Abstract
In recent decades, the phenotype of an organism (i.e. its traits and behaviour) has been studied as the outcome of a developmental ‘programme’ coded in its genotype. This deterministic view is implicit in the Modern Synthesis approach to adaptive evolution as a sorting process among genetic variants. Studies of developmental pathways have revealed that genotypes are in fact differently expressed depending on environmental conditions. Accordingly, the genotype can be understood as a repertoire of potential developmental outcomes or norm of reaction. Reconceiving the genotype as an environmental response repertoire rather than a fixed developmental programme leads to three critical evolutionary insights. First, plastic responses to specific conditions often comprise functionally appropriate trait adjustments, resulting in an individual-level, developmental mode of adaptive variation. Second, because genotypes are differently expressed depending on the environment, the genetic diversity available to natural selection is itself environmentally contingent. Finally, environmental influences on development can extend across multiple generations via cytoplasmic and epigenetic factors transmitted to progeny individuals, altering their responses to their own, immediate environmental conditions and, in some cases, leading to inherited but non-genetic adaptations. Together, these insights suggest a more nuanced understanding of the genotype and its evolutionary role, as well as a shift in research focus to investigating the complex developmental interactions among genotypes, environments and previous environments.
Keywords: norm of reaction, transgenerational plasticity, genetic determinism, genotype by environment interaction
1. Introduction
The concept of genotype is central to both biological and human sciences. New findings at the molecular level have established that it is gene expression as regulated by environmental and cellular factors, rather than DNA sequences per se, that shapes phenotypic variation. This recognition has led to a focus on individual developmental plasticity, a general property of organisms that was known but deemed marginal by mid-twentieth century evolutionists. This essay examines how insights to plasticity destabilize the concept of genotype on which the Modern Synthesis model of evolution was founded, and indicate ways to renew this central concept.
1.1. The genotype as a developmental programme
For the past half-century, biology has been dominated by a gene-based approach in which an organism's DNA sequence is understood to comprise the instructions for that organism's development [1–3]. According to this view, an individual organism's set of genes (its genotype) determines that individual's physical traits and behaviours (its phenotype), so it is possible to know what the organism's features will be just by knowing its DNA sequence. Because gene expression itself is assumed to be under genetic control, the genotype is seen as a self-contained internal developmental ‘programme’ that specifies a single, determinate phenotypic outcome [4]. The interpretive metaphor of the ‘genetic programme’ has become a deeply embedded construct for framing both developmental and evolutionary phenomena [2,5,6].
This view of the genotype has led to three key evolutionary corollaries. First, if genes determine specific traits such as size, structure and behaviour, the organism's adaptation to its environment is set by its genotype. Second, if traits of individuals depend on their genes, then the functional and fitness differences between individuals that cause natural selection are also specified by their genotypes—in other words, fitness differences originate in genetic differences. Third, the DNA sequence inscribed within the nucleus of each cell comprises the developmental information that is passed from one generation to the next. Because this genetically encoded information is impervious to the environment as well as resistant to error, it is faithfully transmitted across a continuous evolutionary trajectory.
Together, these three points form the foundation of the elegantly simple and coherent Modern Synthesis model of adaptation as population-level change over time in the relative frequencies of alternative genetic alleles. It is a commitment to this causal model that lies at the heart of contemporary debate about whether this conceptual framework for adaptive evolution—and thus for contemporary research programmes—remains generally sound [7] or requires revision [8]. This tension reflects the fact that a no longer tenable genetic programme view of phenotypic and hence fitness variation is implicit in the Modern Synthesis approach [5,6,9].
2. Conceptual models and empirical approaches
The idea of the genotype as a set of self-contained developmental specifications was given mechanistic solidity following the work of Watson and Crick in revealing the biochemical ‘code’ of nucleotides in the DNA molecule [1]. Following from this foundational idea, and in marked contrast to the environmentally contextualized view of development that had characterized earlier work [10], the goal of developmental studies has been to reveal this ‘sequestered’ internal information [11]. Similarly, mainstream evolutionary biologists have sought to identify the genetic basis of adaptive variation as if the process of development ‘did not exist’ ([2, p. 18]; see [12]). This is done experimentally by raising genetically different individuals in a single, uniform ‘control’ or ‘common garden’ environment that is meant to be developmentally neutral, in the sense of permitting expression of the phenotype undistorted by environmental effects ([13] and references therein). Since the developmental environment is held constant in these studies, any trait differences among individuals are understood to result from differences in their genotypes.1 As a result, a single-environment experimental design and a determinate view of gene-based variation serve to reinforce each other [14].
Despite the ubiquity of this experimental approach, biologists are well aware that organisms develop not in ‘neutral’ environment-less conditions, but rather in particular environments—whether in nature or in the laboratory—that are characterized by specific physical factors, chemical compositions, resource levels and the presence or absence of biotic interactors. They are equally aware that the exact states of such environmental factors influence the developmental process, and consequently the organism's functional and fitness traits. Indeed, it is precisely because of this influence that researchers employ the ‘control environment’ approach: they do so in order to exclude variability in environmental factors that would otherwise affect phenotypes. By rationalizing this approach, the idea of an internally contained developmental programme led to a neglect of environmental context in studies of gene expression [3].
Unexpectedly, it is the intense focus of contemporary biologists on molecular, presumably internal pathways of developmental regulation that has newly underscored the environment's critical role by providing a mechanistic basis for it. Thanks to a flood of recent observations, it is now clear that genes are differently expressed depending on environmental context, leading to tremendous regulatory diversity and complexity ([15–17] and references therein). In the light of these findings, genes can more accurately be viewed as ‘potential resources’ for developmental pathways [4] than as fixed pieces of information. Even biologists who seek to preserve the Modern Synthesis conceptual framework acknowledge that ‘technological advances in the past decade have revealed an incredible degree of plasticity in gene expression in response to diverse environmental conditions' [7]. These molecular data make clear that phenotypes are not scripted in advance from the nucleus, but instead emerge from regulatory interactions in which environmental factors participate in specific ways. The organism's environment as well as its genotype provides the kind of precise developmental information that guides the cellular and nuclear processes that shape phenotypes, including dynamic traits such as physiology and behaviour [11,18].
This powerful insight requires that biologists replace the ‘genetic programme' model of internal developmental control with one in which each genotype may express different phenotypes depending on its environment—in other words, with a focus on developmental plasticity as expressed in response to specific conditions (figure 1). More broadly, the general term ecological development or eco-devo [20,21] situates the normal developmental process in its environmental context by emphasizing how regulatory pathways integrate environmental signals at the cellular and molecular levels ([17,22–25] and references therein). Under this unified concept, plasticity describes those cases in which outcomes differ appreciably among environments, as distinct from environmentally insensitive or canalized trait expression patterns.
Figure 1.
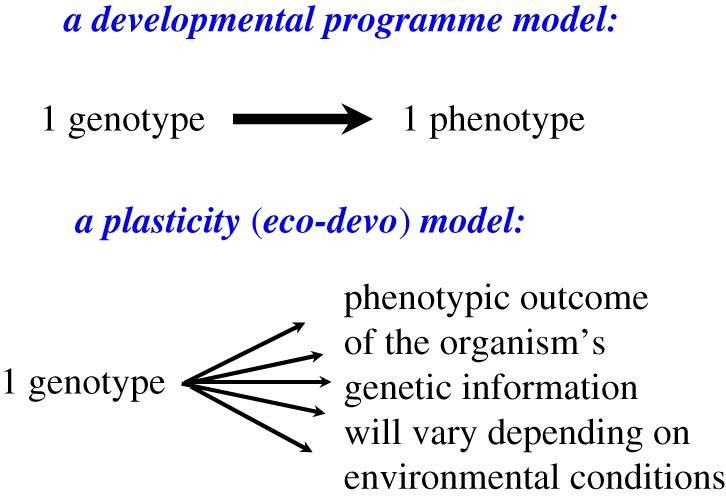
Alternative views of the genotype. Inherent to the Modern Synthesis is a deterministic model of phenotypic expression (above) in which the genotype is seen as a self-contained, internal developmental programme. By contrast, a model that recognizes developmental plasticity (below) views the genotype as a developmental repertoire of varying, environmentally context-dependent outcomes. (Modified from [19].) (Online version in colour.)
An ‘eco-devo’ approach can be implemented by means of a key experimental change: by inverting the design so as to bring in rather than exclude environmental variation. To do this, a researcher generates replicate individuals of each experimental genotype (via cloning or inbreeding) and grows these genetic replicates in each of several distinct environments. The resulting phenotypes can be plotted to visually characterize each genotype's range of environment-specific developmental outcomes, known as its norm of reaction [26–29]. The norm of reaction for any trait in an organism reflects both the particular genotype and the precise set of environmental states in which it is measured.
Note that the idea of characterizing a genotype by its pattern of environmental responses (rather than by the trait it expresses in a single ‘control’ environment) predates the Modern Synthesis, with its emphasis on inborn, genetic determination of phenotypes [30]. Instead, the norm of reaction makes explicit the environmental context-dependency of the phenotypes that a given genotype produces. Once this context-dependency is recognized, the researcher's choice of environmental conditions becomes critically important [31]. Indeed, subtle differences among laboratories in animal handling and rearing techniques may be one reason why bioemedical researchers have often been unable to replicate one another's results, leading to a ‘reproducibility crisis' that is mistakenly attributed to sloppiness or chance [32]. To the extent that experimental environments reflect naturally occurring conditions, norm of reaction studies can provide information about trait expression in real populations. As discussed below, empirically determined norms of reaction illuminate two key evolutionary issues: adaptation and genetic variation.
3. Developmental plasticity as adaptive variation
Based on knowledge of a species’ ecology, it is possible to evaluate whether the phenotypes expressed by a given genotype are functionally adaptive to the alternative environments in which they occur (e.g. [33–36]). The norm of reaction for any developmental, physiological or behavioural trait of interest may be relatively constant across environments or change from one environment to another. Such changes may constitute adaptive adjustments (as indicated by positive ecophysiological or fitness effects in the inducing environment), or may simply reflect inevitable environmental effects on development such as reduced growth in resource-poor conditions. In the many plants, fungi, lichens, invertebrates, amphibians, reptiles, fish, mammals and birds in which norms of reaction have been found to comprise adaptive responses to specific conditions, plasticity provides for an individual, developmental mode of adaptation ([17,24,25,28,37–46] and references therein).
For example, individual plants of the widespread colonizing species Polygonum persicaria grown at reduced light produce far greater photosynthetic leaf surface area relative to their mass than do cloned plants of the same genotypes grown in full sun [44,47,48]. This increase in the plant's ability to catch scarce photons (and hence maintain growth and reproduction) results from two developmental changes expressed in moderate and low light compared with full sun: increased relative allocation of plant tissue to leaves (figure 2), and broader, thinner leaf size and structure (figure 3). Similarly, Polygonum plants raised in dry or nutrient-poor soil invest a higher proportion of their body mass into root tissues, and make the roots themselves longer and thinner, compared with genetically identical individuals grown in moist or rich soil. These plastic responses result in much more extensive root systems that can more effectively collect soil resources that are present in low concentrations [50–53].
Figure 2.
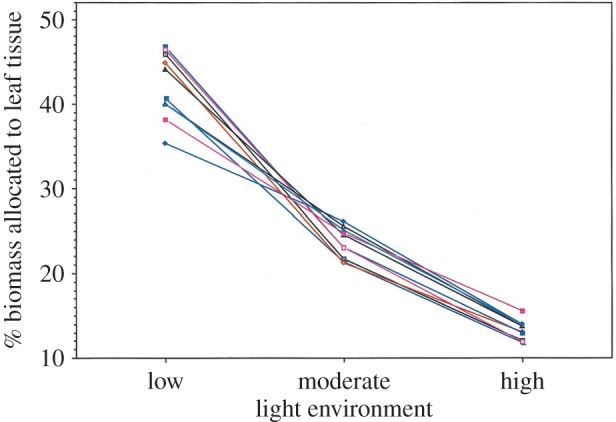
Developmental plasticity for the proportion of total biomass allocated to leaf tissue by Polygonum persicaria plants in response to contrasting light levels. Each line represents the reaction norm of a single plant genotype, based on the mean of six clonal replicates in each of three greenhouse light treatments (low (8%), moderate (37%) and high (100%) levels of incident midsummer sun). Across genotypes, trait change due to plasticity (the effect of light environment) is highly significant (p < 0.001). Genotypes differ in their specific patterns of plastic response, resulting in changes in among-genotype variance and rank order from one environment to another (genotype × environment interaction effect; p < 0.001). Because their norms of reaction cross, there is no consistent effect of genotype on phenotype (the main effect of genotype is non-significant; p > 0.05). (Figure reprinted from [49].) (Online version in colour.)
Figure 3.
Developmental plasticity expressed by genotypes of the common annual plant Polygonum persicaria. Significantly broader and structurally thinner leaves are produced by replicate plants of the same Polygonum genotype when grown in moderate shade (a) compared with full summer sun (b). (Photo courtesy of Dan B. Sloan and S. E. Sultan.)
These findings from cloned Polygonum plants grown in contrasting light and soil conditions exemplify three key points that characterize developmental plasticity across biological systems. First, these plastic responses are not trivial tweaks to a pre-determined developmental programme, but substantial changes in the expression of functionally important traits. Second, the very different phenotypes produced by Polygonum genotypes in different conditions constitute environment-specific adjustments, in this case ones that enhance function by increasing the availability of the most limited resource. Viewed in another way, such functionally adaptive developmental adjustments improve the environment that the plant experiences: plants in low light that increase their surface area experience an environment in which more photons are available, and plants with very high root surface area for water uptake have greater access to moisture. Third, whether adaptive or inevitable, phenotypic changes due to developmental plasticity alter external conditions for that individual as well as for co-occurring plants, animals and microbes in its habitat. For instance, plants in darker microsites produce larger leaves that cast more shade, reducing temperature, light quantity and red:far red spectral quality at the soil surface; these thinner leaves also decompose more rapidly, which increases mineral cycling rates in the soil. Because the particular phenotypes that organisms express will differently influence their experienced and external environments, plastic developmental responses partially shape the selective pressures under which they evolve ([17]; see also [54]), an evolutionary feedback termed niche construction [55–57].
Norm of reaction (eco-devo) studies thus reveal the genotype as a repertoire of possible developmental responses expressed by the organism in specific conditions, rather than as a self-contained set of fixed developmental instructions with a single outcome. As in the case of Polygonum plants, these environmental responses often comprise an immediate, developmental mode of adaptation to contrasting conditions. This mode of adaptation takes place at the level of the individual organism, as distinct from adaptive phenotypes produced by natural selection via population-level allele frequency change. An important evolutionary consequence is that, unlike the random and rare occurrence of favourable new genetic variants, plasticity can provide adaptive variation when it is needed (i.e. in response to a particular environmental challenge or change) and in numerous individuals in a population at once. As noted by Sewall Wright [58], this may buffer selective change by allowing existing genotypes to maintain fitness in altered or diverse conditions (recent models demonstrating this effect include [59–61]).
4. A norm of reaction view of genetic diversity
When genotypes are viewed as determinative, self-contained developmental programmes, they are assumed to be consistently associated with particular outcomes. Accordingly, in this model the functional and fitness trait differences that fuel natural selection directly reflect the genotypic diversity that is present. Just as conceptualizing the genotype as a repertoire of environmentally contingent outcomes reveals new sources of adaptive variation, this conceptual step also leads to a more nuanced view of the genetic diversity necessary for selective evolution.
Owing to sequence differences along pathways of environmental perception and phenotypic response, distinct genotypes exposed to the same range of conditions will express different norms of reaction, for various traits [29,62]. In a classic paper on ‘nature and nurture’, J.B.S. Haldane [63] observed that, in naturally evolved systems, these differing norms of reaction are very rarely parallel. Instead, as numerous quantitative-genetic studies have since confirmed, genotypes are generally characterized by plastic adjustments that differ in magnitude and/or direction in response to a given set of environments ([64–66]; genotype by environment interaction, the statistical term for such non-parallel response patterns, results in 874 000 publication hits on Google Scholar). As a result of non-parallel norms of reaction, the trait differences among a given group of genotypes will depend not only on those genotypes but also on the environments they encounter.
Two evolutionary points follow from this insight, as illustrated by Polygonum norms of reaction for leaf allocational plasticity (figure 2). First, the size of trait differences among genotypes varies from one environment to another: the same set of genotypes may produce phenotypes that are similar or identical in some conditions but quite different in others. For example, the 10 Polygonum genotypes shown (which were originally drawn from a natural population) invested similarly in leaf tissue when they were grown at high and moderate light, but differed considerably at low light, because some genotypes increased leaf allocation more sharply in this more extreme environment than did others. In general, existing genetic variation may be exposed to natural selection only in certain conditions, and hidden from selection or ‘cryptic’ in environments where genotypic norms converge ([67,68]; evolutionary consequences discussed by [69–71]; and references therein). Consequently, a population's potential for selective evolution depends jointly on its genotypic diversity and on the environment(s) that occur (additional references in [17]).
Second, the rank order of phenotypes produced by a given set of genotypes can vary from one environment to another, if non-parallel norms of reaction happen to cross. In the Polygonum data, for example, the genotype with the highest leaf allocation at high light has the second lowest allocation at low light, the two highest-allocation genotypes in low light are the two lowest in both moderate and high light, and the lowest-allocating genotype at low light is the highest at moderate light (figure 2). If environments vary, such ‘crossing over’ of reaction norms can prevent consistent selective change (in this case, for example, selection for genotypes that allocate more to leaf tissue) and instead maintain multiple genotypes in a population [72,73]. Norm of reaction data thus reveal that both the amount and the particular patterns of genetic diversity are environmentally contingent and not intrinsic properties of a population's genotypes. In other words, the surprising answer to two basic questions regarding the potential selective evolution of a functional or fitness trait—how much genetic variation for the trait is present, and which genotype produces the highest trait value—must both be answered, ‘it depends on the environment(s)’. One practical consequence is that evolutionary studies require precise information about environmental as well as genetic variation within natural or experimental populations.
5. Transgenerational plasticity: developmental effects of previous environments
A fully contextualized picture of the genotype includes the recognition that an organism's development may be influenced by its parents' conditions as well as by its own immediate environment (e.g. [74]; see [75] and references therein for examples across 32 biological orders, from Archaea to Mammalia). Effects of parental environment on progeny development are generally considered as a transgenerational form of developmental plasticity, mediated by several distinct and often interacting mechanisms of inheritance (reviewed by [76,77]).
In both animals and plants, maternal individuals can directly transmit environmental influences on progeny development (for instance, due to resource stress or predation) to eggs or seeds, via changes in the amount and composition of cytoplasmic factors including nutrient reserves, hormones, defensive chemicals and small RNAs [78,79]. Molecular epigenetic effects such as DNA methylation and histone modifications can be transmitted to progeny by either paternal or maternal individuals [80,81]. These inherited epigenetic ‘marks’ alter gene expression via effects on DNA transcriptional activity and hence modify developmental outcomes ([82–84] and references therein). Although few data are available as yet, epigenetic variants may comprise a substantial portion of heritable fitness-related differences among individuals in natural populations (e.g. [85]). Once induced—often by specific environmental stresses—epigenetic modifications in plants and animals may be stably transmitted across several or many generations (e.g. [86,87]; additional references in [17,80,88,89]).
Like immediate plastic responses, transgenerational environmental effects on development may comprise either inevitable limits (such as reduced offspring mass due to maternal nutrient stress) or specifically adaptive adjustments [90]. Studies in diverse systems have shown that adaptive transgenerational plasticity may be surprisingly common, and may contribute substantially to individual fitness [75,77,91]. For instance, when Polygonum plants suffered drought stress, their offspring developed more extensive root systems and consequently survived better in dry soil, compared with progeny of isogenic parents that had instead been given ample moisture [92]. In anemonefish (Amphiprion melanopus), juveniles raised in water with a high concentration of carbon dioxide did not exhibit the predicted decrease in growth and survival if their parents had been exposed to the same elevated carbon dioxide conditions [93]. This developmental resilience was evidently mediated by parentally transmitted carbon dioxide-induced epigenetic changes to enzymes that affect acid–base metabolism [93]. Epigenetic mechanisms also mediate adaptive parent–environment effects in Mimulus (monkeyflower) plants: when parent individuals experienced simulated insect attack, their progeny produced leaves with altered gene expression patterns that resulted in an increased density of defensive hairs [94,95]. Interestingly, both maternal and paternal Mimulus plants evidently contribute to this progeny response, via distinct epigenetic mechanisms [96].
6. The multi-generational norm of reaction
Together, cytoplasmic and epigenetic factors provide for a non-genetic source of heritable phenotypic variation that may originate in parental, grandparental or possibly more remote generations [97,98]. These inborn environmental effects show clearly that distinguishing internal from external developmental information is deeply problematic [17,99]. They also add a further layer of complexity to the relationship between an organism's genotype and its realized functional and fitness traits. A given genotype will be to some extent differently expressed in alternative environments, resulting in a specific norm of reaction. Yet this response pattern itself will be influenced by previous conditions due to environmentally induced, inherited regulatory elements.
An example from a transgenerational plasticity experiment in Polygonum serves to illustrate this point (for an animal example, see [100]). Each of the three panels in figure 4 presents the norm of reaction for a single genotype, showing the different sizes of leaves produced by replicate seedlings of that genotype grown in shade versus full sun. However, not one but two norms are shown for each genotype: for seedlings of a given genotype, their plastic response to alternative light conditions was very different depending on whether their parent plant had grown in sun or in shade (figure 4; compare orange and green lines in each panel). Also note that the effect of parental shade on progeny responses was not consistent across the three genotypes (compare the difference between orange and green lines across panels). Rather, the transgenerational effect of shade versus sun was genotype-specific, presumably due to DNA sequence effects on the induction and transmission to offspring of particular cytoplasmic and/or epigenetic factors.
Figure 4.
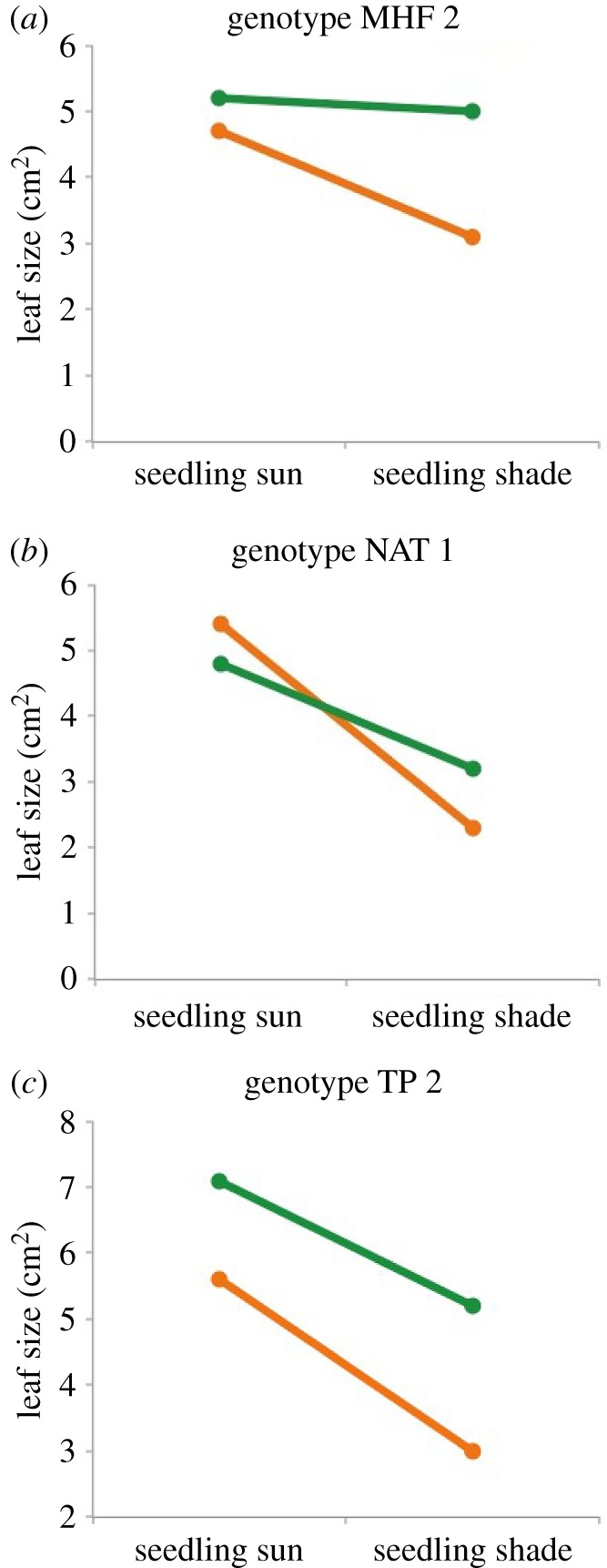
The effect of parental environment on progeny norms of reaction. Data plots show the size of individual leaves that were produced by seedlings growing in either full sun or simulated shade, for three Polyonum persicaria genotypes. Green = norm of reaction showing seedling developmental responses to the two environments when their parent plant had been grown in shade; orange = norm of reaction showing developmental responses of seedlings of the same genotype when their parent plant had been grown in full sun. Norms of reaction are based on mean leaf size for 10 replicate seedlings of each genotype and parental environment in each progeny growth treatment (B.H. Baker, L. Berg and S. E. Sultan 2015, unpublished data).
These data make clear that the norm of reaction is not a determinate property of the genotype, but is itself conditioned by inherited environmental information. Just as a genotype does not specify a single, determinate phenotype, neither does it give rise to one determinate plasticity pattern in response to a given environmental range. Moreover, just as genotypes differ in patterns of immediate environmental response, they also differ in transgenerational environmental effects on development [101,102], because DNA sequence influences the production of heritable regulatory molecules and the dynamics of epigenetic mechanisms (e.g. via differences in potential methylation sites [84,103]). Consequently, an organism's realized phenotype represents not only an active interaction between its evolved genotype and its environment, but a higher-order interaction between genotype, environment and a sequence of previous environments whose developmental effects may themselves interact—an ‘immensely complex web of interactions' or ‘entanglement’ between genotype and environment over several generations [104, p. 7].
As a result of this complexity, developmental plasticity cannot simply be accommodated into a deterministic model of adaptive evolution as a genotype's ‘extended phenotype’—that is, as a unique, genotype-specified response norm. Note that theoretical models that use this simplification have provided valuable insights regarding the environmental heterogeneity, accurate cues and other conditions expected to favour the evolution of plastic versus fixed reaction norms (e.g. [41,72,105–110]). The effects of inherited non-genetic factors on selective dynamics have also been investigated in a number of sophisticated models (e.g. [79,111–115]; reviewed in [116]). A further modelling challenge will be to fully integrate multi-generational influences on adaptive variation and selection. For example, a simulation model by Leimar & McNamara [117] showed that developmental systems can evolve so as to adaptively use genetic, environmental and prior-environmental developmental information. Models that address this complexity may help to frame key questions about the potential impact on selective trajectories of these variably persistent modes of developmental information. Resolving these questions will ultimately depend on empirical studies to illuminate the causal ‘entanglement’ that shapes adaptive variation.
7. Implications for research
The developmental programme view of the genotype has dictated an exclusive focus on heritable genetic information as the basis of phenotypes and hence of selective evolution. As a result of this simplified causal framework, evolutionary biologists have aimed to isolate the genetic component of phenotypic variation in order to track the genetic basis of adaptation, completing an internally sequestered causal circle. Even studies of plasticity and epigenetics have been circumscribed by this view: epigenetic changes are considered to be evolutionarily relevant only if they persist stably across hundreds of generations as ‘epimutations’ [85,118], while a predominant evolutionary question regarding plastically expressed phenotypes is whether they can become constitutive (genetically assimilated sensu [2,119]).
Reconceiving the genotype in the light of developmental plasticity calls for a shift in focus and in research approaches. An essential first step is to recognize the evolutionary relevance of short-term environmental and epigenetic factors. As a result of immediate and inherited effects on gene expression, these transient influences substantially shape the phenotypic variation expressed in each generation, and consequently selective trajectories [17,28,44,64,120–124]. Because genotypes respond differently to these influences, developmental response systems are themselves subject to selection, but as ‘entangled’ evolutionary entities; the impact of selection on genotypes is attenuated by highly complex environmental interactions.
To understand the causes and consequences of natural selection requires focusing directly on this mechanistic and evolutionary complexity. The empirical study of interacting influences on phenotypes (for instance, interactions between sequence variation and epigenetic dynamics) is just beginning [84]. As West-Eberhard has noted, ascribing phenotypic and fitness determination to the genotype has ‘deflected’ attention from the central biological question of how ‘condition-sensitive regulation is organized and evolves' [2, p. 17]; the time has come to take on this compelling question.
First, evolutionary biologists must devote serious attention to the environments of organisms, not only in terms of putative selective pressures, but with respect to both cues and direct influences on development. This requires identifying such factors and characterizing their patterns of spatial and temporal variation, including environmental auto-correlation across generations. Such studies are particularly demanding because developmental cues and influences may involve multiple, covarying aspects of natural environments [31,125].
A related point pertains to empirical research more broadly. Because environmental state affects the expression of phenotypes and of genetic diversity, experimental decisions regarding growth conditions can matter enormously to the results and to their utility for understanding natural systems. Ideally, the design of uniform growth environments, as well as the choice of alternative environmental states in norm of reaction experiments, should reflect conditions that are relevant to the organism in real populations; to the extent that this is not feasible, interpretation of experimental findings should include this point of reference.
Incorporating epigenetics into evolutionary biology will require intensive research activity to illuminate several key issues, including (i) epigenetic effects on functional and fitness variation in natural systems; (ii) induction and persistence dynamics in response to specific environmental cues or stresses; and (iii) genetic variation for induced epigenetic changes and their transmission. Data on these questions will inform experimental and theoretical investigations into the possible role of epigenetic systems as a distinct mode of adaptive variation, longer-term than immediate plasticity yet more labile than selective change ([6,83,126]; e.g. [127]). For technical reasons, initial work has focused on methylation, but it is equally important to investigate the various other epigenetic regulatory mechanisms that have recently come to light.
A developmental plasticity viewpoint can also inform approaches to studying human evolution. Just as genotype and environment cannot meaningfully be isolated from each other as causes of adaptive evolution, nature and culture can be seen as ‘entangled’ causes in the evolution of key human traits: like ecologically meaningful features of other organisms, the traits that characterize human beings take shape only in cultural, i.e. environmental, context [128].
Beyond a more inclusive framework for understanding adaptive evolution, a focus on developmental plasticity may offer new insights to related research areas. One pressing issue is biodiversity conservation. Human activities are increasingly altering natural habitats, from the spread of agrochemicals and other contaminants to the terrestrial and aquatic effects of global change. The near- and long-term prospects of organisms to adaptively withstand these changes will depend critically on existing developmental response norms, because novel conditions will affect the expression of functional phenotypes and of the genetic potential for further selective evolution [22,129–132]. To date, some of this information has proved encouraging. In studies with fish, for instance, parental exposure to both higher water temperatures and elevated carbon dioxide levels caused offspring to express phenotypes that were adaptive to these novel stresses. In these cases, transgenerational plasticity provided for a rapid and substantial increase in offspring tolerance to predicted future conditions [93,133].
In medicine, a shift is partly underway from seeking genetic determinants of disease as such to a less simplistic focus on the role of genetic factors in modulating the effects of physical, nutritional and social environments [134]. For instance, researchers studying the impact of a particular genetic variant on the incidence of depression explicitly described this as differential genetic modulation of stressful life experiences—that is, as an interaction between an individual's environment and his or her genotype [135]. This framework has shaped a productive and important programme of research, leading to the recent identification of epigenetic mechanisms that mediate this interaction [136]. Several of the most prevalent human diseases in modern societies are currently being investigated using a plasticity (i.e. genotype × environment interaction) framework in place of a simple ‘gene for’ hypothesis; these include several cancers (reviewed in [137]), diabetes and cardiovascular disease (reviewed by [138]) and Parkinson's disease (e.g. [139]). Such studies may lead to new therapeutic approaches focused on changing environmental factors to improve health outcomes for individuals or communities [140,141].
8. Conclusion: the evolving genotype
The phenotype emerges from multi-generation interactions between genotype and environment. This complicated picture is concordant with an explosion of recent discoveries regarding extra-genetic inherited factors that transmit environmental information across generations and the regulatory flexibility of gene expression, in general. These data make clear the need to replace a twentieth century understanding of the genotype as a self-contained, deterministic developmental ‘programme’ with a contemporary model that reflects the environmental context-dependency of phenotypic outcomes.
Along with a changed view of the genotype itself, the evolutionary corollaries of the developmental programme model must be revised. To begin with, the notion that an individual's genotype dictates its adaptedness to its environment must be amended. Phenotypes are produced actively through the process of individual development, as shaped by the genotype's interactions with regulatory information that is conditioned by past and present environments. Depending on the organism and trait in question, the environments encountered, and the particular genotype, the plasticity inherent in the developmental process may provide for considerable adaptive adjustment, or alternatively it may lead to inevitable fitness limits; both adaptive and inevitable aspects of plasticity shape phenotypic outcomes at the individual level [44]. As a result, genotypes do not specify trait or fitness differences among individuals. Rather, the differences that fuel natural selection reflect not genotypic diversity alone, but interacting developmental factors including the immediate environment and inherited cytoplasmic and epigenetic elements. Importantly, the developmental impact of these extra-genetic factors, as well as their precise patterns of perception, transduction and transmission, are genotype-specific rather than entirely independent of DNA sequence.
In view of these complex regulatory interactions, an organism's DNA cannot be considered to contain its developmental norm of reaction, much less the complete instructions to specify a particular phenotype. What, then, is the status of the genotype as an evolutionary unit? One way to approach this question is to distinguish between genetic information as evolutionary record and as evolutionary cause [17]. Unquestionably, genotypes evolve: they contain the biochemical material that resulted from a history of transmission and mutation over time, as conditioned by phylogenetic context, selection, random drift and gene flow. The genotype can thus be seen as the product of evolution, as it comprises a uniquely stable repository of these historical events across time. Yet the genotypes in a population do not in themselves determine the adaptive diversity that shapes selective change, because they contain only partial developmental information, and hence only partial information regarding fitness variation. The causes of adaptive evolution include the genotype-specific dynamics of immediate and transgenerational developmental response in the context of environmental distributions. Studying these causes requires changing experimental design and approach so as to directly interrogate these complex interacting sources of variation. This research programme offers a revised and renewed understanding of the genotype that will allow development to be fully integrated into the evolutionary process.
Acknowledgements
I thank the organizers of the joint Royal Society/British Academy meeting on New Trends in Evolutionary Biology for the invitation to participate and to contribute this paper. I also thank four anonymous referees for thoughtful and constructive comments on the manuscript.
Endnote
In such ‘common garden’ studies, inherited effects of previous environments are generally confounded with genotypic differences; see §6.
Data accessibility
This article has no additional data.
Competing interests
I declare I have no competing interests.
Funding
Funding for the data presented in this paper was provided by the US National Science Foundation and Wesleyan University.
References
- 1.Keller EF. 2000. The century of the gene. Cambridge, MA: Harvard University Press. [Google Scholar]
- 2.West-Eberhard MJ. 2003. Developmental plasticity and evolution. New York, NY: Oxford University Press. [Google Scholar]
- 3.Griffiths PE. 2006. Philip Kitcher, genetic determinism, and the informational gene. In Genes in development: re-reading the molecular paradigm (eds Neumann-Held EM, Rehmann-Sutter C), pp. 175–198. Durham, NC: Duke University Press. [Google Scholar]
- 4.Sarkar S. 2006. From genes as determinants to DNA as resource. In Genes in development: re-reading the molecular paradigm (eds Neumann-Held EM, Rehmann-Sutter C), pp. 77–95. Durham, NC: Duke University Press. [Google Scholar]
- 5.Newman SA, Müller GB. 2006. Genes and form: inherency in the evolution of developmental mechanisms. In Genes in development: re-reading the molecular paradigm (eds Neumann-Held EM, Rehmann-Sutter C), pp. 38–73. Durham, NC: Duke University Press. [Google Scholar]
- 6.Noble D. 2015. Evolution beyond neo-Darwinism: a new conceptual framework. J. Exp. Biol. 218, 7–13. ( 10.1242/jeb.106310) [DOI] [PubMed] [Google Scholar]
- 7.Wray GA, et al. 2014. Does evolutionary theory need a rethink? No, all is well. Nature 514, 161–164. ( 10.1038/514161a) [DOI] [PubMed] [Google Scholar]
- 8.Laland K, et al. 2014. Does evolutionary theory need a rethink? Yes, urgently. Nature 514, 161–164. ( 10.1038/514161a) [DOI] [PubMed] [Google Scholar]
- 9.Lynch V, Wagner GP. 2008. Resurrecting the role of transcription factor change in developmental evolution. Evolution 62, 2131–2154. ( 10.1111/j.1558-5646.2008.00440.x) [DOI] [PubMed] [Google Scholar]
- 10.Byrnes WM, Eckberg WR. 2006. Ernest Everett Just (1883–1941)—an early ecological developmental biologist. Dev. Biol. 296, 1–11. ( 10.1016/j.ydbio.2006.04.445) [DOI] [PubMed] [Google Scholar]
- 11.Gottlieb G. 2004. Normally occurring environmental and behavioral influences on gene activity: from central dogma to probabilistic epigenesis. In Nature and nurture: the complex interplay of genetic and environmental influences on human behavior and development (eds Coll CG, Bearer EL, Lerner RM), pp. 85–106. London, UK: Lawrence Erlbaum Associates. [DOI] [PubMed] [Google Scholar]
- 12.Amundsen R. 2001. Adaptation and development: on the lack of common ground. In Adaptationism and optimality (eds Orzack S, Sober E), pp. 303–334. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 13.Neumann-Held EM. 2006. Genes-causes-codes: deciphering DNA's ontological privilege. In Genes in development: re-reading the molecular paradigm (eds Neumann-Held EM, Rehmann-Sutter C), pp. 238–271. Durham, NC: Duke University Press. [Google Scholar]
- 14.Levins R, Lewontin R. 1985. The dialectical biologist. Cambridge, MA: Harvard University Press. [Google Scholar]
- 15.Carroll SB, Grenier JK, Weatherbee SD. 2005. From DNA to diversity: molecular genetics and the evolution of animal design, 2nd edn Malden, MA: Blackwell Publishing. [Google Scholar]
- 16.Lemos B. et al 2008. Evolution of genomic expression. In Evolutionary genomics and proteomics (eds Pagel M, Pomiankowski A), pp. 81–118. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 17.Sultan SE. 2015. Organism and environment: ecological development, niche construction and adaptation. London, UK: Oxford University Press. [Google Scholar]
- 18.Gilbert SF. 2012. Ecological developmental biology: environmental signals for normal animal development. Evol. Dev. 14, 20–28. ( 10.1111/j.1525-142X.2011.00519.x) [DOI] [PubMed] [Google Scholar]
- 19.Sultan SE. 2009. Evolutionary implications of individual plasticity. In Transformations of Lamarckism (eds Jablonka E, Gissis S), pp. 193–203. Vienna Series in Theoretical Biology Cambridge, MA: MIT Press. [Google Scholar]
- 20.Gilbert SF. 2001. Ecological developmental biology: developmental biology meets the real world. Dev. Biol. 233, 1–12. ( 10.1006/dbio.2001.0210) [DOI] [PubMed] [Google Scholar]
- 21.Gilbert SF, Bolker JD. 2003. Ecological developmental biology: preface to the symposium. Evol. Dev. 5, 3–8. ( 10.1046/j.1525-142X.2003.03002.x) [DOI] [PubMed] [Google Scholar]
- 22.Sultan SE. 2007. Development in context: the timely emergence of eco-devo. Trends Ecol. Evol. 22, 575–582. ( 10.1016/j.tree.2007.06.014) [DOI] [PubMed] [Google Scholar]
- 23.Sultan SE. 2010. Plant developmental responses to the environment: eco-devo insights. Curr. Opin. Plant Biol. 13, 96–101. ( 10.1016/j.pbi.2009.09.021) [DOI] [PubMed] [Google Scholar]
- 24.Gilbert SF, Epel D. 2009. Ecological developmental biology: integrating epigenetics, medicine, and evolution. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 25.Gilbert SF, Epel D. 2015. Ecological developmental biology: the environmental regulation of development, health, and evolution. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 26.Woltereck R. 1909. Weitere experimentelle Untersuchungen über Arrveranderung, speziell über das Wesen quntitativer Artuntershiede bei Daphniden. Verh. Dtsch. Zool. Ges. 1909, 110–172. [Google Scholar]
- 27.Gupta AP, Lewontin RC. 1982. A study of reaction norms in natural populations of Drosophila pseudoobscura. Evolution 36, 934–948. ( 10.1111/j.1558-5646.1982.tb05464.x) [DOI] [PubMed] [Google Scholar]
- 28.Stearns SC. 1989. The evolutionary significance of phenotypic plasticity. Bioscience 39, 436–445. ( 10.2307/1311135) [DOI] [Google Scholar]
- 29.Sultan SE, Stearns SC. 2005. Environmentally contingent variation: phenotypic plasticity and norms of reaction. In Variation: a hierarchical examination of a central concept in biology (eds Hall B, Hallgrimsson B), pp. 303–332. New York, NY: Elsevier Academic Press. [Google Scholar]
- 30.Sarkar S. 2004. From the reaktionsnorm to the evolution of adaptive plasticity: a historical sketch, 1909–1999. In Phenotypic plasticity: functional and conceptual approaches (eds DeWitt TJ, Scheiner SM), pp. 10–30. Oxford, UK: Oxford University Press. [Google Scholar]
- 31.Miner BG, Sultan SE, Morgan SG, Padilla DK, Relyea RA. 2005. Ecological consequences of phenotypic plasticity. Trends Ecol. Evol. 20, 685–692. ( 10.1016/j.tree.2005.08.002) [DOI] [PubMed] [Google Scholar]
- 32.Voelkl B, Würbel H. 2016. Reproducibility crisis: are we ignoring reaction norms? Trends Pharmacol. Sci. 37, 509–510. ( 10.1016/j.tips.2016.05.003) [DOI] [PubMed] [Google Scholar]
- 33.Dudley SA, Schmitt J. 1996. Testing the adaptive plasticity hypothesis: density-dependent selection on manipulated stem length in Impatiens capensis. Am. Nat. 147, 445–465. ( 10.1086/285860) [DOI] [Google Scholar]
- 34.Schmitt J, Stinchcombe JR, Heschel MS, Huber H. 2003. The adaptive evolution of plasticity: phytochrome-mediated shade avoidance responses. Integr. Compar. Biol. 43, 459–469. ( 10.1093/icb/43.3.459) [DOI] [PubMed] [Google Scholar]
- 35.Sassi PL, Borghi CE, Bozinovic F. 2007. Spatial and seasonal plasticity in digestive morphology of cavies (Microcavia australis) inhabiting habitats with different plant qualities. J. Mammal. 88, 165–172. ( 10.1644/06-MAMM-A-046R1.1) [DOI] [Google Scholar]
- 36.Chapman LJ, Albert J, Galis F. 2008. Developmental plasticity, genetic differentiation, and hypoxia-induced trade-offs in an African cichlid fish. Open Evol. J. 2, 75–88. ( 10.2174/1874404400802010075) [DOI] [Google Scholar]
- 37.Schmalhausen II. 1949. Factors of evolution. (Transl. by I Dordick.) Philadelphia, PA: Blakiston Press. [Google Scholar]
- 38.Bradshaw AD. 1965. Evolutionary significance of phenotypic plasticity in plants. Adv. Genet. 13, 115–155. ( 10.1016/S0065-2660(08)60048-6) [DOI] [Google Scholar]
- 39.Lively CM. 1986. Predator-induced shell dimorphism in the acorn barnacle Chthamalus anisopoma. Evolution 40, 232–242. ( 10.1111/j.1558-5646.1986.tb00466.x) [DOI] [PubMed] [Google Scholar]
- 40.Schlichting CD. 1986. The evolution of phenotypic plasticity in plants. Annu. Rev. Ecol. Syst. 17, 667–693. ( 10.1146/annurev.es.17.110186.003315) [DOI] [Google Scholar]
- 41.Scheiner SM. 1993. Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Syst. 24, 35–68. ( 10.1146/annurev.es.24.110193.000343) [DOI] [Google Scholar]
- 42.Sultan SE. 1995. Phenotypic plasticity and plant adaptation. Acta Bot. Neerlandica 44, 1–21. ( 10.1111/j.1438-8677.1995.tb00793.x) [DOI] [Google Scholar]
- 43.Sultan SE. 2000. Phenotypic plasticity for plant development, function and life-history. Trends Plant Sci. 5, 537–542. ( 10.1016/S1360-1385(00)01797-0) [DOI] [PubMed] [Google Scholar]
- 44.Sultan SE. 2003. The promise of ecological developmental biology. J. Exp. Zool. B 296B, 1–7. ( 10.1002/jez.b.10) [DOI] [PubMed] [Google Scholar]
- 45.Pigliucci M. 2001. Phenotypic plasticity: beyond nature and nurture. Baltimore, MD: Johns Hopkins University Press. [Google Scholar]
- 46.DeWitt TJ, Scheiner SM (eds). 2004. Phenotypic plasticity: functional and conceptual approaches. Oxford, UK: Oxford University Press. [Google Scholar]
- 47.Sultan SE, Bazzaz FA. 1993. Phenotypic plasticity in Polygonum persicaria. I. Diversity and uniformity in genotypic norms of reaction to light. Evolution 47, 1009–1031. ( 10.1111/j.1558-5646.1993.tb02132.x) [DOI] [PubMed] [Google Scholar]
- 48.Griffith TM, Sultan SE. 2005. Shade tolerance plasticity in response to neutral vs green shade cues in Polygonum species of contrasting ecological breadth. New Phytol. 166, 141–148. ( 10.1111/j.1469-8137.2004.01277.x) [DOI] [PubMed] [Google Scholar]
- 49.Sultan SE. 2003. Phenotypic plasticity in plants: a case study in ecological development. Evol. Dev. 5, 25–33. ( 10.1046/j.1525-142X.2003.03005.x) [DOI] [PubMed] [Google Scholar]
- 50.Sultan SE, Bazzaz FA. 1993. Phenotypic plasticity in Polygonum persicaria II. Norms of reaction to soil moisture and the maintenance of genetic diversity. Evolution 47, 1032–1049. ( 10.1111/j.1558-5646.1993.tb02133.x) [DOI] [PubMed] [Google Scholar]
- 51.Sultan SE, Bazzaz FA. 1993. The evolution of ecological breadth for nutrient environment. Evolution 47, 1050–1071. ( 10.1111/j.1558-5646.1993.tb02134.x) [DOI] [PubMed] [Google Scholar]
- 52.Bell DL, Sultan SE. 1999. Dynamic phenotypic plasticity for root growth in Polygonum: a comparative study. Am. J. Bot. 86, 807–819. ( 10.2307/2656702) [DOI] [PubMed] [Google Scholar]
- 53.Heschel S, Sultan SE, Sloan D, Glover S. 2004. Population differentiation and plastic responses to drought stress in the generalist annual Polygonum persicaria. Int. J. Plant Sci. 165, 817–824. ( 10.1086/421477) [DOI] [Google Scholar]
- 54.Laland KN, Odling-Smee J, Gilbert SF. 2008. Evo-devo and niche construction: building bridges. J. Exp. Zool. 310, 549–566. ( 10.1002/jez.b.21232) [DOI] [PubMed] [Google Scholar]
- 55.Odling-Smee J, Laland KN, Feldman MW. 2003. Niche construction: the neglected process in evolution. Princeton, NJ: Princeton University Press. [Google Scholar]
- 56.Odling-Smee J, Erwin DH, Palkovacs EP, Feldman MW, Laland KN. 2013. Niche construction theory: a practical guide for ecologists. Q. Rev. Biol. 88, 3–28. ( 10.1086/669266) [DOI] [PubMed] [Google Scholar]
- 57.Laland K, Matthews B, Feldman MW. 2016. An introduction to niche construction theory. Evol. Ecol. 30, 191–202. ( 10.1007/s10682-016-9821-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wright S. 1931. Evolution in Mendelian populations. Genetics 16, 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chevin L-M, Lande R, Mace GM. 2010. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. Pub. Libr. Sci. Biol. 8, e1000357 ( 10.1371/journal.pbio.1000357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Draghi JA, Whitlock MC. 2012. Phenotypic plasticity facilitates mutational variance, genetic variance, and evolvability along the major axis of environmental variation. Evolution 66, 2891–2902. ( 10.1111/j.1558-5646.2012.01649.x) [DOI] [PubMed] [Google Scholar]
- 61.Gomez-Mestre I, Jovani R. 2013. A heuristic model on the role of plasticity in adaptive evolution: plasticity increases adaptation, population viability and genetic variation. Proc. R. Soc. B 280, 20131869 ( 10.1098/rspb.2013.1869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moczek AP, Sultan SE, Foster S, Ledón-Rettig C, Dworkin I, Nijhout HF, Abouheif E, Pfennig DW. 2011. The role of developmental plasticity in evolutionary innovation. Proc. R. Soc. B 278, 2705–2713. ( 10.1098/rspb.2011.0971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haldane JBS. 1946. The interaction of nature and nurture. Ann. Eugenics 13, 197–205. ( 10.1111/j.1469-1809.1946.tb02358.x) [DOI] [PubMed] [Google Scholar]
- 64.Barton NH, Turelli M. 1989. Evolutionary quantitative genetics: how little do we know? Annu. Rev. Genet. 23, 337–370. ( 10.1146/annurev.ge.23.120189.002005) [DOI] [PubMed] [Google Scholar]
- 65.Kruuk LEB, Slate J, Wilson AJ. 2008. New answers for old questions: the evolutionary quantitative genetics of wild animal populations. Annu. Rev. Ecol. Evol. Syst. 39, 525–548. ( 10.1146/annurev.ecolsys.39.110707.173542) [DOI] [Google Scholar]
- 66.Des Marais DL, Hernandez KM, Juenger TE. 2013. Genotype-by-environment interaction and plasticity: exploring genomic responses of plants to the abiotic environment. Annu. Rev. Ecol. Evol. Syst. 44, 5–29. ( 10.1146/annurev-ecolsys-110512-135806) [DOI] [Google Scholar]
- 67.Wilson AJ, Pemberton JM, Pilkington JG, Coltman DW, Mifsud DV, Clutton-Brock TH, Kruuk LEB. 2006. Environmental coupling of selection and heritability limits evolution. Pub. Libr. Sci. Biol. 4, e216 ( 10.1371/journal.pbio.0040216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Snell-Rood EC, Van Dyken JD, Cruickshank T, Wade MJ, Moczek AP. 2010. Toward a population genetic framework of developmental evolution: the costs, limits, and consequences of phenotypic plasticity. Bioessays 32, 71–81. ( 10.1002/bies.200900132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Dyken JD, Wade MJ. 2010. The genetic signature of conditional expression. Genetics 184, 557–570. ( 10.1534/genetics.109.110163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ledón-Rettig CC, Pfennig DW, Chunco AJ, Dworkin I. 2014. Cryptic genetic variation in natural populations: a predictive framework. Integr. Comp. Biol. 54, 783–793. ( 10.1093/icb/icu077) [DOI] [PubMed] [Google Scholar]
- 71.Paaby AB, Rockman MV. 2014. Cryptic genetic variation: evolution's hidden substrate. Nat. Rev. Genet. 15, 247–258. ( 10.1038/nrg3688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Via S, Lande R. 1985. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution 39, 505–522. ( 10.1111/j.1558-5646.1985.tb00391.x) [DOI] [PubMed] [Google Scholar]
- 73.Gillespie JH, Turelli M. 1989. Genotype-environment interactions and the maintenance of polygenic variation. Genetics 121, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Falconer DS. 1989. Introduction to quantitative genetics, 3rd edn London, UK: Longman. [Google Scholar]
- 75.Salinas S, Brown SC, Mangel M, Munch SB. 2013. Non-genetic inheritance and changing environments. Non-Genetic Inheritance 1, 38–50. ( 10.2478/ngi-2013-0005) [DOI] [Google Scholar]
- 76.Badyaev A, Uller T. 2009. Parental effects in ecology and evolution: mechanisms, processes and implications. Phil. Trans. R. Soc. B 364, 1169–1177. ( 10.1098/rstb.2008.0302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herman JJ, Sultan SE. 2011. Adaptive transgenerational plasticity: case studies, mechanisms, and implications for natural populations. Front. Plant Genet. Genom. 2, 102 ( 10.3389/fpls.2011.00102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roach DA, Wulff RD. 1987. Maternal effects in plants. Annu. Rev. Ecol. Syst. 18, 209–235. ( 10.1146/annurev.es.18.110187.001233) [DOI] [Google Scholar]
- 79.Kirkpatrick M, Lande R. 1989. The evolution of maternal characters. Evolution 43, 485–503. ( 10.1111/j.1558-5646.1989.tb04247.x) [DOI] [PubMed] [Google Scholar]
- 80.Jablonka E, Raz G. 2009. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84, 131–176. ( 10.1086/598822) [DOI] [PubMed] [Google Scholar]
- 81.Soubry A, Hoyo Jirtle CRL, Murphy SK. 2014. A paternal environmental legacy: evidence for epigenetic inheritance through the male germ line. Bioessays 36, 359–371. ( 10.1002/bies.201300113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duncan EJ, Gluckman PD, Dearden PK. 2014. Epigenetics, plasticity, and evolution: how do we link epigenetic change to phenotype? J. Exp. Zool. B 322, 208–220. ( 10.1002/jez.b.22571) [DOI] [PubMed] [Google Scholar]
- 83.Gugger PF, Fitz Gibbon S, Pellegrini M, Sork VL. 2016. Species-wide patterns of DNA methylation variation in Quercus lobata and its association with climate gradients. Mol. Ecol. 25, 1665–1680. ( 10.1111/mec.13563) [DOI] [PubMed] [Google Scholar]
- 84.Kawakatsu T, et al. 2016. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166, 492–505. ( 10.1016/j.cell.2016.06.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cortijo S, et al. 2014. Mapping the epigenetic basis of complex traits. Science 343, 1145–1148. ( 10.1126/science.1248127) [DOI] [PubMed] [Google Scholar]
- 86.Remy J-J. 2010. Stable inheritance of an acquired behavior in Caenorhabditis elegans. Curr. Biol. 20, R877–R878. ( 10.1016/j.cub.2010.08.013) [DOI] [PubMed] [Google Scholar]
- 87.Schmitz RJ, Schultz MD, Lewseyet MG, O'Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. 2011. Transgenerational epigenetic instability is a source of novel methylation variants. Science 334, 369–373. ( 10.1126/science.1212959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jaenisch R, Bird A. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254. ( 10.1038/ng1089) [DOI] [PubMed] [Google Scholar]
- 89.Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM. 2014. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 17, 667–669. ( 10.1038/nn.3695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uller T. 2008. Developmental plasticity and the evolution of parental effects. Trends Ecol. Evol. 23, 432–438. ( 10.1016/j.tree.2008.04.005) [DOI] [PubMed] [Google Scholar]
- 91.Mousseau TA, Fox C. 1998. Maternal effects as adaptations. New York, NY: Oxford University Press. [Google Scholar]
- 92.Herman JJ, Sultan SE, Horgan-Kobelski T, Riggs CE. 2012. Adaptive transgenerational plasticity in an annual plant: grandparental and parental drought stress enhance performance of seedlings in dry soil. Integr. Compar. Biol. 52, 1–12. ( 10.1093/icb/ics041) [DOI] [PubMed] [Google Scholar]
- 93.Miller GM, Watson S-A, Donelson JM, McCormick MI, Munday PL. 2012. Parental environment mediates impacts of increased carbon dioxide on a coral reef fish. Nat. Clim. Change 2, 858–861. ( 10.1038/nclimate1599) [DOI] [Google Scholar]
- 94.Scoville AG, Barnett LL, Bodbyl-Roels S, Kelly JK, Hileman LC. 2011. Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol. 191, 251–263. ( 10.1111/j.1469-8137.2011.03656.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Colicchio JM, Monnahan PJ, Kelly JK, Hileman LC. 2015. Gene expression plasticity resulting from parental leaf damage in Mimulus guttatus. New Phytol. 205, 894–906. ( 10.1111/nph.13081) [DOI] [PubMed] [Google Scholar]
- 96.Akkerman KC, Sattarin A, Kelly JK, Scoville AG. 2016. Transgenerational plasticity is sex-dependent and persistent in yellow monkeyflower (Mimulus guttatus). Environ. Epigenet. 2, 1–8. ( 10.1093/eep/dvw003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bonduriansky R. 2012. Rethinking heredity, again. Trends Ecol. Evol. 27, 330–336. ( 10.1016/j.tree.2012.02.003) [DOI] [PubMed] [Google Scholar]
- 98.English S, Pen I, Shea N, Uller T. 2015. The information value of non-genetic inheritance in plants and animals. PLoS ONE 10, e0116996 ( 10.1371/journal.pone.0116996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bateson P, Gluckman P. 2011. Plasticity, robustness, development and evolution. New York, NY: Cambridge University Press. [Google Scholar]
- 100.Plaistow SJ, Shirley C, Collin H, Cornel SJ, Harney ED. 2015. Offspring provisioning explains clone-specific maternal age effects on life history and life span in the water flea, Daphnia pulex. Am. Nat. 186, 376–389. ( 10.1086/682277) [DOI] [PubMed] [Google Scholar]
- 101.Vu WT, Chang PL, Moriuchi KS, Friesen ML. 2015. Genetic variation of transgenerational plasticity of offspring germination in response to salinity stress and the seed transcriptome of Medicago truncatula. BMC Evol. Biol. 15, 1 ( 10.1186/s12862-014-0274-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Herman JJ, Sultan SE. 2016. DNA methylation mediates genetic variation for adaptive transgenerational plasticity. Proc. R. Soc. B 283, 20160988 ( 10.1098/rspb.2016.0988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meaney MJ, Ferguson-Smith AC. 2010. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat. Neurosci. 13, 1313–1318. ( 10.1038/nn1110-1313) [DOI] [PubMed] [Google Scholar]
- 104.Keller EF. 2010. The mirage of a space between nature and nurture. Durham, NC: Duke University Press. [Google Scholar]
- 105.Scheiner SM. 2013. The genetics of phenotypic plasticity. XII. Temporal and spatial heterogeneity. Ecol. Evol. 3, 4596–4609. ( 10.1002/ece3.792) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moran NA. 1992. The evolutionary maintenance of alternative phenotypes. Am. Nat. 139, 971–989. ( 10.1086/285369) [DOI] [Google Scholar]
- 107.Tufto J. 2000. The evolution of plasticity and nonplastic spatial and temporal adaptations in the presence of imperfect environmental cues. Am. Nat. 156, 121–130. ( 10.1086/303381) [DOI] [PubMed] [Google Scholar]
- 108.Sultan SE, Spencer HG. 2002. Metapopulation structure favors plasticity over local adaptation. Am. Nat. 160, 271–283. ( 10.1086/341015) [DOI] [PubMed] [Google Scholar]
- 109.Berrigan D, Scheiner SM. 2004. Modeling the evolution of phenotypic plasticity. In Phenotypic plasticity: functional and conceptual approaches (eds DeWitt TM, Scheiner SM), pp. 82–97. New York, NY: Oxford University Press. [Google Scholar]
- 110.Scheiner SM, Holt RD. 2012. The genetics of phenotypic plasticity. X. Variation versus uncertainty. Ecol. Evol. 2, 751–767. ( 10.1002/ece3.217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Danchin É, Charmantier A, Champagne FA, Mesoudi A, Pujol B, Blanchet S. 2011. Beyond DNA: integrating inclusive inheritance into an extended theory of evolution. Nat. Rev. Genet. 12, 475–486. ( 10.1038/nrg3028) [DOI] [PubMed] [Google Scholar]
- 112.Day T, Bonduriansky R. 2011. A unified approach to the evolutionary consequences of genetic and nongenetic inheritance. Am. Nat. 178, E18–E36. ( 10.1086/660911) [DOI] [PubMed] [Google Scholar]
- 113.Bonduriansky R, Crean AJ, Day T. 2012. The implications of non-genetic inheritance for evolution in changing environments. Evol. Appl. 5, 192–201. ( 10.1111/j.1752-4571.2011.00213.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Danchin E. 2013. Avatars of information: towards an inclusive evolutionary synthesis. Trends Ecol. Evol. 20, 1–8. ( 10.1016/j.tree.2013.02.010) [DOI] [PubMed] [Google Scholar]
- 115.Geoghegan J, Spencer HG. 2013. The adaptive invasion of epialleles in a heterogeneous environment. Theor. Popul. Biol. 88, 1–8. ( 10.1016/j.tpb.2013.05.001) [DOI] [PubMed] [Google Scholar]
- 116.Van Dooren TJM, Hoyle RB, Plaistow SJ. 2016. Maternal effects. Encycl. Evol. Biol. 2, 444–452. [Google Scholar]
- 117.Leimar O, McNamara JM. 2015. The evolution of transgenerational integration of information in heterogeneous environments. Am. Nat. 185, E55–E69. ( 10.1086/679575) [DOI] [PubMed] [Google Scholar]
- 118.Haig D. 2007. Weismann rules! Ok? Epigenetics and the Lamarckian temptation. Biol. Philos. 22, 415–428. ( 10.1007/s10539-006-9033-y) [DOI] [Google Scholar]
- 119.Ehrenreich IM, Pfennig DW. 2016. Genetic assimilation: a review of its potential proximate causes and evolutionary consequences. Ann. Bot. 117, 769–779. ( 10.1093/aob/mcv130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wade MJ, Kalisz S. 1990. The causes of natural selection. Evolution 44, 1947–1955. ( 10.1111/j.1558-5646.1990.tb04301.x) [DOI] [PubMed] [Google Scholar]
- 121.Sultan SE. 1992. Phenotypic plasticity and the neo-Darwinian legacy. Evol. Trends Plants 6, 61–71. [Google Scholar]
- 122.Nager RG, Keller LF, Van Noordwijk AJ. 2000. Understanding natural selection on traits that are influenced by environmental conditions. In Adaptive genetic variation in the wild (eds Mousseau T, Sinervo B, Endler JA), pp. 95–115. New York, NY: Oxford University Press. [Google Scholar]
- 123.Kingsolver JG, Diamond SE, Siepielski AM, Carlson SM. 2012. Synthetic analyses of phenotypic selection in natural populations: lessons, limitations and future directions. Evol. Ecol. 26, 1101–1118. ( 10.1007/s10682-012-9563-5) [DOI] [Google Scholar]
- 124.Anderson JT, Wagner MR, Rushworth CA, Prasad KVSK, Mitchell-Olds T. 2014. The evolution of quantitative traits in complex environments. Heredity 112, 4–12. ( 10.1038/hdy.2013.33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chevin L-M, Lande R. 2015. Evolution of environmental cues for phenotypic plasticity. Evolution 69, 2767–2775. ( 10.1111/evo.12755) [DOI] [PubMed] [Google Scholar]
- 126.Herman JJ, Spencer HG, Donohue K, Sultan SE. 2014. How stable ‘should’ epigenetic modifications be? Insights from adaptive plasticity and bet hedging. Evolution 68, 632–643. ( 10.1111/evo.12324) [DOI] [PubMed] [Google Scholar]
- 127.Houri-Ze'evi L, et al. 2016. A tunable mechanism determines the duration of the transgenerational small RNA inheritance in C. elegans. Cell 165, 88–99. ( 10.1016/j.cell.2016.02.057) [DOI] [PubMed] [Google Scholar]
- 128.Laland KN, O'Brien MJ. 2011. Cultural niche construction: an introduction. Biol. Theory 6, 191–202. ( 10.1007/s13752-012-0026-6) [DOI] [Google Scholar]
- 129.Nussey DH, Postma E, Gienapp P, Visser ME. 2005. Selection on heritable phenotypic plasticity in a wild bird population. Science 310, 304–306. ( 10.1126/science.1117004) [DOI] [PubMed] [Google Scholar]
- 130.Ghalambor CK, McKay JS, Carroll SP, Reznick DN. 2007. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct. Ecol. 21, 394–407. ( 10.1111/j.1365-2435.2007.01283.x) [DOI] [Google Scholar]
- 131.Visser ME. 2008. Keeping up with a warming world; assessing the rate of adaptation to climate change. Proc. R. Soc. B 275, 649–659. ( 10.1098/rspb.2007.0997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Carroll SP, et al. 2014. Applying evolutionary biology to address global challenges. Science 346, 1245993 ( 10.1126/science.1245993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Salinas S, Munch SB. 2012. Thermal legacies: transgenerational effects of temperature on growth in a vertebrate. Ecol. Lett. 15, 159–163. ( 10.1111/j.1461-0248.2011.01721.x) [DOI] [PubMed] [Google Scholar]
- 134.Gluckman PD, Hanson M. 2005. The fetal matrix: evolution, development and disease. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 135.Caspi A, et al. 2003. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301, 386–389. ( 10.1126/science.1083968) [DOI] [PubMed] [Google Scholar]
- 136.Wankerl M, Miller R, Kirschbaum C, Hennig J, Stalder T, Alexander N. 2014. Effects of genetic and early environmental risk factors for depression on serotonin transporter expression and methylation profiles. Transl. Psychiatry 4, e402 ( 10.1038/tp.2014.37) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ghazarian AA, Simonds NI, Bennett K, Pimentel CB, Ellison GL, Gillanders EM, Schully SD, Mechanic LE. 2013. Identifying opportunities for future research in genes and environment in cancer. Cancer Epidemiol. Biomarkers Prevention 22, 501–507. ( 10.1158/1055-9965.EPI-13-0156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee YC, Lai CQ, Ordovas JM, Parnell LD. 2011. A database of gene-environment interactions pertaining to blood lipid traits, cardiovascular disease and type 2 diabetes. J. Data Mining Genomics Proteomics 2, 106 ( 10.4172/2153-0602.1000106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ritz BR Paul KC, Bronstein JM. 2016. Of pesticides and men: a California story of genes and environment in Parkinson's disease. Curr. Environ. Health Rep. 3, 40–52. ( 10.1007/s40572-016-0083-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gluckman PD, et al. 2009. Towards a new developmental synthesis: developmental plasticity and human disease. Lancet 373, 1654–1657. ( 10.1016/S0140-6736(09)60234-8) [DOI] [PubMed] [Google Scholar]
- 141.Lock M. 2015. Comprehending the body in the era of the epigenome. Curr. Anthropol. 56, 151–177. ( 10.1086/680350) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.



