, HDAC inhibition could cause off-target effects with systemic administration. The accessibility at the appropriate central nervous system (CNS) location of agents targeting epigenetic modifiers should be assessed in well-designed toxicological studies. The challenge for HDAC inhibitors will be to achieve effective concentrations able to inhibit the target, which would require either direct CNS administration or sufficient penetration of the blood-brain barrier with systemic administration, with a large enough therapeutic window and an acceptable toxicity profile, considering the ongoing normal development of pediatric patients. To define the adverse effect and maximum tolerated dose, phase I clinical trials of this compound are currently being tested in children with DIPG (NCT02717455, NCT02899715). NCT02717455 is sponsored phase 1 clinical trial of panobinostat in children with recurrent and progressive DIPG by Pediatric Brain Tumor Consortium (PBTC) and currently recruiting the patient with age from 2 to 21 year olds. Panobinostat will be administered orally every other day, 3 times a week for three weeks, followed by one week off of therapy. Three weeks of therapy plus the one-week rest period (total 4 weeks) will constitute one course. Treatment will continue for up to 26 courses (approximately 2 years) barring progressive disease or unacceptable toxicity. NCT02899715 is sponsored phase 1 clinical trial of panobinostat in children with DIPG by National Cancer Institute (NCI) and currently recruiting the patient. Patients will receive panobinostat orally thrice weekly for 3 weeks. Treatment repeats every 28 days for 26 courses (2 years) in the absence of disease progression or unacceptable toxicity. In addition to the pharmacokinetic (PK) analysis of panobinostat, this trial will measure changes of H3K27M mutations in cell-free DNA from peripheral blood and urine samples.
Targeting BET proteins
In addition to pharmacologic inhibition of histone acetylation by HDAC inhibitors, highly selective BET protein inhibitors, such as JQ-1,39,43,88,89) I-BET151,90,91) I-BET762,92) INCB054329,42) and OTX-015,93) have been generated to target specifically the recognition of acetylated lysine residues (Fig. 1) (Table 1). The BET bromodomain BRD4 binds to acetylated histones through its conserved bromodomains and activates transcription.74) Competitive binding of BET inhibitors to the bromodomain pocket results in displacement of BRD4 from acetylated histones in active chromatin, leading to transcriptional inactivation (Fig. 1).74,88,89,91,94,95) Several preclinical studies with the BET inhibitors JQ1 and I-BET151 have been conducted in glioma models including DIPG, with encouraging antitumor activity.43,96,97) Moreover, a novel BET inhibitor, OTX-015 (MK-8628), was tested at the clinical level in dose-finding studies in glioblastoma (GBM) patients (NCT02296476), having shown antitumor effects in vitro and in vivo as monotherapy and in combination with conventional treatment in GBM models42) (Table 1). A phase 1/2 study of another BET inhibitor, INCB057643, is also currently being tested in advanced solid tumors (NCT02711137) (Table 1). Of note, the anti-proliferative effects of BET inhibitors in GBM may be mediated, at least in part, by reduced expression of the long non-coding RNA HOX transcript antisense RNA (HOTAIR), which has tumor-promoting effects.98)
Conclusion
Recently, our knowledge of DIPG genetics and epigenetics has significantly improved in association with the discovery of histone gene mutations. Current investigations are underway to define the function of histone mutations and to determine how they interact with other known oncogenes. Further investigation is needed to understand the mechanisms of action of the mutations observed DIPG and the corresponding alterations in chromatin machinery, in addition to their complex interactions with other known oncogenes. Therapeutic targeting of histone modifiers responsible for chromatin formation and epigenetic regulation of gene expression is a relatively new strategy and an area of increased interest in pediatric neuro-oncology. Ongoing clinical trials are evaluating the anti-tumor activity of histone modifier inhibitors, and also any interactions with conventional genotoxic therapies, with the goal of improving outcomes for children with DIPG. Given the important functions of histone modifications in normal physiology, optimizing the dose and frequency of administration of inhibitors of histone modifiers to maximize the therapeutic window and limit toxicity will be a key factor for reaching a successful clinical outcome. The combination of agents with different mechanisms of action may offer a better chance for tumor control with limited toxicity and a reduced likelihood of acquired resistance compared with single agent therapy. Clinical trials should include approaches for monitoring tumor treatment response, which will need the identification and validation of non-invasive biomarkers for residual tumors, and for assessing the molecular effects of treatment through analysis of tumor tissue or other tissue samples obtained during and/or after treatment.
Acknowledgment
R.H. was supported by US National Institutes of Health (NIH) grant NS093079, and the Bear Necessities Pediatric Cancer Foundation and Rally Foundation, and the John McNicholas Pediatric Brain Tumor Foundation. We thank Michael Gallagher for illustration of the figure and Accixx Biomedical Consulting (www.Accixx.com) for editorial assistance
Footnotes
Conflicts of Interest Disclosure
The author declares that there is no conflict of interest with the content of this report.
References
- 1).Wong ET, Hess KR, Gleason MJ, et al. : Outcomes and prognostic factors in recurrent glioma patients enrolled onto phase II clinical trials. J Clin Oncol 17: 2572–2578, 1999 [DOI] [PubMed] [Google Scholar]
- 2).Buckner JC: Factors influencing survival in high-grade gliomas. Semin Oncol 30: 10–14, 2003 [DOI] [PubMed] [Google Scholar]
- 3).Louis DN, Ohgaki H, Wiestler OD, et al. : The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114: 97–109, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Freeman CR, Farmer JP: Pediatric brain stem gliomas: a review. Int J Radiat Oncol Biol Phys 40: 265–271, 1998 [DOI] [PubMed] [Google Scholar]
- 5).Berger MS, Edwards MS, LaMasters D, Davis RL, Wilson CB: Pediatric brain stem tumors: radiographic, pathological, and clinical correlations. Neurosurgery 12: 298–302, 1983 [DOI] [PubMed] [Google Scholar]
- 6).Littman P, Jarrett P, Bilaniuk LT, et al. : Pediatric brain stem gliomas. Cancer 45: 2787–2792, 1980 [DOI] [PubMed] [Google Scholar]
- 7).Hargrave D, Bartels U, Bouffet E: Diffuse brainstem glioma in children: critical review of clinical trials. Lancet Oncol 7: 241–248, 2006 [DOI] [PubMed] [Google Scholar]
- 8).Donaldson SS, Laningham F, Fisher PG: Advances toward an understanding of brainstem gliomas. J Clin Oncol 24: 1266–1272, 2006 [DOI] [PubMed] [Google Scholar]
- 9).Frazier JL, Lee J, Thomale UW, Noggle JC, Cohen KJ, Jallo GI: Treatment of diffuse intrinsic brainstem gliomas: failed approaches and future strategies. J Neurosurg Pediatr 3: 259–269, 2009 [DOI] [PubMed] [Google Scholar]
- 10).Robison NJ, Kieran MW: Diffuse intrinsic pontine glioma: a reassessment. J Neurooncol 119: 7–15, 2014 [DOI] [PubMed] [Google Scholar]
- 11).Roos DE, Smith JG: Randomized trial on radiotherapy for paediatric diffuse intrinsic pontine glioma (DIPG). Radiother Oncol 113: 425, 2014 [DOI] [PubMed] [Google Scholar]
- 12).Warren KE: Diffuse intrinsic pontine glioma: poised for progress. Front Oncol 2: 205, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Fangusaro J: Pediatric high-grade gliomas and diffuse intrinsic pontine gliomas. J Child Neurol 24: 1409–1417, 2009 [DOI] [PubMed] [Google Scholar]
- 14).Kebudi R, Cakir FB: Management of diffuse pontine gliomas in children: recent developments. Paediatr Drugs 15: 351–362, 2013 [DOI] [PubMed] [Google Scholar]
- 15).Barkovich AJ, Krischer J, Kun LE, et al. : Brain stem gliomas: a classification system based on magnetic resonance imaging. Pediatr Neurosurg 16: 73–83, 1990 [DOI] [PubMed] [Google Scholar]
- 16).Roujeau T, Machado G, Garnett MR, et al. : Stereotactic biopsy of diffuse pontine lesions in children. J Neurosurg 107: 1–4, 2007 [DOI] [PubMed] [Google Scholar]
- 17).Puget S, Philippe C, Bax DA, et al. : Mesenchymal transition and PDGFRA amplification/mutation are key distinct oncogenic events in pediatric diffuse intrinsic pontine gliomas. PLoS One 7: e30313, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Geoerger B, Hargrave D, Thomas F, et al. : ITCC (Innovative Therapies for Children with Cancer) European Consortium: Innovative Therapies for Children with Cancer pediatric phase I study of erlotinib in brainstem glioma and relapsing/refractory brain tumors. Neuro-oncology 13: 109–118, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, et al. : Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet 46: 462–466, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Schwartzentruber J, Korshunov A, Liu XY, et al. : Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231, 2012 [DOI] [PubMed] [Google Scholar]
- 21).Wu G, Broniscer A, McEachron TA, et al. St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project : Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44: 251–253, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Chan KM, Fang D, Gan H, et al. : The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27: 985–990, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Lewis PW, Müller MM, Koletsky MS, et al. : Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Bender S, Tang Y, Lindroth AM, et al. : Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24: 660–672, 2013 [DOI] [PubMed] [Google Scholar]
- 25).Venneti S, Garimella MT, Sullivan LM, et al. : Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol 23: 558–564, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Hashizume R, Andor N, Ihara Y, et al. : Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med 20: 1394–1396, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Khuong-Quang DA, Buczkowicz P, Rakopoulos P, et al. : K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 124: 439–447, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Churchman LS, Weissman JS: Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C: Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325: 626–628, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Furey TS, Sethupathy P: Genetics. Genetics driving epigenetics. Science 342: 705–706, 2013 [DOI] [PubMed] [Google Scholar]
- 31).Strahl BD, Allis CD: The language of covalent histone modifications. Nature 403: 41–45, 2000 [DOI] [PubMed] [Google Scholar]
- 32).Henikoff S: Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet 9: 15–26, 2008 [DOI] [PubMed] [Google Scholar]
- 33).Rivera CM, Ren B: Mapping human epigenomes. Cell 155: 39–55, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Bird A: Perceptions of epigenetics. Nature 447: 396–398, 2007 [DOI] [PubMed] [Google Scholar]
- 35).Kaelin WG, McKnight SL: Influence of metabolism on epigenetics and disease. Cell 153: 56–69, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Black JC, Van Rechem C, Whetstine JR: Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 48: 491–507, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Arcipowski KM, Martinez CA, Ntziachristos P: Histone demethylases in physiology and cancer: a tale of two enzymes, JMJD3 and UTX. Curr Opin Genet Dev 36: 59–67, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Wang Z, Zang C, Cui K, et al. : Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Filippakopoulos P, Knapp S: Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 13: 337–356, 2014 [DOI] [PubMed] [Google Scholar]
- 40).Herz HM, Morgan M, Gao X, et al. : Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 345: 1065–1070, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Lin CY, Erkek S, Tong Y, et al. : Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 530: 57–62, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Wadhwa E, Nicolaides T: Bromodomain Inhibitor Review: Bromodomain and Extra-terminal Family Protein Inhibitors as a Potential New Therapy in Central Nervous System Tumors. Cureus 8: e620, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Piunti A, Hashizume R, Morgan MA, et al. : Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med 23: 493–500, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Barski A, Cuddapah S, Cui K, et al. : High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837, 2007 [DOI] [PubMed] [Google Scholar]
- 45).Heintzman ND, Hon GC, Hawkins RD, et al. : Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Müller J, Hart CM, Francis NJ, et al. : Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208, 2002 [DOI] [PubMed] [Google Scholar]
- 47).Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V: Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196, 2002 [DOI] [PubMed] [Google Scholar]
- 48).Ezponda T, Licht JD: Molecular pathways: deregulation of histone h3 lysine 27 methylation in cancer-different paths, same destination. Clin Cancer Res 20: 5001–5008, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Agger K, Cloos PA, Christensen J, et al. : UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734, 2007 [DOI] [PubMed] [Google Scholar]
- 50).Deb G, Singh AK, Gupta S: EZH2: not EZHY (easy) to deal. Mol Cancer Res 12: 639–653, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Simon JA, Lange CA: Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res 647: 21–29, 2008 [DOI] [PubMed] [Google Scholar]
- 52).Kleer CG, Cao Q, Varambally S, et al. : EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 100: 11606–11611, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Varambally S, Dhanasekaran SM, Zhou M, et al. : The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419: 624–629, 2002 [DOI] [PubMed] [Google Scholar]
- 54).Hübner MR, Spector DL: Role of H3K27 demethylases Jmjd3 and UTX in transcriptional regulation. Cold Spring Harb Symp Quant Biol 75: 43–49, 2010 [DOI] [PubMed] [Google Scholar]
- 55).Kooistra SM, Helin K: Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 13: 297–311, 2012 [DOI] [PubMed] [Google Scholar]
- 56).Cloos PA, Christensen J, Agger K, Helin K: Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 22: 1115–1140, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Jones DT, Jäger N, Kool M, et al. : Dissecting the genomic complexity underlying medulloblastoma. Nature 488: 100–105, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Ntziachristos P, Tsirigos A, Van Vlierberghe P, et al. : Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 18: 298–301, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59).Pugh TJ, Weeraratne SD, Archer TC, et al. : Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488: 106–110, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60).Imielinski M, Berger AH, Hammerman PS, et al. : Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150: 1107–1120, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61).Martinez-Garcia E, Licht JD: Deregulation of H3K27 methylation in cancer. Nat Genet 42: 100–101, 2010 [DOI] [PubMed] [Google Scholar]
- 62).Sturm D, Witt H, Hovestadt V, et al. : Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22: 425–437, 2012 [DOI] [PubMed] [Google Scholar]
- 63).Buczkowicz P, Hoeman C, Rakopoulos P, et al. : Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet 46: 451–456, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64).Waldmann T, Schneider R: Targeting histone modifications—epigenetics in cancer. Curr Opin Cell Biol 25: 184–189, 2013 [DOI] [PubMed] [Google Scholar]
- 65).Taylor KR, Mackay A, Truffaux N, et al. : Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet 46: 457–461, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Sturm D, Bender S, Jones DT, et al. : Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer 14: 92–107, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67).Zadeh G, Aldape K: ACVR1 mutations and the genomic landscape of pediatric diffuse glioma. Nat Genet 46: 421–422, 2014 [DOI] [PubMed] [Google Scholar]
- 68).Funato K, Major T, Lewis PW, Allis CD, Tabar V: Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346: 1529–1533, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69).Bax DA, Mackay A, Little SE, et al. : A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin Cancer Res 16: 3368–3377, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70).Paugh BS, Qu C, Jones C, et al. : Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol 28: 3061–3068, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).Becher OJ, Wechsler-Reya RJ: Cancer. For pediatric glioma, leave no histone unturned. Science 346: 1458–1459, 2014 [DOI] [PubMed] [Google Scholar]
- 72).Zahnow CA, Topper M, Stone M, et al. : Inhibitors of DNA methylation, histone deacetylation, and histone demethylation: a perfect combination for cancer therapy. Adv Cancer Res 130: 55–111, 2016 [DOI] [PubMed] [Google Scholar]
- 73).Song Y, Wu F, Wu J: Targeting histone methylation for cancer therapy: enzymes, inhibitors, biological activity and perspectives. J Hematol Oncol 9: 49, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74).Popovic R, Licht JD: Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov 2: 405–413, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75).McGrath J, Trojer P: Targeting histone lysine methylation in cancer. Pharmacol Ther 150: 1–22, 2015 [DOI] [PubMed] [Google Scholar]
- 76).Liu Y, Liu K, Qin S, Xu C, Min J: Epigenetic targets and drug discovery: part 1: histone methylation. Pharmacol Ther 143: 275–294, 2014 [DOI] [PubMed] [Google Scholar]
- 77).Liu K, Liu Y, Lau JL, Min J: Epigenetic targets and drug discovery Part 2: Histone demethylation and DNA methylation. Pharmacol Ther 151: 121–140, 2015 [DOI] [PubMed] [Google Scholar]
- 78).Kruidenier L, Chung CW, Cheng Z, et al. : A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488: 404–408, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79).Ramaswamy V, Remke M, Taylor MD: An epigenetic therapy for diffuse intrinsic pontine gliomas. Nat Med 20: 1378–1379, 2014 [DOI] [PubMed] [Google Scholar]
- 80).Morales La Madrid A, Hashizume R, Kieran MW: Future Clinical Trials in DIPG: Bringing Epigenetics to the Clinic. Front Oncol 5: 148, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81).Lulla RR, Saratsis AM, Hashizume R: Mutations in chromatin machinery and pediatric high-grade glioma. Sci Adv 2: e1501354, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82).Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, et al. : ETV6 mutations in early immature human T cell leukemias. J Exp Med 208: 2571–2579, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83).Valk PJ, Verhaak RG, Beijen MA, et al. : Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 350: 1617–1628, 2004 [DOI] [PubMed] [Google Scholar]
- 84).Mohammad F, Weissmann S, Leblanc B, et al. : EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med 23: 483–492, 2017 [DOI] [PubMed] [Google Scholar]
- 85).Ellis L, Pan Y, Smyth GK, et al. : Histone deacetylase inhibitor panobinostat induces clinical responses with associated alterations in gene expression profiles in cutaneous T-cell lymphoma. Clin Cancer Res 14: 4500–4510, 2008 [DOI] [PubMed] [Google Scholar]
- 86).Grasso CS, Tang Y, Truffaux N, et al. : Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med 21: 555–559, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87).Hennika T, Hu G, Olaciregui NG, et al. : Pre-clinical study of panobinostat in xenograft and genetically engineered murine diffuse intrinsic pontine glioma models. PLoS One 12: e0169485, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88).Filippakopoulos P, Qi J, Picaud S, et al. : Selective inhibition of BET bromodomains. Nature 468: 1067–1073, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89).Delmore JE, Issa GC, Lemieux ME, et al. : BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90).Nicodeme E, Jeffrey KL, Schaefer U, et al. : Suppression of inflammation by a synthetic histone mimic. Nature 468: 1119–1123, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91).Dawson MA, Prinjha RK, Dittmann A, et al. : Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478: 529–533, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92).Mirguet O, Gosmini R, Toum J, et al. : Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J Med Chem 56: 7501–7515, 2013 [DOI] [PubMed] [Google Scholar]
- 93).Berenguer-Daizé C, Astorgues-Xerri L, Odore E, et al. : OTX015 (MK-8628), a novel BET inhibitor, displays in vitro and in vivo antitumor effects alone and in combination with conventional therapies in glioblastoma models. Int J Cancer 139: 2047–2055, 2016 [DOI] [PubMed] [Google Scholar]
- 94).Puissant A, Frumm SM, Alexe G, et al. : Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov 3: 308–323, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95).Lovén J, Hoke HA, Lin CY, et al. : Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96).Pastori C, Daniel M, Penas C, et al. : BET bromodomain proteins are required for glioblastoma cell proliferation. Epigenetics 9: 611–620, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97).Cheng Z, Gong Y, Ma Y, et al. : Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res 19: 1748–1759, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98).Pastori C, Kapranov P, Penas C, et al. : The bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci USA 112: 8326–8331, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]