Abstract
Stochasticity is harnessed by organisms to generate functionality. Randomness does not, therefore, necessarily imply lack of function or ‘blind chance’ at higher levels. In this respect, biology must resemble physics in generating order from disorder. This fact is contrary to Schrödinger's idea of biology generating phenotypic order from molecular-level order, which inspired the central dogma of molecular biology. The order originates at higher levels, which constrain the components at lower levels. We now know that this includes the genome, which is controlled by patterns of transcription factors and various epigenetic and reorganization mechanisms. These processes can occur in response to environmental stress, so that the genome becomes ‘a highly sensitive organ of the cell’ (McClintock). Organisms have evolved to be able to cope with many variations at the molecular level. Organisms also make use of physical processes in evolution and development when it is possible to arrive at functional development without the necessity to store all information in DNA sequences. This view of development and evolution differs radically from that of neo-Darwinism with its emphasis on blind chance as the origin of variation. Blind chance is necessary, but the origin of functional variation is not at the molecular level. These observations derive from and reinforce the principle of biological relativity, which holds that there is no privileged level of causation. They also have important implications for medical science.
Keywords: evolution and physiology, Schrödinger's error, biological relativity, stochasticity, neo-Darwinism, modern synthesis
1. Introduction: the original formulation of the neo-Darwinist modern synthesis
The theory of evolution by natural selection was formulated by Charles Darwin and Alfred Russel Wallace who presented their ideas to the Linnean Society of London in 1858, followed by Darwin's book On the Origin of Species in 1859. Darwin was cautious in the presentation of his ideas. He wrote ‘Natural Selection has been the main, but not the exclusive means of modification’. He was concerned that he did not know the origin of variation and he acknowledged the existence of other mechanisms, including the inheritance of acquired characteristics. Ernst Mayr wrote in 1962: ‘Curiously few evolutionists have noted that, in addition to natural selection, Darwin admits use and disuse as an important evolutionary mechanism. In this he is perfectly clear’ [1]. Although Darwin disagreed with Lamarck on whether evolution had a direction (what Lamarck called le pouvoir de la vie [2,3]), he nevertheless acknowledged ‘this justly celebrated naturalist … who upholds the doctrine that all species, including man, are descended from other species' [4]. However, Darwin's multi-mechanism approach to evolution became significantly narrowed with the rise of neo-Darwinism.
Weismann's formulation of neo-Darwinism involved three major assumptions. First, that all genetic variation is random. Second, that the germline is isolated from variations in the soma. This is the Weismann barrier. Third, together with these two assumptions, that natural selection is then all-sufficient (allmacht) to explain evolution [5]. The subsequent integration of Mendelian genetics into this scheme led to the formulation of the modern synthesis [6].
Several important consequences followed. First, genetic variation is not itself viewed as functional. It becomes so only through the operation of natural selection to weed out harmful variations and promote helpful ones. The origin of variation is therefore completely blind. If this view is correct, we should not explain genetic variation in terms of existing or anticipated functionality. As physiology is the study of functional processes in organisms, physiology is thereby excluded from any direct role in the source of variation. Second, the inheritance of acquired characteristics, often called Lamarckism, cannot occur because it would require either that the germ line is not isolated from influences of somatic variations and/or that some forms of functional genetic reorganization can be triggered as a response to environmental stress. In an 1896 publication [7], Weismann added his theory of germinal selection, involving competition and selection among the hereditary units within the germplasm but, as Charlotte Weissman shows, this change in Weismann's view did not make any real concessions to the Lamarckians [8].
The neo-Darwinist modern synthesis was therefore both an extension and a simplification of Darwin's ideas. It was an extension through the incorporation of Mendelian genetics, about which Darwin unfortunately knew nothing. It was a simplification because it excluded the inheritance of acquired characteristics, whereas Darwin not only included this form of inheritance, he even proposed a theory for how it could happen, his pangenesis theory of gemmules [9], which resembles some forms of such inheritance discovered recently (see §6).
2. Purpose of this article
A central thesis of this paper is that blind stochasticity is a misconceived idea as it has been used in evolutionary biology. Stochasticity is used by organisms to generate new functional responses to environmental challenges. Far from proving that evolution is necessarily blind, randomness is the clay from which higher level order can be crafted. But it necessarily works the other way too: higher levels then organize the molecular level through many forms of constraint. The reason we do not necessarily see that organization from the molecular level is that the difference of scale is vast. If we focus on particular molecular events, such as gene mutations at particular loci, they will still appear stochastic. Blind chance can then seem to be the sole determinant of variation even when, in fact, the variation is directed in response to environmental challenges.
I will present the case for the following theses, which run counter to neo-Darwinism and the modern synthesis. With respect to neo-Darwinism, the view in this paper is a replacement more than an extension.
1. Randomness (stochasticity) is what one should generally expect at the molecular level even if determinate functionality rules at higher (cellular, tissue, organ, systems, organisms, sociological) levels. Randomness and functionality necessarily coexist at different levels.
2. Organisms can and do harness stochasticity in generating function.
3. Functional genome reorganization can occur in response to environmental stress.
4. Non-DNA information can be transmitted across generations.
5. By using diverse higher level processes, organisms can resist potentially harmful effects of many random genetic variations, at lower levels of function.
6. Physical constraints can and must influence both development and evolution.
7. The gene-centric view has so far been very disappointing from the viewpoint of medicine.
3. Stochasticity and order coexist at different levels
Physics teaches us that at a molecular level, there must be stochasticity. At any temperature above a value near absolute zero, below which a Bose–Einstein condensate becomes possible [10], molecules have kinetic energy which generates random movement. But physics also teaches us that, once there is a constraint at a higher level, e.g. a gas in a container, thermodynamics can describe determinate behaviour arising from the averaged behaviour within the constraint. This is the reason why Schrödinger argued correctly in What is life? that physics generates order from disorder [11].
Yet he contrasted this with biology, which he described as generating order at a high level from order at a molecular level, i.e. that the functional order at a high level actually results directly from order at the molecular level. But this is highly problematic from a physical viewpoint. Why then did he propose a theory that even he initially characterized as ridiculous? The reason is that following Delbrück [12], he predicted that the genetic material would be found to be an aperiodic crystal, which is a good description of DNA sequences if one thinks of a polymer as a kind of crystal. Crystal structure can be investigated accurately using diffraction. I believe he saw the ‘read-out’ of genetic sequences as determinate in the same kind of way. In this respect, he anticipated the formulation of Crick's central dogma of molecular biology [13]. Francis Crick and James Watson both acknowledged Schrödinger's influence in their thinking about the central dogma.
There are two fatal problems with this approach, as noted by Kupiec [14,15]. The first is that, as is clear from Crick's original statement, the central dogma refers only to the fact that sequence information passes one way, from DNA to proteins:
The central dogma of molecular biology deals with the detailed residue-by-residue transfer of sequential information. It states that such information cannot be transferred back from protein to either protein or nucleic acid. [16, p. 561]
I have italicized ‘such information’ and ‘from protein’ because it is evident that the statement does not say that no information can pass from the organism to the genome. In fact, it is obvious that it must do so to produce many different patterns of gene expression, which enable many different phenotypes (e.g. many different cell types in the same body) to be generated from the same genome. In addition to controlling relative expression levels, the organism also makes use of protein-mediated protein processing to add yet another layer of control following transcription.
This information from organisms is conveyed to their genomes by patterns of transcription factors, genome marking, histone marking and many RNAs, which in turn control the patterns of gene expression. These controls are exerted through preferential targeted binding to the genome or histone proteins. For example, methylation of cytosines preferentially occurs at CpG sites. Binding to histones preferentially occurs at the histone tails. Even though these are the targeted molecular mechanisms by which the functional control is exerted, there is no guarantee that the functionality will be evident at the molecular level. It would require many correlations between the patterns of binding and the functional processes at a higher level to identify the functionality involved. Without that correlation, the binding patterns will appear random. There are simply far too many sites. There are millions of CpG sites in the whole genome and tens of thousands of CpG clusters, which significantly are located near gene regulatory sites [17].
The second problem is that, as Schrödinger must have understood as a physicist, there is no way in which the molecules in an organism can avoid stochasticity. He wrote:
We seem to arrive at the ridiculous conclusion that the clue to understanding of life is that it is based on a pure mechanism, a ‘clock-work’ in the sense of Planck's paper. [18, p. 101]
But he then confuses the logic by continuing: ‘The conclusion is not ridiculous and is, in my opinion, not entirely wrong, but it has to be taken “with a very big grain of salt”’. He then explains the ‘big grain of salt’ by showing that even clock work is, ‘after all statistical’ (p.103). This seriously compromises the logic because the stochasticity in clockwork has to be negligible. We now know that the stochasticity in biology is far from negligible.
Schrödinger realizes that something is far from right but is struggling to identify what it might be. We would now say that the molecules involved (DNA) are subject to frequent statistical variations (copying errors, chemical and radiation damage, etc.), which are then corrected by the cell's protein and lipid machinery that enables DNA to become a highly reproducible molecule [19]. This is a three-stage process that reduces the copy error rate from 1 in 104 to around 1 in 1010, which is an astonishing degree of accuracy. In a genome of 3 billion bp, this works out as less than 1 error in copying a complete genome, compared to millions of errors without error correction. The order at the molecular scale is therefore actually created by the system as a whole, including lipid components that are not encoded by DNA sequences [20]. This requires energy, of course, which Schrödinger called negative entropy. Perhaps therefore this is what Schrödinger was struggling towards, but we can only see this clearly in retrospect. He could not have known how much the genetic molecular material experiences stochasticity and is constrained to be highly reproducible by the organism itself. The order at the molecular (DNA) level is actually imposed by higher level constraints.
4. Organisms can and do harness stochasticity in generating function
4.1. Stochasticity is a population-level attractor
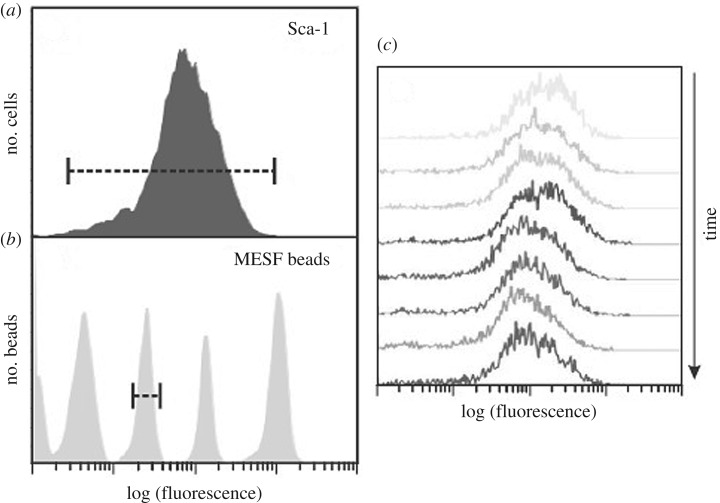
Experiments on the stochasticity of gene expression in cell populations show that, at least in some cases, it is the population as a whole that controls the stochasticity. Figure 1 is taken from Chang et al. [21].
Figure 1.
The robustness of heterogeneity of expression of Sca-1 protein expression in a cloned cell population. Heterogeneity detected by immunofluorescence flow cytometry (a) was significantly larger than the resolution limit of the method (b). (c) The stability of the clonal heterogeneity over a period of three weeks. Note that the spread of gene expression levels is three orders of magnitude [21].
The results show that in this case, the range of gene expression is 1000-fold and it follows a simple bell-shaped curve. The range is a population-level attractor, which is stable over long periods of time. That the population controls the heterogeneity is shown by experiments of the kind illustrated in figure 2. In a cell population showing a bimodal distribution, new populations of cells were cloned from one of the peaks (left), while in a monomodal distribution, cells were cloned from outliers. In both cases, after a few days, the original heterogeneity became re-established.
Figure 2.
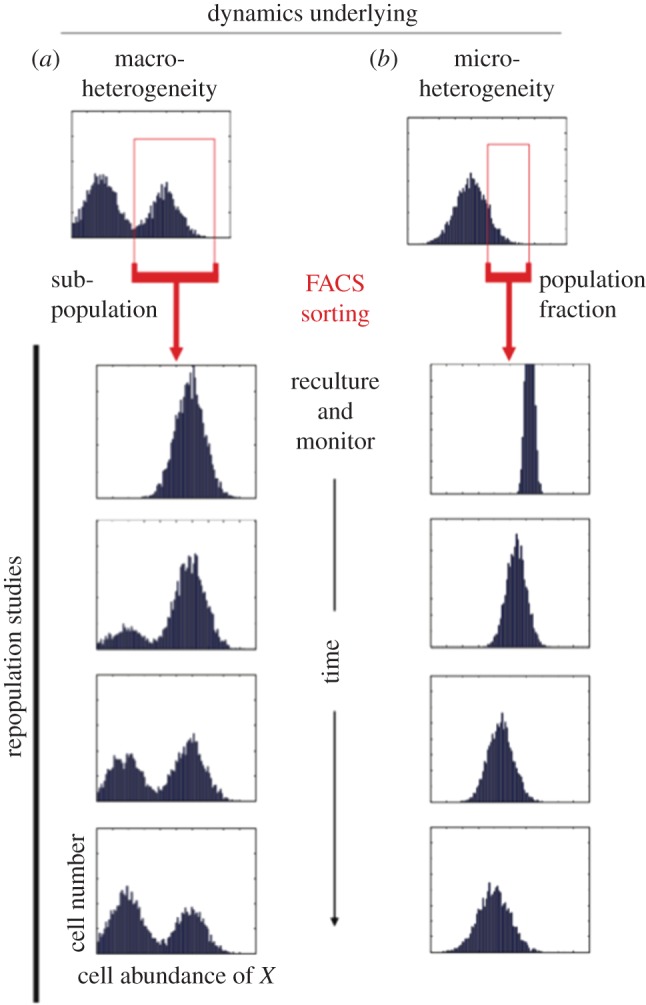
Two examples illustrating experiments in which populations were produced by cloning either from one of the peaks in a bimodal distribution (a) or from outliers in a monomodal distribution (b). In both cases, the new population initially exhibits the range of expression of the parent subpopulation. Over time (several days), however, the heterogeneity reverts to the original distribution [22]. (Online version in colour.)
Cell populations can therefore control stochasticity.
4.2. Cells can harness stochasticity to generate function
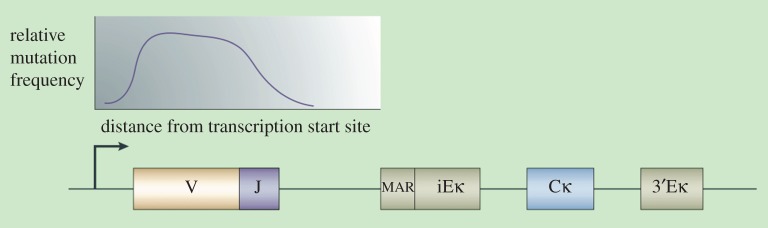
That cells can also harness stochasticity to generate specific function is known from experiments on the cells of the immune system that show the phenomenon of somatic hypermutation. Figure 3 summarizes what we know. Faced with a new antigen challenge, the mutation rate in the variable part of the genome can be accelerated by as much as 1 million times. So far as we know, the mutations occur randomly. But the location in the genome is certainly not random. The functionality in this case lies precisely in the targeting of the relevant part of the genome. The mechanism is directed, because the binding of the antigen to the antibody itself activates the proliferation process.
Figure 3.
Schematic diagram of gene-specific targeted hypermutation in immunoglobulin gene loci. The mutation rate is greatly increased only in the variable part of the genome, which is an approximately 1.5 kb region in each of the three immunoglobulin loci. In this figure, the graph above the rearranged variable (V) and joining (J) gene segments that form the variable region of Igκ depicts the mutation domain in the κ-light chain (Igκ) locus. 3′Eκ, Igκ 3′ enhancer; Cκ, Igκ constant; iEκ, Igκ intronic enhancer; MAR, matrix attachment region [23]. (Online version in colour.)
This example from the immune system shows that functionally significant targeted hypermutation can occur in the lifetime of an individual organism. There is no reason why this kind of mechanism should not be used in evolutionary change, as shown in the next example.
A well-known functionally driven form of genome change is the response to starvation in bacteria. Starvation can increase the targeted reorganizations of the genome by five orders of magnitude, i.e. by a factor of over 100 000 [24,25]. This is one of the mechanisms by which bacteria can evolve very rapidly and in a functional way in response to environmental stress.
A similar targeting of location where genomic change can occur has been found in experiments on genetically modified fruit flies. One of the common ways in which genetic modification is achieved is to use a particular kind of mobile genetic element that can move around the genome using a cut-and-paste mechanism that does not require an RNA intermediate. Most often, the insertions occur in a random way. But when DNA sequences from certain regulatory regions are used, they get inserted preferentially near the gene from which the sequence was derived [26]. This process targets the changes in a way that is clearly not random with respect to possible function.
5. Functional genome reorganization can occur in response to stress
5.1. Barbara McClintock and the genome as an organ of the cell
Barbara McClintock first observed that whole domains of genetic material move around the genome, even from one chromosome to another. She was working on Indian corn in the 1930s and 1940s, but it was much later, in 1983, that she was recognized with the award of a Nobel Prize. In her Prize lecture, she was very clear about the functional significance of her discovery. She described the genome ‘as a highly sensitive organ of the cell, monitoring genomic activities and correcting common errors, sensing the unusual and unexpected events, and responding to them, often by restructuring the genome’ [27].
She could not have anticipated the extent to which her idea would be confirmed by the sequencing of whole genomes. From the 2001 Nature paper on the first draft sequence of the human genome, we have comparisons between sequences in completely different species of eukaryotes for two classes of proteins, transcription factor proteins and chromatin binding proteins [28]. These show that the evolution of these proteins must have involved the movement of whole functional domains. This is far from the idea of slow progressive accumulation of point mutations. And it has much greater evolutionary significance because the rearrangement of whole domains including the functionality of those domains in response to stress could have been the origin of creativity in the evolutionary process. It is obvious that combining two or more domains each of which already has functionality is much more likely to produce a viable solution to a problem than waiting for random sorting of point mutations. This is why McClintock characterized the genome as a highly sensitive organ of the cell.
5.2. Can we observe genome reorganization happening in evolutionary experiments?
We can now observe organisms making use of this ability to reorganize their genomes. Bos et al. have observed the emergence of antibiotic resistance from multi-nucleated bacterial filaments. They write:
the strategy of generating multiple mutant chromosomes within a single cell may represent a widespread and conserved mechanism for the rapid evolution of genome change in response to unfavorable environments (i.e. chemo-therapy drugs and antibiotics). [29, p. 182]
Jack et al. [30] have shown that
signaling pathways that sense environmental nutrients control genome change at the ribosomal DNA. This demonstrates that not all genome changes occur at random and that cells possess specific mechanisms to optimize their genome in response to the environment. (my italics) [30, p. 9674]
How can genomes know about what is happening at the cell surface? The physiological mechanisms by which events in tiny micro-domains near the cell surface signal to the nucleus to control specific gene expression levels have now been studied in fine detail [31,32]. There is no longer any mystery in understanding the highly specific transmission of information to the nucleus that can control gene expression. There is no reason why genomes should not use similar communication pathways in response to stress signals received by cells and organisms.
6. Non-DNA information can be transmitted across generations
Recent experiments have demonstrated that non-DNA information can be transmitted between generations [33], and this rapidly growing field has been reviewed in an important paper in Science [34]. Two quotations from that review are relevant:
Many phenomena and mechanisms of nongenetic and/or non–DNA sequence–based inheritance have been described in a range of model organisms, challenging our perception of the well-established relationship between transmitted genotype and phenotype. [34, p. 59]
They conclude
The idea of certain sequences that might be refractory to germline epigenetic reprogramming provides a compelling mechanism for the inheritance of modulated epigenetic states. [34, p. 63]
To illustrate the range of processes that can be involved, I will briefly describe three examples.
Rechavi et al. [35] investigated the inheritance of resistance to viral infection in the nematode worm, Caenorhabditis elegans. The resistance is acquired when infected worms have the DNA required to make a viral-silencing RNAi, which is triggered by viral replication. They cross-bred these worms with a wild-type population, including worms that do not have the required DNA. Some of the later generations have the required DNA, others do not. Yet subsequent generations inherited the acquired silencing response irrespective of whether they had the required DNA. The RNAi is inherited through the germline, and is then amplified by RNA polymerase in each generation. This non-DNA inheritance was followed successfully for 100 generations. It resembles Darwin's gemmule theory (see Introduction).
Nelson et al. [36] found robust inheritance of epigenetic marking in mice with Apobec1 deficiency. They found that ‘these [epigenetic] effects persist for many generations and are as strong as conventional genetic inheritance’. The journal, PNAS, published a commentary article in the same issue, which concludes: ‘the belief that the soma and germline do not communicate is patently incorrect’ [37].
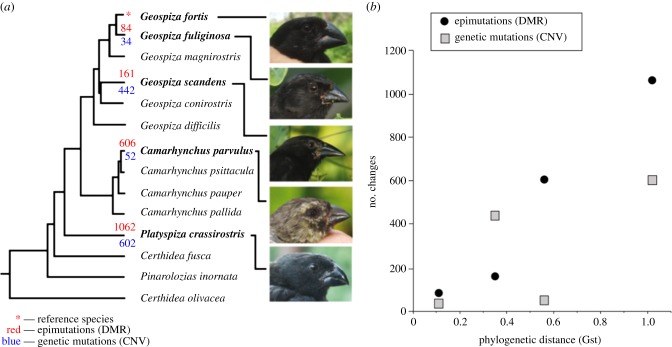
The question whether epigenetic transmission of acquired characteristics could have been responsible for the evolution of separate species has been answered by Skinner et al. [38] who investigated the DNA mutations and non-DNA epigenetic changes in one of the icons of Darwinian speciation, the Galapagos finches. Five species were studied with different phylogenetic distances between them. Figure 4 shows the results. Both DNA mutations and epigenetic variations increase with the phylogenetic distance, with the epigenetic changes correlating better with distance. The authors conclude that both changes were involved in speciation and that they must have interacted.
Figure 4.
(a) Five of the Galapagos finch species were studied, the reference species Geospiza fortis and four others. The graph in (b) shows the number of genetic and epigenetic changes plotted as a function of phylogenetic distance. The epigenetic changes correlate well with phylogenetic distance, the genetic mutations do not correlate as strongly [38]. (Online version in colour.)
7. Organisms can resist the harmful effects of many molecular-level variations
One of my own fields of research is cardiac rhythm and arrhythmias. The main pacemaker in the heart, the sinus node, is an example of a robust functional process. Several different ionic transporter circuits are involved, any one of which could generate rhythm. The evolutionary advantage of this situation is obvious: if one mechanism fails, another can take over the function. In 1992, we investigated this robustness by reverse engineering an experimentally based computer model. We found that removing a transporter that could carry as much as 80% of the ionic current necessary for generating the rhythm would change the overall frequency by only around 10–15% [39]. Reverse engineering studies using a physiological model reveals the mechanism of the substitution. The small voltage changes that occur when one component is knocked out are sufficient to activate the substituting mechanism. This discovery formed the basis of the development of a safe heart slowing medication, ivabradine [40].
This kind of ‘back up’ of important physiological functions is ubiquitous. A systematic study of gene knockouts in yeast showed that 80% of knockouts have little or no effect on physiological functions under normal physiological conditions [41]. Metabolic stress was needed to reveal the functional roles of most of the genes involved.
These studies pose a serious problem for bottom-up gene-centric theories of biology. The functionality will simply not be seen at that level or may be far from quantitatively accurate. Organisms seem to be very resourceful when challenged with knockouts, blockers or absence of nutrients. If we look for that ingenuity at the molecular level, we may not find it.
Again, we can ask the question whether such processes can be demonstrated in actual evolutionary time. This was done recently by Taylor et al. [42] who have shown that bacteria that have lost their flagella through deletion of the relevant DNA sequence can evolve the regulatory networks required to restore flagella and so restore motility in response to a stressful environment within just 4 days. Specifically, Taylor et al. show that deletion of FleQ (Flagellar transcriptional regulator) in Pseudomonas fluorescens, and starvation of the bacteria, produces mutations that enable the regulatory role to be taken over by a different pathway, normally involved in nitrogen uptake and assimilation. The genes required to produce flagellae are then reactivated by the new regulatory pathway. The authors interpret their work as showing how selection can rapidly produce this kind of substitution to restore activation of flagella genes. But, equally clearly, the mutations are targeted in a remarkably precise way. They are not randomly occurring anywhere in the genome. This example is therefore somewhat comparable to the cardiac pacemaker example I discuss earlier in this section, in that one network takes over the lost function when another network is no longer functional. That ability is a property of the bacterium regulatory networks and of the ability of the organism to signal the environment pressure to the genome to activate mutation.
It is important to note that such examples, and the earlier ones I quoted above in §5, involve what, so far as we know, are random mutations. At each location on the DNA sequence level, this will therefore appear as ‘blind’ variation. At that level, there will also be a form of Darwinian selection operating [14]. But the targeting of particular locations, which is what enables the response to the environmental challenge to be effective, is not blind. Nor does targeting necessarily require differential mutation rates in the genome. Buffering of non-functional genome changes by regulatory networks can also ensure the preservation of existing functionality, just as the regulatory networks involved in cardiac rhythm can ensure insensitivity to molecular-level changes, as I described at the beginning of this section.
Differential mutation rates have been extensively investigated by Moxon et al. [43] who use the term ‘contingency locus’ to characterize the targeted loci of hypermutable DNA. In bacteria, these loci are simple sequence repeats in which the repeating unit is one to several nucleotides. In eukaryotes, these loci are called microsatellites and often consist of hundreds of repeats. As ‘mutation rates vary significantly at different locations within the genome’, they propose that ‘it is precisely in the details of these differences and how they are distributed that major contributions to fitness are determined’. In an earlier article, Moxon & Thaler [44] write ‘This phenotypic variation, which is stochastic with respect to the timing of switching but has a programmed genomic location, allows a large repertoire of phenotypic solutions to be explored, while minimizing deleterious effects on fitness’.
8. Physical constraints can and must influence both development and evolution
Natarajan et al. [45], in a paper significantly entitled ‘Predictable convergence in hemoglobin function has unpredictable molecular underpinnings', have examined the molecular basis of convergence in haemoglobin function involving 56 avian taxa that have contrasting altitudinal range limits. They found that ‘Convergent increases in hemoglobin–oxygen affinity were pervasive among high-altitude taxa, but few such changes were attributable to parallel amino acid substitutions at key residues. Thus, predictable changes in biochemical phenotype do not have a predictable molecular basis’. This article beautifully illustrates the main point I am making in this paper, which is that unpredictability at the molecular level, which would lead one to think the changes are random, can be perfectly compatible with predictability and functionality at a higher level. This is biology's equivalent of the physical principle that determinate thermodynamics can coexist with unpredictable stochastic behaviour at a molecular level. The difference is that, in biological systems, through the process of evolution, the higher level becomes functional. That is the level at which the functionality can be seen. It is then the level from which the lower level stochasticity can be understood, including the functional constraints.
If physics can be so important by using stochasticity in convergent evolution, can it also be important in a similar way in constraining development? It is tempting to think so because early embryonic development is similar in all multi-cellular eukaryotes, despite many differences in genome sequences. Edelman et al. [46] have explored this question by showing graphically how some simple physical constraints might be sufficient to explain certain aspects of embryonic development without having to assume that there must always be a specific DNA basis for all such processes. Their images are speculative and would require computational modelling to develop and test the ideas. Stuart Newman, Santiago Schnell and Philip Maini have led the way on this approach [47,48]. There must be interaction between overall physical constraints and molecular-level specifications. Ehrlich et al. [49] show how modelling such physical constraints can account for the evolution of shell form in ammonites.
These examples illustrate a general point. Nature does not need to write to the ‘hard disc’ of the organism, its DNA, when it can get functions automatically from physical ‘free rides’, i.e. by letting physics do what it will do naturally. There is no need for DNA to be involved, for example, in ensuring that lipid membranes naturally fuse and form vesicles and many of the other properties of thin oily bilayers. And, of course, there is no DNA forming templates for the wide variety of lipids in organisms.
9. The gene-centric view has so far been very disappointing from the viewpoint of medicine
There is another field of science where focusing on the molecular level has blinded us to functional processes at higher levels. That is the field of medicine. But before I explain why that is the case, I want to make it quite clear that I fully recognize the great scientific value of genome sequencing.
Sequencing whole genomes has been of immense value in evolutionary biological studies. The benefits for phylogeny and in discovering new parts of the ‘trees’ or ‘networks’ of life are obvious. It was sequencing that enabled Carl Woese to make his fundamental discovery of the archaea and how they differ from bacteria and eukaryotes [50]. Sequencing also enabled us to identify the extent to which mobile genetic elements must have been involved in the evolution of many proteins. In this sense, describing the genome as the ‘book of life’ has been a useful metaphor. But, as a metaphor used to publicize the health benefits that would accrue from genome sequencing it has been misinterpreted. The promise was that by a decade or so following sequencing of the human genome, the ‘book of life’ would reveal how to treat cancer, heart disease, nervous diseases, diabetes and many others through the discovery of many new pharmaceutical targets. This did not happen. An editorial in Nature in 2010 spelt this out:
But for all the intellectual ferment of the past decade, has human health truly benefited from the sequencing of the human genome? A startlingly honest response can be found on pages 674 and 676, where the leaders of the public and private efforts, Francis Collins and Craig Venter, both say ‘not much’. [51]
The targets were identified all right. At least 200 new possible pharmaceutical targets are now known and there may be more to come, but we simply do not understand how to use them. The problem does not therefore lie in the absence of knowledge about the sequences. The problem is that we neglected to do the relevant physiology at the higher levels. A valuable critique of genotype–phenotype relations as a basis for the common disease–common variant hypothesis has been published by Joyner & Prendergast [52].
Before the shift towards genomic approaches to pharmacology, we did in fact have reasonably adequate methods for developing new drugs against specific diseases. The method was to work initially at a phenotype level to identify possible active compounds, and then to drill down towards individual protein or other molecular targets. This was the approach used so successfully by Sir James Black, the Nobel laureate discoverer of β-blockers and H2 receptor blockers [53]. It is the method by which the work of collaborators in my laboratory eventually led to the successful heart drug, ivabradine, to which I have already referred.
But the consequence of diverting large-scale funding towards the search for new drugs via genomics has been that the Black approach is now much less common and that the pharmaceutical industry is producing fewer new medications at vastly greater cost. Of course, the Black approach could and should be complemented by genomics, and there are successful cases where protein targets found by classical methods were later also identified as coding templates formed by particular genes. A good example is Duchenne muscular dystrophy, where the gene for the protein utrophin that can substitute, in mice at least, to cure the disease was discovered before the DNA sequence was identified [54].
10. Conclusion
There has been much debate about whether the neo-Darwinist modern synthesis needs extending or replacing. Both views are correct. It depends on the context in which they are assessed. Theories in biology, as in any branch of science, can be judged by several criteria.
10.1. Falsifiability
The original neo-Darwinist assumptions of the modern synthesis have been clearly falsified. I will consider the three basic assumptions outlined in the Introduction.
10.2. The Weismann barrier
The Weismann barrier should be seen as a relative not an absolute barrier. Strict isolation of the genome was required in order to exclude the inheritance of acquired characteristics. As we now know that acquired characteristics can be inherited, I believe it is more honest to admit that this reason for departing from Darwinism is no longer valid. In any case, the barrier could only apply in those organisms that have a separate germ line. For the great majority of the duration of life on the Earth, there was no separate germ line. And plants can reproduce separately from their germ line. Quite simply, then, two of the original basic assumptions, isolation of the germ line and the impossibility of inheritance of acquired characteristics, can be seen to be incorrect.
Some criticisms of this conclusion refer to the rarity of experiments showing intergenerational transmission of epigenetic mutations. Originally, this was based on the idea that the genome was always wiped clean of epigenetic marking, so that it was thought that the idea was misconceived and impossible. As I have shown, this is simply not correct.
Another criticism was that it would not be robust. It has been demonstrated to persist for as many as 100 generations, and that it can, in some cases, be as robust as DNA transmission. Moreover, it does not need to be robust in all cases. As the review by Burggren [55] shows, the softness and therefore reversibility of epigenetic inheritance is one of its evolutionary virtues. Sultan and co-workers [56] have also identified the factors that may determine the transience or persistence of epigenetic variation.
The third criticism is that it is observed in only rare cases. My reply is that so is speciation. Speciation is such a rare event that in thousands of years of selective breeding of cats, dogs, fish, etc., we have not succeeded in producing new species, as defined by reproductive isolation.
Note also that these criticisms obviously do not apply to functionally significant reorganization or hypermutation of genomes.
10.3. Blind stochasticity
The other basic assumption is blind stochasticity, meaning that what are seen as random genetic variations are not functionally directed. The concept of randomness is a major topic of research in philosophy, mathematics and physics. One way to by pass these highly technical issues is to ask the question ‘random with respect to what’? The key in relation to evolutionary biology is whether variations are random with respect to function and whether they can be seen to be so. Even if the molecular-level variations do in fact represent functional order at a higher level, we will almost certainly require insight from the functional level to appreciate the functional nature of the molecular variations. The randomness I am referring to is therefore epistemological: without knowing the constraints by higher levels, the variations will appear to be random and unpredictable. Once we know those constraints the possibility of prediction at the molecular level begins to exist. Whether it is computable is a very different question. Given the huge differences of scale, e.g. between molecular and cellular, it is implausible to expect molecular-level computation alone to reveal the functionality.
Even before we consider whether a theory based on blind stochasticity has been falsified, we have to examine its conceptual status. A very basic lesson from physics is that stochasticity at lower, such as molecular, levels is not only inevitable as a consequence of molecular kinetic energy, it is also perfectly compatible with regular law-like behaviour at higher levels, a fact that was appreciated long ago by one of the founders of population genetics, Fisher [57]. Even if behaviour at a high level is directed, stochasticity is what we can expect at lower levels. The example in this paper concerning the evolution in different species of haemoglobins at high altitude illustrates that point perfectly. As the authors of that paper say ‘predictable changes in biochemical phenotype do not have a predictable molecular basis’ [45]. It is the physics of oxygen transport in organisms living at low partial pressures of oxygen that dictates the changes that occur to adapt to such environments, not specific changes in the genome.
From a gene-centric viewpoint, it could be objected that the genome changes are nevertheless those that enable the beneficial changes in oxygen transport to happen. That is certainly true. But it is precisely the higher level perspective that enables us to show that fact. What we can see here is that a conceptual issue, which is the question of the level at which functionality occurs, interacts with an empirical issue, which is whether the changes at the molecular level are predictable, from that level alone. Another way to put the conceptual issue is to say that, in any information transmission system, whether languages or genomes, sequences by themselves do not have meaning. They acquire meaning through their context, which can only be understood at a much higher level. As a linguistic example, the three letter alphabetic sequence ‘but’ has two totally different meanings and pronunciations in English and French. Similarly, genome sequences acquire meaning in their context. Sequences enabling arms, legs and eyes derive from organisms that had none of these.
10.4. Unravelling the problem
My paper unravels this problem by showing where some aspects of biological thought went wrong in the twentieth century. Schrödinger's book, What is life?, was a landmark in predicting correctly that the genetic material would be found to be an aperiodic crystal. But it contained the seeds of a major misunderstanding, leading Schrödinger, and then Crick and Watson, to maintain that, like a crystal, the genetic material could be read in a determinate way. That could be true only if the ‘crystal’, that is the linear polymer DNA, could be read and copied faithfully, with few or no copy errors. As we can now see, that is not an inherent property of DNA alone. On the contrary, it is a property of the complex system by which the copy error rate can be reduced from an unacceptable frequency of millions per genome to less than 1. That is a higher level systems property of cells, including an army of proteins and lipids, not of DNA alone. In life as we know it on the Earth, this process occurs only in the context of living cells.
A possible objection to this conclusion is that all proteins have DNA templates that determine their amino acid sequences. That includes the proteins that contribute to the error-correcting systems for DNA. That is true, but it is usually taken a step further to mean that therefore the genome determines everything. That is not true. The error-correcting systems operate within cellular structures that contain molecular elements, such as lipid membranes, that do not require DNA templates in order to exist. Elsewhere, I have shown that the structural information in cells can be represented as comparable to that in the genome [58]. Organisms always inherit both. In one of the rare examples of a successful clone from the nucleus of one species inserted into the enucleated but fertilized egg cell from another species, both the cell and the nucleus contribute to the final structure of the adult. Reproductive hybridization between species has also been shown to produce intermediate forms which can generate speciation [59].
Experimentally, we need to re-examine the way in which functional change in organisms can harness stochasticity at lower levels to create new functionality. Huang and his co-workers have shown the way forward here by demonstrating that stochasticity in gene expression is an attractor produced by a cell population. The many studies of targeted hypermutation, e.g. by Moxon's group, also show the way forward. Organisms in their evolution had to harness stochasticity because at a low enough level, this is an inevitable property of the physics of molecular-level systems that have kinetic energy.
We can now return to the question whether the assumption of blind stochasticity has been falsified. If the case presented in this paper is correct, then one answer would be that it is very difficult for it to be falsified because stochasticity necessarily reigns at a low enough level, even if functionality reigns at higher levels. The constraints may have too subtle an effect at the molecular level. The falsifiability then depends on a prior conceptual question, which is whether one accepts multi-level functionality. A purely gene-centric theory does not accept multi-level functionality and can therefore maintain its view of everything being ‘blind chance followed by natural selection’.
To a physiologist or a medical scientist, this is not a useful viewpoint. Functionality arises in organisms at many different levels. This is one of the bases of my formulation of the principle of biological relativity, first proposed in a previous article in this journal, and developed more completely in a book, Dance to the tune of life. Biological relativity [60].
10.5. Utility
These points naturally lead to the other main criterion for judging a theory, which is its utility. Theories can be useful, even if they are false. Indeed, on a Popperian view of the logic of science, that must always be true. We can only ever falsify theories about the natural world, never conclusively prove them. I want therefore to acknowledge the fact that the neo-Darwinist modern dynthesis was very useful. Whole fields of mathematical biology, such as population genetics, would not have flourished in the twentieth century without the modern synthesis as a framework.
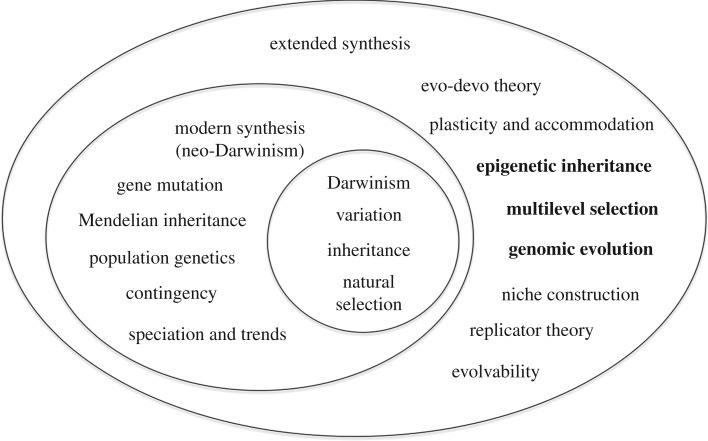
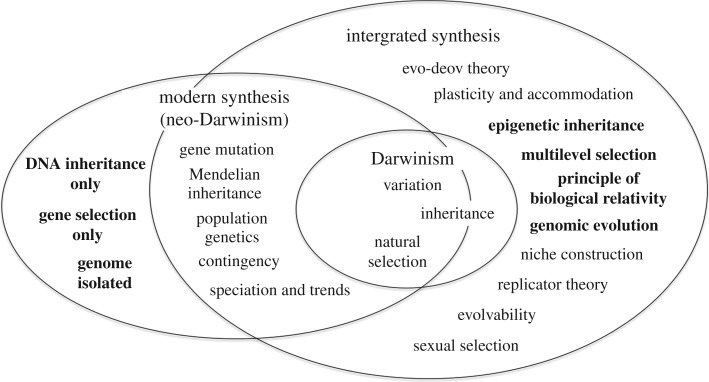
But, I also think that we have reached a watershed in relation to the issue of the utility of the neo-Darwinist modern synthesis. As I have argued in detail elsewhere, there are too many experimental breaks with the original theory as formulated by Weismann & Wallace [61]. Moreover the metaphorical language of neo-Darwinism is a problem. The metaphors used strongly reinforce a simplistic gene-centric view. The time has come to see that evolutionary biology would progress faster if we used a different framework to develop a more inclusive theory, as illustrated in figures 5 and 6.
Figure 5.
The extended evolutionary synthesis representing the extension as extensions of Darwinism and then of the neo-Darwinist modern synthesis (from [62]).
Figure 6.
The integrated synthesis representing the extensions as extensions of Darwinism but only partially from neo-Darwinism. Darwin's view of inheritance is also represented as extending outside the boundary of neo-Darwinism/(developed for this article from [61], based on [62]).
Figure 5 shows the extended evolutionary synthesis, which is represented as a development from the neo-Darwinist modern synthesis, in turn developed from Darwinism.
Figure 6 shows the version of this diagram that better represents the conclusions of this paper. There are several important differences. First, it represents the fact that Darwin's view of inheritance included the inheritance of acquired characteristics, which was excluded by neo-Darwinism. Darwin's concept of inheritance is therefore shown as being partly outside the neo-Darwinist modern synthesis. Second, it represents the features of the extended synthesis (highlighted in bold in both figures 5 and 6) that lie outside the range of neo-Darwinism as defined by Weismann and Wallace. The features of that theory that were excluded are shown as corresponding bold-face items. The highlighted items on the far left correspond with the highlighted items at the far right. Also included as a bold-face item is the principle of biological relativity. Although beyond the scope of this paper, I have included sexual selection.
In spirit, this approach inherits an important part of Darwin's more nuanced philosophical approach. I emphasize philosophical here because it is obvious that we have moved way beyond what Darwin knew experimentally, as figures 5 and 6 also show. But we can learn from his approach. Darwin was cautious in acknowledging the limits of what he knew. He was even unsure whether he had discovered the title of his book, because he did not know what produced variations in organisms, and he did not exclude the inheritance of acquired characteristics. Unjustified certainty is not the best way forward in scientific research. It remains open to further experimentation to clarify the extent of the many mechanisms now known to be available to nature, and to determine how she used them, alone or more probably in various combinations, to evolve life as we now know it.
Acknowledgements
I wish to thank: Raymond Noble for discussion of evolutionary biology over many years; Sir Anthony Kenny for introducing me to the argument that a symbolic sequence in itself is meaningless out of its context, in a debate with Richard Dawkins in 1976; Jean-Jacques Kupiec for first pointing out the error in Schrödinger's What is life?; Charles Taylor for his insights into the conceptual nature of teleology in a debate with me published in Analysis in 1967; James Shapiro for introducing me to the significance of mobile genetic elements; Lynn Margulis for introducing me to the importance of symbiogenesis in her debate with Richard Dawkins in 2009; Eva Jablonka for discussions on Lamarckism; Michael Joyner for his insights on the deficiencies of the gene-centric view in medicine; David Paterson for arranging and chairing the EB2015 Boston Conversation (https://www.youtube.com/watch?v=A_q_bOWc8i0); the co-organizers of a Balliol Interdisciplinary Institute (BII) project on the conceptual foundations of System Biology, Jonathan Bard, Tom Melham and Eric Werner; Nicholas Beale for valuable comments on a draft of this paper; Susan Noble for great support and criticism of my articles and books during the last years of her life—with Hilary Brown and Dario DiFrancesco she was responsible for the discovery that led to the life-saving drug ivabradine which depends precisely on the ability of the heart's pacemaker function to adapt to molecular-level changes.
Data accessibility
This article has no additional data.
Competing interests
I declare I have no competing interests.
Funding
We received no funding for this study.
References
- 1.Mayr E. 1964. Introduction. In On the origin of species (ed. Darwin C.), pp. xxv–xxvi. Cambridge, MA: Harvard. [Google Scholar]
- 2.Lamarck J-B. 1815–1822 Histoire Naturelle des animaux sans vertèbres. Paris, France: Verdière. [Google Scholar]
- 3.Lamarck J-B. 1994. Philosophie Zoologique, original edition of 1809 with introduction by Andre Pichot. Paris, France: Flammarion. [Google Scholar]
- 4.Darwin C. 1869. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life, 3rd edn London, UK: John Murray. [PMC free article] [PubMed] [Google Scholar]
- 5.Weismann A. 1893. Die Allmacht der Naturzüchtung; eine Erwiderung an Herbert Spencer. Jena, Germany: Fischer. [Google Scholar]
- 6.Huxley JS. 1942. Evolution: the modern synthesis. London, UK: Allen & Unwin. [Google Scholar]
- 7.Weismann A. 1896. On germinal selection as a source of definite variation. Chicago, IL: Open Court. [Google Scholar]
- 8.Weissman C. 2011. Germinal selection: a Weismannian solution to Lamarckian problematics. In Transformations of Lamarckism from subtle fluids to molecular biology (eds Gissis SB, Jablonka E), pp. 57–66. Cambridge, MA: MIT. [Google Scholar]
- 9.Darwin C. 1868. The variation of animals and plants under domestication. London, UK: John Murray. [Google Scholar]
- 10.Whitfield J. 2003. Molecules form new state of matter. Nature 408, 361–365. ( 10.1038/news031110-16) [DOI] [Google Scholar]
- 11.Schrödinger E. 1944. What is life? Cambridge, UK: Cambridge University Press. [Google Scholar]
- 12.Dronamraju KR. 1999. Erwin Schrödinger and the origins of molecular biology. Genetics 153, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crick FHC. 1958. On protein synthesis. Symp. Soc. Exp. Biol. 12, 138–163. [PubMed] [Google Scholar]
- 14.Kupiec J-J. 2009. The origin of individuals: a Darwinian approach to developmental biology. London, UK: World Scientific Publishing Company. [Google Scholar]
- 15.Kupiec J-J. 2014. Cell differentiation is a stochastic process subjected to natural selection. In Towards a theory of development (eds Minelli A, Pradeu T), pp. 155–173. Oxford, UK: OUP. [Google Scholar]
- 16.Crick FHC. 1970. Central dogma of molecular biology. Nature 227, 561–563. ( 10.1038/227561a0) [DOI] [PubMed] [Google Scholar]
- 17.Deaton AM, Bird A. 2011. CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022. ( 10.1101/gad.2037511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Planck M. 1917. Dynamische und statistische Gesetzmäßigkeit. Zeitschrift fur Elektrochemie und angewandte physikalische. Chemie 23, 63. [Google Scholar]
- 19.McElhinny SAN, Pursell ZF, Kunkel TA. 2009. Mechanisms for high fidelity DNA replication. In Molecular themes in DNA replication (ed. Cox LS.), pp. 86–111. London, UK: RSC Publishing. [Google Scholar]
- 20.Pani A, Dessi S. 2004. Cell growth and cholesterol esters. Dordrecht, Netherlands: Kluwer/Plenum. [Google Scholar]
- 21.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. 2008. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 453, 544–547. ( 10.1038/nature06965) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S. 2009. Non-genetic heterogeneity of cells in development: more than just noise. Development 136, 3853–3862. ( 10.1242/dev.035139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Odegard VH, Schatz DG. 2006. Targeting of somatic hypermutation. Nat. Rev. Immunol. 8, 573–583. ( 10.1038/nri1896) [DOI] [PubMed] [Google Scholar]
- 24.Jablonka E, Lamb M. 2005/2014 Evolution in four dimensions. Boston, MA: MIT Press. [Google Scholar]
- 25.Bridges BA. 1997. Hypermutation under stress. Nature 387, 557–568. ( 10.1038/42370) [DOI] [PubMed] [Google Scholar]
- 26.Bender W, Hudson A. 2000. P element homing to the Drosophila bithorax complex. Development 127, 3981–3992. [DOI] [PubMed] [Google Scholar]
- 27.McClintock B. 1984. The significance of responses of the genome to challenge. Science 226, 792–801. ( 10.1126/science.15739260) [DOI] [PubMed] [Google Scholar]
- 28.Landers ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921. ( 10.1038/35057062) [DOI] [PubMed] [Google Scholar]
- 29.Bos J, Zhang Q, Vyawahare S, Rogers E, Rosenberg SM, Austin R. 2015. Emergence of antibiotic resistance from multinucleated bacterial filaments. Proc. Natl Acad. Sci. USA 112, 178–183. ( 10.1073/pnas.1420702111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jack CV, Cruz C, Hull RM, Ralser M, Houseley J. 2015. Regulation of ribosomal DNA amplification by the TOR pathway. Proc. Natl Acad. Sci. USA 112, 9674–9679. ( 10.1073/pnas.1505015112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kar P, Mirams GR, Christian HC, Parekh AB. 2016. Control of NFAT isoform activation and NFAT-dependent gene expression through two coincident and spatially segregated intracellular Ca2+ signals. Mol. Cell 64, 746–759. ( 10.1016/j.molcel.2016.11.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma H, et al. 2014. γCaMKII shuttles Ca2+/CaM to the nucleus to trigger CREB phosphorylation and gene expression. Cell 159, 281–294. ( 10.1016/j.cell.2014.09.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tollefsbol T. (ed.). 2014. Transgenerational epigenetics: evidence and debate. Waltham, MA: Academic Press. [Google Scholar]
- 34.Miska EA, Ferguson-Smith AC. 2016. Transgenerational inheritance: models and mechanisms of non-DNA sequence-based inheritance. Science 354, 59–63. ( 10.1126/science.aaf4945) [DOI] [PubMed] [Google Scholar]
- 35.Rechavi O, Minevish G, Hobert O. 2011. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 147, 1248–1256. ( 10.1016/j.cell.2011.10.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson VR, Heaney JD, Tesar PJ, Davidson NO, Nadeau JH. 2012. Transgenerational epigenetic effects of Apobec1 deficiency on testicular germ cell tumor susceptibility and embryonic viability. Proc. Natl Acad. Sci. USA 109, E2766–E2773. ( 10.1073/pnas.1207169109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattick JS. 2012. Rocking the foundations of molecular genetics. Proc. Natl Acad. Sci. USA 109, 16400 ( 10.1073/pnas.1214129109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skinner MK, Gurerrero-Bosagna C, Haque MM, Nilsson EE, Koops JAH, Knutie SA, Clayton DH. 2014. Epigenetics and the evolution of Darwin's finches. Genome Biol. Evol. 6, 1972–1989. ( 10.1093/gbe/evu158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noble D, Denyer JC, Brown HF, DiFrancesco D. 1992. Reciprocal role of the inward currents ib,Na and if in controlling and stabilizing pacemaker frequency of rabbit sino-atrial node cells. Proc. R. Soc. Lond. B 250, 199–207. ( 10.1098/rspb.1992.0150) [DOI] [PubMed] [Google Scholar]
- 40.DiFrancesco D, Camm JA. 2004. Heart rate lowering by specific and selective If current inhibition with ivabradine: a new therapeutic perspective in cardiovascular disease. Drugs 64, 1757–1765. ( 10.2165/00003495-200464160-00003) [DOI] [PubMed] [Google Scholar]
- 41.Hillenmeyer ME, et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320, 362–365. ( 10.1126/science.1150021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor TB, et al. 2015. Evolutionary resurrection of flagellar motility via rewiring of the nitrogen regulation system. Science 347, 1014–1017. ( 10.1126/science.1259145) [DOI] [PubMed] [Google Scholar]
- 43.Moxon R, Bayliss C, Hood D. 2006. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 40, 307–333. ( 10.1146/annurev.genet.40.110405.090442) [DOI] [PubMed] [Google Scholar]
- 44.Moxon ER, Thaler DS. 1997. The tinkerer's evolving tool-box. Nature 387, 659–662. ( 10.1038/42607) [DOI] [PubMed] [Google Scholar]
- 45.Natarajan C, Hoffmann FG, Weber RE, Fago A, Witt CC, Storz JF. 2016. Predictable convergence in hemoglobin function has unpredictable molecular underpinnings. Science 354, 336–339. ( 10.1126/science.aaf9070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edelman DB, McMenamin M, Sheesley P, Pivar S. 2016. Origin of the vertebrate body plan via mechanically biased conservation of regular geometrical patterns in the structure of the blastula. Prog. Biophys. Mol. Biol. 121, 212–244. ( 10.1016/j.pbiomolbio.2016.06.007) [DOI] [PubMed] [Google Scholar]
- 47.Müller G, Newman SA (eds). 2003. Origination of organismal form: beyond the gene in developmental and evolutionary biology. Cambridge MA: MIT Press. [Google Scholar]
- 48.Schnell S, Maini PK, Newman SA, Newman T, Schatten GP (eds). 2007. Multiscale modeling of developmental systems. London, UK: Academic Press. [Google Scholar]
- 49.Ehrlich A, Moulton DE, Goriely A, Chirat R. 2016. Morphomechanics and developmental constraints in the evolution of ammonites shell form. J. Exp. Zool. (Mol. Dev. Evol.) 00B, 1–14. [DOI] [PubMed] [Google Scholar]
- 50.Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. USA 74, 5088–5090. ( 10.1073/pnas.74.11.5088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Editorial. 2010. The human genome at ten. Nature 464, 649–650. ( 10.1038/464649a) [DOI] [PubMed] [Google Scholar]
- 52.Joyner MJ, Prendergast FG. 2014. Chasing Mendel: five questions for personalized medicine. J. Physiol. 592, 2381–2388. ( 10.1113/jphysiol.2014.272336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Black JW, Crowther AF, Shanks RG, Smith LH, Dornhorst AC. 1964. A new adrenergic betareceptor antagonist. Lancet 283, 1080–1081. ( 10.1016/S0140-6736(64)91275-9) [DOI] [PubMed] [Google Scholar]
- 54.Fairclough RJ, Wood MJ, Davies KE. 2013. Therapy for Duchenne muscular dystrophy: renewed optimism from genetic approaches. Nat. Rev. Genet. 14, 373–378. ( 10.1038/nrg3460) [DOI] [PubMed] [Google Scholar]
- 55.Burggren W. 2016. Epigenetic inheritance and its role in evolutionary biology: re-evaluation and new perspectives. Biology 5, 24 ( 10.3390/biology5020024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herman JJ, Spencer HG, Donohue K, Sultan SE. 2013. How stable ‘should’ epigenetic modifications be? Insights from adaptive plasticity and bet hedging. Evolution 68, 632–643. ( 10.1111/evo.12324) [DOI] [PubMed] [Google Scholar]
- 57.Fisher RA. 1934. Indeterminism and natural selection. Philos. Sci. 1, 99–117. ( 10.1086/286308) [DOI] [Google Scholar]
- 58.Noble D. 2011. Differential and integral views of genetics in computational systems biology. Interface Focus 1, 7–15. ( 10.1098/rsfs.2010.0444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Y-H, Zhu Z-Y. 2014. Cross-species cloning: influence of cytoplasmic factors on development. J. Physiol. 592, 2375–2379. ( 10.1113/jphysiol.2014.272138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noble D. 2016. Dance to the tune of life. Biological relativity. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 61.Noble D. 2015. Evolution beyond neo-Darwinism: a new conceptual framework. J. Exp. Biol. 218, 7–13. ( 10.1242/jeb.106310) [DOI] [PubMed] [Google Scholar]
- 62.Pigliucci M, Müller GB. 2010. Evolution—the extended synthesis. Cambridge, MA: MIT Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.