Abstract
The idea that development is the expression of information accumulated during evolution and that heredity is the transmission of this information is surprisingly hard to cash out in strict, scientific terms. This paper seeks to do so using the sense of information introduced by Francis Crick in his sequence hypothesis and central dogma of molecular biology. It focuses on Crick's idea of precise determination. This is analysed using an information-theoretic measure of causal specificity. This allows us to reconstruct some of Crick's claims about information in transcription and translation. Crick's approach to information has natural extensions to non-coding regions of DNA, to epigenetic marks, and to the genetic or environmental upstream causes of those epigenetic marks. Epigenetic information cannot be reduced to genetic information. The existence of biological information in epigenetic and exogenetic factors is relevant to evolution as well as to development.
Keywords: genetic information, epigenetics, specificity
1. Genetic information
That the development of evolved characteristics is the expression of information accumulated in the genome during evolution and that heredity is the transmission of this information from one generation to the next will strike most biologists as common sense. But it is surprisingly difficult to cash out this statement in a way that is grounded in the detailed theory and practice of the biosciences.1 Biology today is certainly an ‘information science’, both because it is a science of big data and because many specific models are inspired by the information sciences, but these applications and models do not seem to be unified by a single conception of biological information. If the actual science straightforwardly corresponded to that opening statement, we would expect to find that instructions written in the genetic code are read by gene regulatory networks to make an organism. But the genetic code runs out of steam when it has specified the linear structure of proteins [2]. It is impossible to describe higher levels of biological organization in the genetic code for the same reason that I cannot write literature using a geodetic coordinate system: the language does not have the expressive power. Nor is it easy to see how the expressive power of the genetic code could be expanded to describe something beyond the order of amino acids in a polypeptide. The ‘histone codes’ [3] and ‘splicing codes’ [4] that have been proposed as supplements to the genetic code are not integrated with the genetic code through a shared measure of coded information. As things stand, histone modification and mRNA splicing are molecular mechanisms that interact with the mechanisms of transcription and translation in the same straightforward way as any combination of physical mechanisms can. This paper outlines a measure of information that allows us to compare the contributions made by each of these mechanisms to determine a final product in a shared, informational currency.
Turning our attention to gene regulatory networks, these are productively modelled as computing Boolean functions and/or differential equations, but these computational operations are not specified in any of the three ‘codes’ to which we just referred. Instead, these operations are specified by the stereochemical affinities of genomic regions and gene products. The science that connects the ‘codes’ with the ‘computing networks’ is the physics of how stereochemical properties emerge from the linear structure of biomolecules and the cellular contexts in which those biomolecules mature and function. The same is true of the other molecular networks that are at the heart of our understanding of the cell—when we model these networks as performing computations, those formal operations do not take as inputs representations written in the genetic code.
All this suggests that perhaps ‘biology is an information science’ only in the sense that it uses many models that start with analogies to some aspect of communication or computing, and makes many direct applications of formalisms from the information sciences. Each of these models or applications stands or falls on its own scientific merits. They do not link together to form a single theory of biological information or a theory of life as an informational phenomenon [2,5–7]. On this sceptical view, the ubiquity of information talk in biology is only evidence of the power and generality of theories of information and computation, something we can observe in many other areas of science.
This paper defends a more robust view of biological information, however. It argues that there is an important sense of ‘information’ which is related very closely to the older notion of biological ‘specificity’. Biological information in this sense gives scientific substance to the claim that development is the expression of information accumulated during evolution, and that heredity is the transmission of this information from one generation to the next. These claims turn out to be more or less equivalent to the idea that heredity is the ability of one cell to transmit biological specificity to another and that development is the expression of that specificity in a controlled manner.
The paper builds on Paul Griffiths and Karola Stotz's ‘bottom-up’ approach to biological information, starting with a simple concept of information that plays a straightforward role at the heart of molecular biology and seeing how many other aspects of biology can be clarified by applying this sense of information. That starting point is what they termed ‘Crick information’, the sense of information introduced by Crick [8] in his ‘sequence hypothesis’ and ‘central dogma of molecular biology’ [9,10].2
Given the central role of Crick's ideas in molecular biology, it is surprising that previous efforts to explicate the idea of biological information have not adopted Crick's straightforward approach. Instead, they have mostly focused on the richer connotations of the term ‘information’: ideas like meaning, representation and semiosis.3 Some authors have even attributed this rich sense of information to Crick: ‘The sense of information relevant to the central dogma is of course the sort which requires “intentionality”, “aboutness”, “content”, the representation of other states of affairs … ’ [14, pp. 550–551]. As we will see in the next section, nothing could be further from Crick's intentions. The problem with rich approaches to biological information is that we do not have developed, technical theories of information in this sense. The various terms used in the passage just cited are, as the author admits, merely ‘one or another facet of a philosophically vexed concept’ [14, p. 151]. So the approach amounts to taking this vexed concept, for which we have no developed theory, and placing it at the foundations of an account of living systems. In this paper, in contrast, we will use only the standard formalism of information theory and the idea of biological specificity.
2. Crick's conception of information
The key move made by Crick in his work on protein synthesis was to supplement the existing idea of stereochemical specificity, embodied in the three-dimensional structure of biomolecules and underlying the well-known lock-and-key model of interaction between enzymes and their substrates, with the idea of informational specificity, embodied in the linear structure of nucleic acids that determine the linear structure of a gene product [5,15]. This idea is present in Crick's statements of both the sequence hypothesis, and the central dogma (figure 1):
The Sequence Hypothesis … In its simplest form it assumes that the specificity of a piece of nucleic acid is expressed solely by the sequence of its bases, and that this sequence is a (simple) code for the amino acid sequence of a particular protein.
The Central Dogma This states that once ‘information’ has passed into a protein it cannot get out again. In more detail, the transfer of information from nucleic acid to protein may be possible, but transfer from protein to protein, or from protein to nucleic acid is impossible. Information means here the precise determination of sequence, either of bases in the nucleic acid or of amino-acid residues in the protein. [8, pp. 152–153, italics in original]
Figure 1.
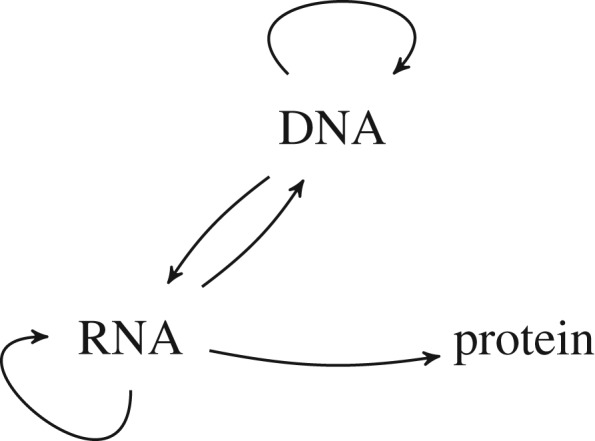
The central dogma, as it is held today. After [16], with modifications. In particular, an arrow from DNA to protein has been removed.
According to Crick, the process of protein synthesis involves ‘the flow of energy, the flow of matter, and the flow of information’. While noting the importance of the ‘exact chemical steps’, he separated this transfer of matter and energy from what he regarded as ‘the crux of the problem’, namely how to join the amino acids in the right order—‘the crucial act of sequentialization’. His solution to this problem would ‘particularly emphasize the flow of information’ where ‘By information I mean the specification of the amino acid sequence of the protein’ [8, p. 144].
Crick maintained the same, straightforward view of information throughout his career. In his well-known paper clarifying the central dogma, he reiterated that his key achievement in 1958 was to reduce the problem of protein synthesis to ‘the formulation of the general rules for information transfer from one polymer with a defined alphabet to another’ [16, p. 561]. Information is a causal concept, referring simply to precise determination. Crick reiterated this 40 years later: ‘ … “Information” in the dna, rna, protein sense is merely a convenient shorthand for the underlying causal effect’. (Crick to Morgan, 20 March 1998). ‘As to “information,” I imagine one could avoid the word if one didn't like it and say “detailed residue-by-residue determination”’ (Crick to Morgan, 3 April 1998). Moreover, ‘As to “meaning” … I would keep away from the term.’ (Crick to Morgan, 3 April 1998).4
So if we take Crick at his word, then information is about (i) precise determination and (ii) transfer of biological specificity from one biomolecule to another (in both development and in heredity).
These two aspects of Crick's ideas about information can be made precise using Shannon information measures and algorithmic information measures, respectively. This paper concentrates on the first aspect of information and on Shannon measures of information.5
3. Information as precise determination
When Crick said that he would emphasize information in his account of protein synthesis, rather than matter and energy, he meant that he would focus on the precise determination of the structure of one biomolecule by another. There are variables through which the cell exercises this precise determination, notably coding sequences of nucleic acid, and other variables through which it does not, such as the presence or absence of an RNA polymerase in the transcription process. Variables of this second kind are absolutely required to construct the downstream biomolecule: without them nothing will happen. But they do not precisely determine the structure of that biomolecule: their role will remain the same no matter what particular structure is produced. Crick's distinction between ‘matter and energy’, on the one hand, and ‘information’, on the other, thus corresponds to the standard distinction between the efficiency and specificity of a molecular process. The efficiency of a molecular process is a matter of how much product is obtained for a given quantity of inputs. The specificity of the process is the extent to which the process produces just one output, rather than other energetically equivalent outputs. A well-designed polymerase chain reaction, for example, will produce just one DNA product (specificity) but many copies of that product (efficiency).
Biological specificity is explained by locating the variables through which cells exercise precise determination of outcomes. In philosophy, these variables are known (coincidentally as far as the author can discover) as ‘specific causes’ [17,18]. In earlier work, the present author and collaborators have developed an information-theoretic approach to measuring the specificity of causal relationships [19,20].
This work was a contribution to the so-called ‘interventionist’ approach to causation [21,22], which is based on the insight that ‘causal relationships are relationships that are potentially exploitable for purposes of manipulation and control’ [17, p. 314]. Interventionists treat causation as relationships between the variables that characterize an organized system. These relationships can be represented by a directed acyclic graph. In such a graph, variable C is a cause of variable E when a suitably isolated manipulation of C would change the value of E. With suitable restrictions on the idea of ‘manipulation’, this test provides a criterion of causation, distinguishing causal relationships between variables from merely correlational relationships [21, pp. 94–107].
Using this definition, most events have many, many causes. But only some of these causal relationships are highly specific. The presence of oxygen in the atmosphere was one cause of the bushfire, but the arsonist was a more specific cause. The intuitive idea of specificity is that interventions on C can be used to produce any one of a large number of values of E, so that the cause variable has what Woodward terms ‘fine-grained influence’ over the effect variable [17, p. 302]. This idea can be quantified using Shannon information theory with the addition of an intervention operator that allows us to isolate the causal component of the correlation between variables:
SPEC: the specificity of a causal variable is obtained by measuring how much mutual information interventions on that variable carry about the effect variable [19,20].6
Formally, the specificity (I) of C for E against a background of other variables B is
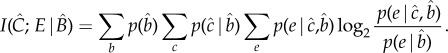 |
3.1 |
The above equation is a variant on the equation for Shannon's mutual information, which measures the overlap, or redundancy, in the probability distributions of two variables. The  (hat) on a variable denotes Judea Pearl's intervention operator [22] and indicates that the value of that variable is determined by intervention rather than observation. These interventions transform the symmetrical mutual information measure into an asymmetric measure of causal influence, because it now represents not the observed correlation between the variables, but the effect on E of experimentally intervening on C while controlling for background variables B. If two variables are not causally connected, then however strongly they are correlated,
(hat) on a variable denotes Judea Pearl's intervention operator [22] and indicates that the value of that variable is determined by intervention rather than observation. These interventions transform the symmetrical mutual information measure into an asymmetric measure of causal influence, because it now represents not the observed correlation between the variables, but the effect on E of experimentally intervening on C while controlling for background variables B. If two variables are not causally connected, then however strongly they are correlated, 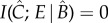 .
.
A more intuitive way to think about the specificity measure is that it measures the extent to which an agent can reduce their uncertainty about the value of the effect variable if they can change the value of the cause, that is, the extent to which the agent can precisely determine the value of E by intervening on C.
SPEC can be used to measure either how specifically two variables are connected (potential causal influence) or how much of the actual variation in E in some data are causally explained by variation in C (actual causal influence) [19,20]. While the use of Shannon information theory means that the measure is restricted to discrete variables, equivalent measures of metric variables are possible. None of these additional complexities need concern us in the present discussion, however. Instead, we will briefly see how SPEC can be used to elucidate the difference between sources of specificity, such as coding sequences of DNA, on the one hand, and sources of efficiency, such as RNA polymerase, on the other. We will then turn our attention to generalizing this approach to sequence specificity.
4. Genetic and epigenetic information
If biological information is precise determination, as measured by SPEC, then it is easy to see that DNA is a rich source of information in the production of biomolecules in a way that distinguishes it from many other causes of those biomolecules. Varying the sequence of DNA exerts fine-grained control over the structure of the molecules produced. Griffiths and collaborators [19, pp. 539–540] constructed a toy causal model of transcription with three variables: RNA polymerase (POL), which is either present or absent, DNA, whose values are alternative DNA sequences, and RNA, whose values are alternative RNA sequences. The value of RNA depends on both POL and DNA. Nothing is transcribed if POL = absent and when POL = present, each value of DNA determines a unique value of RNA. This is roughly how Crick imagined transcription, although, of course, the chemical nature of the transcription machinery was unknown. Assuming for simplicity a maximum entropy distribution over both POL and DNA, the specificity of POL for RNA can never exceed 1 bit, because POL has an entropy of 1 bit and the mutual information between two variables cannot exceed the lowest maximum entropy of either variable. However, once the number of possible values of DNA each determining a unique RNA product exceeds 4, then DNA will always have >1 bit of specificity for RNA.7
Calculations on a toy model are of limited interest. However, the approach that lies behind them has some immediate exciting consequences. The first is that this measure can be applied to both coding and non-coding regions in the genome to allow a quantitative comparison of the contribution of variables of both kinds to the precise determination of the sequence of a biomolecule. For example, mutations to any of the many well-characterized intronic splicing enhancer (ISE) or silencer (ISS) regions change the probability that one or more exons will be removed from the resulting transcript [27]. We could introduce this process into our toy model by replacing the variable DNA with two variables, INT and EXO, whose values would be the intronic and exonic content of the original DNA sequences, respectively. The existence of intronic splicing control regions would be represented by the specificity of INT for RNA. This is an absolutely natural extension of the moves Crick himself made in his 1958 paper in the light of what we now know about how biomolecules are synthesized from the genome. There is sequence specificity in non-coding regions.
Our approach has vindicated the idea that biological information is not restricted to the coding regions of the genome, but can be found also in other functional regions. But we can go further. Our measure can be extended to variables representing epigenetic (narrow sense, see box 1) modifications of DNA, insofar as they make a difference to the precise sequence of biomolecules through their role in the regulation of transcription and post-transcriptional and post-translational processing.
Box 1. Definitions of epigenetic: from Griffiths & Stotz [9, p. 112].
Epigenesis: the idea that the outcomes of development are created in the process of development, not performed in the inputs to development; epigenetic can be used in these senses:
Epigenetics (broad sense [28]): the study of the causal mechanisms by which genotypes give rise to phenotypes;the integration of the effects of individual genes in development to produce the epigenotype.
Epigenetics (narrow sense [29]): the study of the mechanisms that determine which genome sequences will be expressed in the cell; the control of cell differentiation and of mitotically and sometimes meiotically heritable cell identity.
Epigenetic inheritance (narrow sense): the inheritance of genome expression patterns across generations (e.g. through meiosis) in the absence of a continuing stimulus.
Epigenetic inheritance (broad sense): the inheritance of phenotypic features via causal pathways other than the inheritance of nuclear DNA. We refer to this as exogenetic inheritance [30].
Numerous mechanisms have been suggested by which epigenetic marks could determine which exons will be included in a mature mRNA. RNA splicing is frequently co-transcriptional, either by splicing actually occurring while the pre-mRNA is still being transcribed or by the recruitment of factors that determine later splicing while the pre-mRNA is being transcribed. This creates many opportunities for interaction between the splicing machinery and chromatin. The strongest direct evidence to date of epigenetic determination of alternative splicing is by alternative methylation states of histones. Indirect evidence suggests multiple significant roles for chromatin in determining alternative splicing [31–33].
Epigenetic regulation of splicing is another missing variable in the toy model described above. If we extended the model to include it, variable(s) representing the methylation and acetylation state of histones would have some specificity for the RNA product variable. So, by a direct application of Crick's original reasoning, there is both genetic and epigenetic information in Crick's original sense: both genes and epigenes can have sequence specificity.
Epigenetic modifications of chromatin can have sequence specificity. This will seem unsurprising to many biologists, given the number of papers that described the discovery of such mechanisms as the discovery of ‘missing information’ for splicing [27,33]. This way of speaking need not be regarded in the deflationary manner described in §1. The approach to information outlined here shows that it can be taken literally as a step towards a unified theory of biological information. Sequence specificity is a measurable quantity that plays a causal role in the production of biomolecules, namely the precise determination of their linear structure.
5. Why epigenetic information cannot be reduced to genetic information
A common thought about why epigenetics cannot be a distinct source of information is worth considering, because it throws light on why Crick needed to introduce the idea of information. The thought is that, because the machinery that creates epigenetic modifications consists of molecules transcribed from the genome, the information in the epigenetic marks must ultimately be derived from the genome.
The problem with this kind of hair splitting is that ultimately the extra information (e.g. methylation) is provided by enzymes (methylases) encoded by genes in the genome. Epigenetics, per se, doesn't add any new information. It's just a consequence, or outcome, of the information already in the DNA.8
This informal comment is significant precisely because it is a typical first response to the idea that epigenetic marks contain information that supplements the information in the genome. This response makes it clearer why Crick needed to distinguish ‘the flow of energy, the flow of matter, and the flow of information’. (1958, p. 144) The concept of specificity is a causal concept, not a material one, and identifying the sources of biological specificity requires measuring causal control, not material contributions. Once we look at the matter in this light, it becomes clear that some epigenetic modifications are specified by genomes, while others are not.
To see why the ‘matter and energy’ side of how epigenetic marks are created is not relevant, consider a case in which epigenetic marks are a site of conflict between multiple genomes. In cases of parental imprinting of genes, it is biological common sense that the parent, not merely the offspring, is a source of the biological information expressed in the offspring's phenotype. If this genetic conflict is mediated by epigenetic mechanisms that contribute to the precise determination of the sequence of gene products, for example by affecting which exons are included in a transcript [34], then it makes no sense to say that the information specifying the splice variant all comes from the offspring genome. The fact that the coding sequences for the enzymes involved in establishing and maintaining the methylation pattern are in the offspring genome is irrelevant. The relevant issue is where causal control is being exercised over the transcription and processing of those sequences. When parental imprints are established, the offspring provides the efficiency of the reaction, but the parent provides at least part of the specificity of the reaction.
Now, consider a case where the epigenetic mechanism that contributes to the precise determination of phenotype is influenced by the offspring's environment. For example, regulation of alternative splicing by temperature seems to be an important mechanism for maintaining circadian rhythms in a wide range of species [35,36]. It seems reasonable to describe this as a mechanism for conveying environmental information to the genome, so that genome expression can be correctly matched to the environment. After all, the adaptive problem facing the organism is to reduce its uncertainty about where it is in the diurnal cycle and it does this by responding to an environmental cue. Our account of information vindicates this idea—we could, at least in principle, measure the contribution of the environmental variable to the precise determination of sequence, just as we did the contribution of the epigenetic marks further along in the causal graph. The fact that the coding sequences for the enzymes involved are in the genome is irrelevant. The real issue is where causal control is being exercised over the transcription and processing of those sequences. In this case, evolution has designed a mechanism that detects and responds to information from the environment.
In this section, we have seen that our measure can be used to identify sequence specificity in both coding and non-coding sequences, in epigenetic marks, and in the causes of those marks, whether that is other genomes in cases of genetic conflict, or the environment in cases of plasticity. Information in Crick's sense is about precise determination. We have expanded the class of things that do the determining beyond those Crick originally envisaged. In the following section, we will also expand the class of things that get determined.
6. Sequence specificity and other biological information
Crick used ‘information’ to label the distinctive relationship of precise determination that holds between coding sequences of nucleic acids and the order of elements in their products, a relationship that does not hold between those products and many of their other causes. However, in §§4 and 5 we saw that some other causes do have this relationship to the order of elements in gene products. In this section, we ask whether this distinctive relationship of precise determination exists for phenotypes more distal than the primary structure of RNAs or proteins. In this context, we will not talk of ‘sequence specificity’, reserving that term for the precise determination of sequence, which was Crick's original concern. We will use the more general term ‘biological information’ to refer to the precise determination of phenotypes that are causally downstream of the primary structure of gene products, phenotypes such as the tertiary structure of proteins, and still more distally, morphology and behaviour.
As we noted in §1, the expressive power of the genetic code is limited to specifying the linear order of elements in a polypeptide. Changes to DNA coding sequences cause a whole chain of events, but they do not code for the more distal events in that chain [2]. The use of ‘code’ in this extended sense is metaphorical, like saying that when Richard Nixon literally ordered the Watergate cover-up, he also ‘ordered’ his own downfall.
But while the genetic triplet code is limited in this way, the broader idea of information as precise determination is not. The idea of information as precise determination, whether measured using SPEC or another measure, can be applied to any set of variables arranged in a causal graph. In principle, therefore, our approach can be used to measure biological information in a gene (or an epigene) with respect to any downstream variable affected by that gene. In fact, a range of causal Shannon information measures related to the one introduced here are already used in complex systems science to study a wide spectrum of living and non-living systems [37]. Genes or epigenes may not literally ‘code’ for morphology and behaviour, but they do literally contain biological information that specifies to some measurable degree morphology and behaviour.
It is now possible to extend our approach to biological information to mechanisms of exogenetic heredity (broad-sense epigenetic inheritance; see box 1). We have already seen that environmental factors can have sequence specificity, because they can be specific causes of epigenetic modifications of chromatin and thus contribute to the precise determination of the structure of biomolecules. But there are broader mechanisms of environmental heredity, such as habitat or host imprinting, in which the phenotype of the offspring is influenced by the parental phenotype but where no epigenetic mark is transmitted through meiosis, so there is no epigenetic inheritance in the standard, narrow sense. These broader mechanisms are still usually referred to as ‘epigenetic inheritance’ but we will refer to them as exogenetic inheritance to avoid confusion. The question of whether such environmental variables contribute information to development becomes the considerably more precise question of how specific is the causal relationship between those variables and variables representing morphology and behaviour.
At this point, we have something like a general theory of biological information. Information refers to a distinctive relationship of precise determination, which we can identify with the older concept of biological specificity. The phenomenon of biological specificity is explained by the existence of causes through which organisms exercise precise determination of outcomes, and the functional expression of this specificity is explained by natural selection acting on those causes. Central to organisms' ability to exercise this highly specific control is the relationship of precise determination originally identified by Crick between the sequence of DNA and the sequences of RNA and protein. Heredity is the transfer of biological specificity from one generation to the next. Central to organisms' ability to transfer specificity in this way is the existence of coding sequences of DNA which contain the information to determine the specificity of their products.9
7. Development and evolution
We have seen that there can be genetic, epigenetic and exogenetic sources of biological information in development. How significant the later two sources are in development is an empirical question. But even biologists who find it plausible that epigenetic and exogenetic factors are significant in development are often sceptical about whether they are significant in evolution. The most common reason for this scepticism is that epigenetic marks are relatively unstable when compared to genetic mutations.
The key point is that if epigenetic states are important to evolution, they are important through stable changes in these states, namely transmissible epimutations. And if epimutations are not transmitted with reasonable stability over generations, they cannot have any long-term evolutionary potential [39]. If an epimutation is to have evolutionary importance, it must persist [40, p. 391]
The stability of epigenetic marks is certainly an important question. But whether their evolutionary significance turns on their stability depends on what is meant by ‘evolutionary significance’. In at least one important sense of that phrase, epigenetic marks do not need to be stable to be significant. It is surely reasonable to regard a biological phenomenon as having evolutionary significance if it has widespread and substantial impact on the dynamics of evolution, or, to put it another way, if models that do not include this phenomena are unlikely to correctly predict the course of evolution. But we already know that this is the case from work on the evolutionary genetics of maternal effects [41]. Maternal effects can be defined as the causal influence of maternal genotype or phenotype on offspring phenotype independent of offspring genotype [42], which is in line with the approach taken here to defining epigenetic and exogenetic information. Maternal effects may be either epigenetic or exogenetic, depending on the specific causal pathway by which maternal influence is exerted.
Maternal effects, and parental effects generally, are recognized as a significant factor in evolution [43]. But any form of epigenetic or exogenetic heredity that is a significant source of biological information in the sense defined above will be significant in the same way because it substantially alters the mapping from parent phenotype to offspring phenotype. In this sense, epigenetic and exogenetic heredity is significant for evolution for the same reason that Mendelian models of heredity were significant. The primary significance of Mendelism for the theory of natural selection was that it specified the form of the transmission phase. Epigenetic and exogenetic heredity change this form, and even in the most conventional cases, where maternal effects are simply a one-generation time-lag in the expression of an allele, this has substantial impact on the dynamics of natural selection.
As Wilkins is well aware of all these points, we can infer that this is not the sense in which he is asking ‘if epigenetic states are important to evolution’. Another valid sense of that question is whether epigenetic or exogenetic mutations can be the basis of cumulative adaptation. It is plausible that an unstable inheritance system cannot play this role, but that does not mean that it cannot play an important role in a process of cumulative adaptation that also involves the genetic heredity system [44]. Finally, an important perspective on the relative evolutionary significance of genetic, epigenetic and exogenetic heredity is that they may play complementary roles. For example, it is plausible that genetic and epigenetic heredity allow organisms to adapt themselves to changing environments on different timescales [45].
Other authors have argued that to suppose epigenetic inheritance implies anything for evolutionary theory is to conflate ‘proximate’ or mechanistic with ‘ultimate’ or evolutionary biology. Scott-Phillips et al. [46] draw a useful comparison between the discovery of epigenetic inheritance and the discovery of Mendelian genetics. In the first years of the twentieth century, some Mendelians saw Mendelian inheritance as a theory of evolutionary change and presented it as a challenge to the Darwinian theory of natural selection. They suggest that authors who present epigenetic inheritance as a challenge to conventional neo-Darwinism are like those early Mendelians: they are confusing a proximate, mechanistic theory of heredity with an ultimate theory of the causes of evolutionary change. Scott-Phillips et al. [47] are engaged in a wider dispute with authors who question the value of the proximate/ultimate distinction and I will not address that wider dispute here. However, with respect to the specific issue of whether epigenetic inheritance has implications for evolutionary theory, their analogy seems to establish exactly the opposite of their intended conclusion. The founders of modern neo-Darwinism did not dismiss Mendelism as a merely proximal mechanism; they used it to derive the form of the transmission phase in the process of natural selection. As I pointed out above, epigenetic and exogenetic heredity shows up in quantitative genetics as parental effects, and the incorporation of parental effects into evolutionary models has a significant effect on evolutionary dynamics. In this way, both Mendelian heredity and epigenetic heredity are part of ultimate, not merely proximate biology.
An interesting aspect of Scott-Phillips et al.'s argument is their insistence that, ‘Put simply, if we wish to offer an ultimate explanation for the existence of some trait, we must make reference to how that trait contributes to inclusive fitness’ [46, p. 40]. They base this conclusion on the results of Grafen's ‘formal Darwinism’ project [48], which seeks to show that evolutionary dynamics are in important respects equivalent to the maximization of inclusive fitness. But what is done in this very impressive programme of work is to rigorously compare optimization models to population genetic models, where the latter models simply assume that there is no epigenetic heredity. This is not a problem for the formal Darwinism programme.10 But it is a problem for Scott-Phillips et al, who are effectively arguing that epigenetic inheritance cannot contribute to the ultimate explanation because maximizing (genetic) inclusive fitness fully represents evolutionary dynamics in models which assume there is no epigenetic inheritance.
Dickins & Rahman [50] suggest that while epigenetic inheritance may play a role in evolution, those who present it as a challenge to conventional neo-Darwinism have only presented evidence that it is a significant proximate mechanism. They have failed to present evidence that it is significant in ultimate biology. Once again, this seems to overlook the way that epigenetic and exogenetic heredity show up in conventional, quantitative genetic models, namely as parental effects, and the known impact of such effects on evolutionary dynamics.
8. Conclusion
We set out to define a sense of ‘information’ that can make sense of the idea that development is the expression of information that accumulated during evolution and that heredity is the transmission of this information. While compelling at a metaphorical level, this is surprisingly hard to cash out in serious, scientific terms. We began with a simple conception of information that plays a straightforward role at the heart of molecular biology and explored how many other aspects of biology can be clarified using this sense of information. Our starting point was the sense of information introduced by Crick [8]. We identified two aspects of Crick's conception of information: (i) precise determination and (ii) the transfer of biological specificity from one molecule to another. This paper concentrated on the first aspect. We analysed the idea of precise determination using an information-theoretic measure of causal specificity. Using this measure, we showed that coding sequences of DNA have a distinctive relationship of precise determination to RNAs and polypeptides. This distinguishes coding sequences from many other causes of the same outcomes, such as the presence of an RNA polymerase. This is what Crick meant when he identified coding sequences as containing information and the other causes as not doing so. His distinction is closely related to the distinction between the specificity and efficiency of a biochemical process.
Since 1958, however, a great deal has been learnt about the production of biomolecules. We saw that Crick's approach to information has natural extensions to non-coding regions of DNA, to epigenetic marks, and to the genetic or environmental upstream causes of those epigenetic marks. Any of these variables may have sequence specificity, that is, they may contribute substantially to the precise determination of the linear structure of biomolecules. Moreover, we saw that it is a mistake to suppose that the sequence specificity of epigenetic marks must always derive from sequence specificity elsewhere in the genome, or in other genomes. Finally, we generalized to a broader concept of ‘biological information’ that is applicable to more distal phenotypes, and not merely to the linear structure of biomolecules. Relationships of precise determination can exist between genetic, epigenetic and exogenetic factors in development and distal phenotypes, such as morphology and behaviour. This gives us a general theory of biological information that can be used to restate more precisely the idea with which we started. Development is the expression of biological specificity, or biological information conceived as precise determination and measured using causal information theory. In heredity, factors that are able to exercise this precise determination are passed on from previous generations. These factors may be genetic, epigenetic or exogenetic. In the penultimate section of the article, we argued that the existence of biological information in epigenetic and exogenetic factors is relevant to evolution as well as to development.
Acknowledgements
I thank the members of the Theory and Method in Biosciences group at the Charles Perkins Centre for feedback on an earlier draft, Arnaud Pocheville for drawing figure 1 and Stefan Gawronski for his assistance with the preparation of the manuscript.
Endnotes
In his final book, the influential evolutionary theorist George C. Williams called for a new, ‘codical’ biology founded on the concept of information, precisely because that is not the biology we actually have [1].
Griffiths and Stotz used the phrase ‘Crick information’ to refer to what, in this article, will be called ‘sequence specificity’. In more recent work, I and my collaborators have reserved the term ‘Crick information’ for a measure of the intrinsic information content of a sequence, rather than for the measure of the relationship between a sequence and its causes that is the subject of this article.
Sahotra Sarkar [5] gives a brief history of efforts by molecular biologists to construct a theory of biological information. Key papers in philosophical literature are [11,12]. For ‘biosemiotics’, see [13].
Philosopher Gregory Morgan received two letters from Crick in response to questions about how and why Crick came to use the concept of information in his work. These were kindly made available to us by Morgan. Crick also states that the inspiration for his use of ‘code’ in the sequence hypothesis was the Morse Code's purely syntactic mapping between two alphabets (Crick to Morgan, 3 April 1998).
A treatment of the second aspect of Crick's ideas about information using algorithmic information measures is in preparation.
This measure has been independently proposed in neuroscience [23] and in the computational sciences [24]. For other related measures, see Ay & Polani [25] and Janzing et al. [26].
The entropy of RNA is H(RNA) > 2, we have just seen that 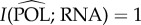 , and DNA accounts for all the remaining entropy:
, and DNA accounts for all the remaining entropy: 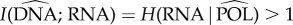 .
.
Larry Moran, Sandwalk Blog: http://sandwalk.blogspot.com.au/2016/10/extending-evolutionary-theory-paul-e.html (accessed 8 December 2016). This was a response to the abstract of the conference presentation from which this article is derived.
Comparison of causal roles need not be reduced to a simple ‘more or less specific’. For example, elucidating the distinction between permissive and instructive induction events in development requires a more complex application of the tools used here [38].
Lu & Bourrat [49] have recently discussed how this programme can be extended to include epigenetic inheritance and suggest that because of this, epigenetic inheritance does not require any radical revision of conventional neo-Darwinism.
Data accessibility
This article has no additional data.
Competing interests
I declare I have no competing interests.
Funding
This publication was made possible through the support of a grant from the Templeton World Charity Foundation.
Disclaimer
The opinions expressed in this publication are those of the author and do not necessarily reflect the views of the Templeton World Charity Foundation.
References
- 1.Williams GC. 1992. Natural selection: domains, levels and challenges. New York, NY: Oxford University Press. [Google Scholar]
- 2.Godfrey-Smith P. 2000. On the theoretical role of ‘genetic coding’. Philos. Sci. 67, 26–44. ( 10.1086/392760) [DOI] [Google Scholar]
- 3.Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293, 1074–1080. ( 10.1126/science.1063127) [DOI] [PubMed] [Google Scholar]
- 4.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. 2010. Deciphering the splicing code. Nature 465, 53–59. ( 10.1038/nature09000) [DOI] [PubMed] [Google Scholar]
- 5.Sarkar S. 1996. Biological information: a sceptical look at some central dogmas of molecular biology. In The philosophy and history of molecular biology: new perspectives (ed. Sarkar S.). Boston Studies in the Philosophy of Science, vol. 183, pp. 187–232. Dordrecht, The Netherlands: Kluwer Academic Publishers. [Google Scholar]
- 6.Levy A. 2011. Information in biology: a fictionalist account. Nous 45, 640–657. ( 10.1111/j.1468-0068.2010.00792.x) [DOI] [Google Scholar]
- 7.Griffiths PE. 2001. Genetic information: a metaphor in search of a theory. Philos. Sci. 68, 394–412. ( 10.1086/392891) [DOI] [Google Scholar]
- 8.Crick FHC. 1958. On protein synthesis. Symp. Soc. Exp. Biol. 12, 138–163. [PubMed] [Google Scholar]
- 9.Griffiths PE, Stotz K. 2013. Genetics and philosophy: an introduction. New York, NY: Cambridge University Press. [Google Scholar]
- 10.Stotz K, Griffiths PE. 2017 Biological Information, causality and specificity—an intimate relationship. In From matter to life: information and causality (eds SI Walker, P Davies, G Ellis), pp. 366–390 Cambridge, UK: Cambridge University Press. [Google Scholar]
- 11.Maynard Smith J. 2000. The concept of information in biology. Philos. Sci. 67, 177–194. ( 10.1086/392768) [DOI] [Google Scholar]
- 12.Shea N. 2007. Representation in the genome and in other inheritance systems. Biol. Philos. 22, 313–331. ( 10.1007/s10539-006-9046-6) [DOI] [Google Scholar]
- 13.Witzany G, Baluŝka F. 2012. Life's code script does not code itself. EMBO. Rep. 13, 1054–1056. ( 10.1038/embor.2012.166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg A. 2006. Is epigenetic inheritance a counterexample to the central dogma? Hist. Philos. Life. Sci. 28, 549–565. [PubMed] [Google Scholar]
- 15.Morange M. 1998. A history of molecular biology. Cambridge, MA: Harvard University Press. [Google Scholar]
- 16.Crick FHC. 1970. Central dogma of molecular biology. Nature 227, 561–563. ( 10.1038/227561a0) [DOI] [PubMed] [Google Scholar]
- 17.Woodward J. 2010. Causation in biology: stability, specificity, and the choice of levels of explanation. Biol. Philos. 25, 287–318. ( 10.1007/s10539-010-9200-z) [DOI] [Google Scholar]
- 18.Waters CK. 2007. Causes that make a difference. J. Philos. 104, 551–579. ( 10.5840/jphil2007104111) [DOI] [Google Scholar]
- 19.Griffiths PE, Pocheville A, Calcott B, Stotz K, Kim H, Knight RD. 2015. Measuring causal specificity. Philos. Sci. 82, 529–555. ( 10.1086/682914) [DOI] [Google Scholar]
- 20.Pocheville A, Griffiths PE, Stotz K. 2017. Comparing causes—an information-theoretic approach to specificity, proportionality and stability. In Proc. of the 15th Congress of Logic, Methodology and Philosophy of Science (eds Leitgeb H, Niiniluoto I, Sober E, Seppälä P), pp. 250–275. London, UK: College Publications. [Google Scholar]
- 21.Woodward J. 2003. Making things happen: a theory of causal explanation. New York, NY: Oxford University Press. [Google Scholar]
- 22.Pearl J. 2009. Causality: models, reasoning, and inference, 2nd edn Cambridge, UK: Cambridge University Press. [Google Scholar]
- 23.Tononi G, Sporns O, Edelman GM. 1999. Measures of degeneracy and redundancy in biological networks. Proc. Natl Acad. Sci. USA 96, 3257–3262. ( 10.1073/pnas.96.6.3257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korb KB, Hope LR, Nyberg EP. 2009. Information-theoretic causal power. In Information theory and statistical learning (eds Emmert-Streib F, Dehmer M), pp. 231–265. Boston, MA: Springer US. [Google Scholar]
- 25.Ay N, Polani D. 2008. Information flows in causal networks. Adv. Complex Syst. 11, 17–41. ( 10.1142/S0219525908001465) [DOI] [Google Scholar]
- 26.Janzing D, Balduzzi D, Grosse-Wentrup M, Schölkopf B. 2013. Quantifying causal influences. Ann. Stat. 41, 2324–2358. ( 10.1214/13-AOS1145) [DOI] [Google Scholar]
- 27.Wang Z, Burge CB. 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14, 802–813. ( 10.1261/rna.876308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waddington CH. 1940. Organisers and genes. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 29.Nanney DL. 1958. Epigenetic control systems. Proc. Natl. Acad. Sci. 44, 712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West MJ, King AP. 1987. Settling nature and nurture into an ontogenetic niche. Develop. Psychobiol. 20, 549–562. [DOI] [PubMed] [Google Scholar]
- 31.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. 2011. Epigenetics in alternative pre-mRNA splicing. Cell 144, 16–26. ( 10.1016/j.cell.2010.11.056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorenson MR, Jha DK, Ucles SA, Flood DM, Strahl BD, Stevens SW, Kress TL. 2016. Histone H3k36 methylation regulates pre-mRNA splicing in Saccharomyces cerevisiae. RNA Biol. 13, 412–426. ( 10.1080/15476286.2016.1144009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Almeida SF, Carmo-Fonseca M. 2012. Design principles of interconnections between chromatin and pre-mRNA splicing. Trends. Biochem. Sci. 37, 248–253. ( 10.1016/j.tibs.2012.02.002) [DOI] [PubMed] [Google Scholar]
- 34.Cowley M, Wood AJ, Bohm S, Schulz R, Oakey RJ. 2012. Epigenetic control of alternative mRNA processing at the imprinted Herc3/Nap1l5 locus. Nucleic Acids Res. 40, 8917–8926. ( 10.1093/nar/gks654) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez SE, et al. 2010. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468, 112–116. ( 10.1038/nature09470) [DOI] [PubMed] [Google Scholar]
- 36.Syed NH, Kalyna M, Marquez Y, Barta A, Brown JWS. 2012. Alternative splicing in plants—coming of age. Trends. Plant. Sci. 17, 616–623. ( 10.1016/j.tplants.2012.06.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bossomaier T, Barnett L, Harré M, Lizier JT. 2016. An introduction to transfer entropy. Cham, Germany: Springer International Publishing. [Google Scholar]
- 38.Calcott B. In press Causal specificity and the instructive-permissive distinction. Biol. Philos. 1–25. ( 10.1007/s10539-017-9568-0) [DOI] [Google Scholar]
- 39.Slatkin M. 2009. Epigenetic Inheritance and the Missing Heritability Problem. Genetics 182, 845–850. ( 10.1534/genetics.109.102798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkins A. 2011. Epigenetic inheritance: where does the field stand today? What do we still need to know? In Transformations of Lamarckism: from subtle fluids to molecular biology (eds Gissis SB, Jablonka E), pp. 389–393. Cambridge, MA: The MIT Press. [Google Scholar]
- 41.Wolf JB, Wade MJ. 2016. Evolutionary genetics of maternal effects. Evolution 70, 827–839. ( 10.1111/evo.12905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf JB, Wade MJ. 2009. What are maternal effects (and what are they not)? Phil. Trans. R. Soc. B 364, 1107–1115. ( 10.1098/rstb.2008.0238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Badyaev AV, Uller T. 2009. Parental effects in ecology and evolution: mechanisms, processes, and implications. Phil. Trans. R. Soc. B 364, 1169–1177. ( 10.1098/rstb.2008.0302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Badyaev AV, Hill GE, Beck ML, Dervan AA, Duckworth RA, McGraw KJ, Nolan PM, Whittingham LA. 2002. Sex-biased hatching order and adaptive population divergence in a Passerine bird. Science 295, 316–318. ( 10.1126/science.1066651) [DOI] [PubMed] [Google Scholar]
- 45.Danchin E, Pocheville A. 2014. Inheritance is where physiology meets evolution. J. Physiol. 592, 2307–2317. ( 10.1113/jphysiol.2014.272096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott-Phillips TC, Dickins TE, West SA. 2011. Evolutionary theory and the ultimate-proximate distinction in the human behavioral sciences. Perspect. Psychol. Sci. 6, 38–47. ( 10.1177/1745691610393528) [DOI] [PubMed] [Google Scholar]
- 47.Laland KN, Sterelny K, Odling-Smee J, Hoppitt W, Uller T. 2011. Cause and effect in biology revisited: is Mayr's proximate-ultimate dichotomy still useful? Science 334, 1512–1516. ( 10.1126/science.1210879) [DOI] [PubMed] [Google Scholar]
- 48.Grafen A. 2006. Optimization of inclusive fitness. J. Theor. Biol. 238, 541–563. ( 10.1016/j.jtbi.2005.06.009) [DOI] [PubMed] [Google Scholar]
- 49.Lu Q, Bourrat P. 2017. The evolutionary gene and the extended evolutionary synthesis. Br. J. Philos. Sci. ( 10.1093/bjps/axw035) [DOI] [Google Scholar]
- 50.Dickins TE, Rahman Q. 2012. The extended evolutionary synthesis and the role of soft inheritance in evolution. Proc. R. Soc. B 279, 2913–2921. ( 10.1098/rspb.2012.0273) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.


