The study findings provide evidence that administration of butyrate in a dose-dependent manner can improve the weight gain of infected mice, enhance clearance of the infection, reduce inflammation through altered cytokine expression, and enhance tissue repair and mucus secretion. Moreover, butyrate treatment also affected the abundance of bacterial populations in both noninflamed and inflamed intestines. Notably, this investigation provides foundational information that can be used to determine the effects of prebiotics and other functional foods on the production of butyrate by enteric bacteria and their impact on intestinal health and host well-being.
KEYWORDS: butyrate, Citrobacter rodentium, inflammation, intestine, mice, microbiota
ABSTRACT
Butyrate is a short-chain fatty acid by-product of the microbial fermentation of dietary fermentable materials in the large intestine; it is the main energy source for enterocyte regeneration, modulates the enteric microbial community, and contributes to increasing host health via mechanisms that are relatively poorly defined. Limited research has examined the therapeutic potential of butyrate using models of enteric inflammation incited by pathogenic organisms. We used Citrobacter rodentium to incite acute Th1/Th17 inflammation to ascertain the impact of butyrate on the host-microbiota relationship. Rectal administration of 140 mM butyrate to mice increased fecal concentrations of butyrate and increased food consumption and weight gain in mice infected with C. rodentium. Histological scores of colonic inflammation were lower in infected mice administered 140 mM butyrate. Expression of Il10, Tgfβ, and Muc2 was elevated in noninfected mice administered butyrate in comparison to mice not administered butyrate. Infected mice administered butyrate displayed elevated expression of genes necessary for pathogen clearance (i.e., Il17A and Il1β) and of genes involved in epithelial barrier repair and restoration (i.e., Relmβ, Tff3, and Myd88). Butyrate supplemented to inflamed colons increased the abundances of Proteobacteria and Lachnospiraceae and reduced the abundance of Clostridiaceae species. Mice with enteritis that were administered butyrate also exhibited an increased abundance of mucus-associated bacteria. In summary, rectal administration of butyrate increased feed consumption and weight gain, ameliorated C. rodentium-induced cell injury through enhanced expression of immune regulation and tissue repair mechanisms, and increased the abundance of butyrate-producing bacteria in mice with enteritis.
IMPORTANCE The study findings provide evidence that administration of butyrate in a dose-dependent manner can increase weight gain in infected mice, enhance clearance of the infection, reduce inflammation through altered cytokine expression, and enhance tissue repair and mucus secretion. Moreover, butyrate treatment also affected the abundance of bacterial populations in both noninflamed and inflamed intestines. Notably, this investigation provides foundational information that can be used to determine the effects of prebiotics and other functional foods on the production of butyrate by enteric bacteria and their impact on intestinal health and host well-being.
INTRODUCTION
Butyrate is a short-chain fatty acid (SCFA) that is produced by the fermentation of dietary fiber in the large intestine and is purported to confer a vareity of health benefits. The role of butyrate is to provide energy to colonocytes, as it is the preferred energy source compared to other SCFAs produced within the colon (1). In people, concentrations of colonic butyrate can range from 10 to 20 mM (2), and approximately 95% to 99% of all SCFAs produced in the colon are rapidly absorbed and metabolized into energy sources by colonic cells (3). A failure to utilize butyrate as an energy source can enhance the symptoms of intestinal disease. As an example, individuals with ulcerative colitis often have metabolic deficiencies in butyrate transport systems, and often these systems play a role in butyrate absorption and usage, suggesting a reduced ability to absorb butyrate (4). The optimal concentration of butyrate within the host has long been disputed. Studies that use experimental models of colitis in rodents have reported administering butyrate at concentrations ranging from 40 mM to 130 mM (5–9). Furthermore, concentrations of butyrate administered to people with ulcerative colitis via enemas have ranged from 80 mM to 150 mM (10). A limited number of researchers have investigated changes in mucus secretion resulting from butyrate enemas in animal models or have administered butyrate at concentrations as high as 100 mM in their models. Therefore, the high absorptive rate of intestinal SCFA necessitates that higher concentrations be used in scientific investigations (6, 11).
The addition of butyrate directly to human colons (12), colonic epithelial cells (13), and carcinoma epithelial cells (14) has been shown to be beneficial in reducing intestinal inflammation (15–17). In vitro analyses have shown that butyrate can reduce expression of proinflammatory cytokines and enhance epithelial barrier function, and in vivo studies conducted in mice without enteritis suggest that butyrate increases mucus synthesis in goblet cells (6, 10). Furthermore, several studies using intestinal cell lines have shown that the administration of butyrate to intestinal cells can downregulate NF-κB and reduce the expression of proinflammatory cytokines (18–20), and this has also been observed in mouse models of chemically induced enteritis (21, 22). The degree and mechanisms by which butyrate influences pathogen-induced inflammation, including its impact on dysbiosis and intestinal injury, have not be extensively studied in animal models.
Some evidence indicates that butyrate plays a role in maintaining the intestinal barrier by increasing the expression of mucins such as MUC2 and enhancing mucus production (23). Intestinal mucins are composed of transmembrane or secretory glycoproteins released from goblet cells and are involved in forming two protective mucus layers, with MUC2 being the major secretory glycoprotein in the colon (24, 25). Although the mucus layer is thought to inhibit bacterial binding to the epithelium, thereby limiting bacterial entry into the gut-associated lymphoid tissue (GALT) and the subsequent induction of proinflammatory responses (26), there are bacterial species that utilize mucus as an energy source, readily colonize mucus layers, and act as indicators of mucus abundance. Akkermansia muciniphila is a well-known mucus-degrading organism and has often been isolated from the colons of mice and people (27), whereas Mucispirillum schaedleri has been shown to selectively colonize intestinal mucus in a variety of organisms (28). More recently, studies investigating the effect of butyrogenic bacteria on the intestinal mucus barrier showed that changes in intestinal bacterial populations influenced the production and secretion of mucins from goblet cells (29). Moreover, treatment with butyrate also altered bacterial populations of Bacteroidetes, Firmicutes, Deferribacteres, and Proteobacteria in the large intestine (30–33). We hypothesized that butyrate supplemented to acutely inflamed colons would contribute to a temporal and spatial increase in mucus secretion and would concomitantly reduce proinflammatory signaling and inflammation. To test this hypothesis, we incited acute inflammation with Citrobacter rodentium (with or without rectally administered butyrate) and temporally measured a variety of variables, including food consumption and weight gain, intestinal SCFA concentrations, colonic cell damage and injury, the expression of genes involved in proinflammatory immune responses and repair, and the overall changes in the colonic bacterial community structure.
RESULTS
Citrobacter rodentium incited enteritis in C57BL/6 mice.
To confirm the presence of C. rodentium in mice exhibiting symptoms of enteritis and to ascertain whether the butyrate concentration affected colonization by the bacterium, fecal samples were collected and densities of the bacterium were quantified. In control mice administered phosphate-buffered saline (PBS) via gavage, C. rodentium was not isolated from fecal samples. In contrast, all mice with enteritis shed the bacterium in their feces (Fig. 1; see also Fig. S1 in the supplemental material). Butyrate administration had no impact (P = 0.965) on temporal densities of C. rodentium in feces; shedding of C. rodentium peaked on day 14 postinoculation (p.i.), and densities of the bacterium decreased (P < 0.050) thereafter. To confirm that mice administered C. rodentium via gavage exhibited phenotypic signs of enteritis, colonic tissues were examined histopathologically. Mice that were not administered C. rodentium via gavage did not show symptoms of infection, exhibit overt evidence of enteric epithelial cell hyperplasia or epithelial cell injury, or show changes in mitotic activity, goblet cell presence, or crypt height, with total average histopathologic scores of less than 1.30 ± 0.25. In contrast, all mice administered C. rodentium via gavage presented symptoms and signs of enteritis and exhibited substantially higher (P ≤ 0.001) histopathologic scores (Fig. 2) than mice without enteritis (data not presented) for all of the categories examined. The degree of tissue injury was highest on day 14 p.i. (i.e., peak infection), decreased by day 21 (i.e., late infection), and was minimal by day 28 (i.e., clearance).
FIG 1 .
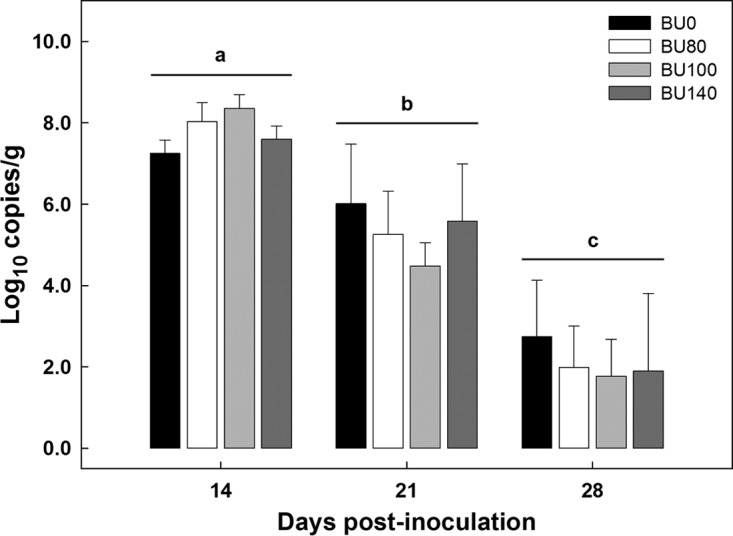
Densities of C. rodentium in feces from mice inoculated with the bacterium and rectally administered PBS (BU0) or butyrate at a concentration of 80 mM (BU80), 100 mM (BU100), or 140 mM (BU140) from the point of inoculation to day 28 postinoculation. No C. rodentium was detected in feces from mice not inoculated with the bacterium. Vertical lines associated with bars represent standard errors of the means (n = 4). There was no difference (P = 0.965) in the densities of C. rodentium among the butyrate treatments, but densities differed (P < 0.001) among the three sample times. Time groups not indicated by the same letter differ (P ≤ 0.012).
FIG 2 .
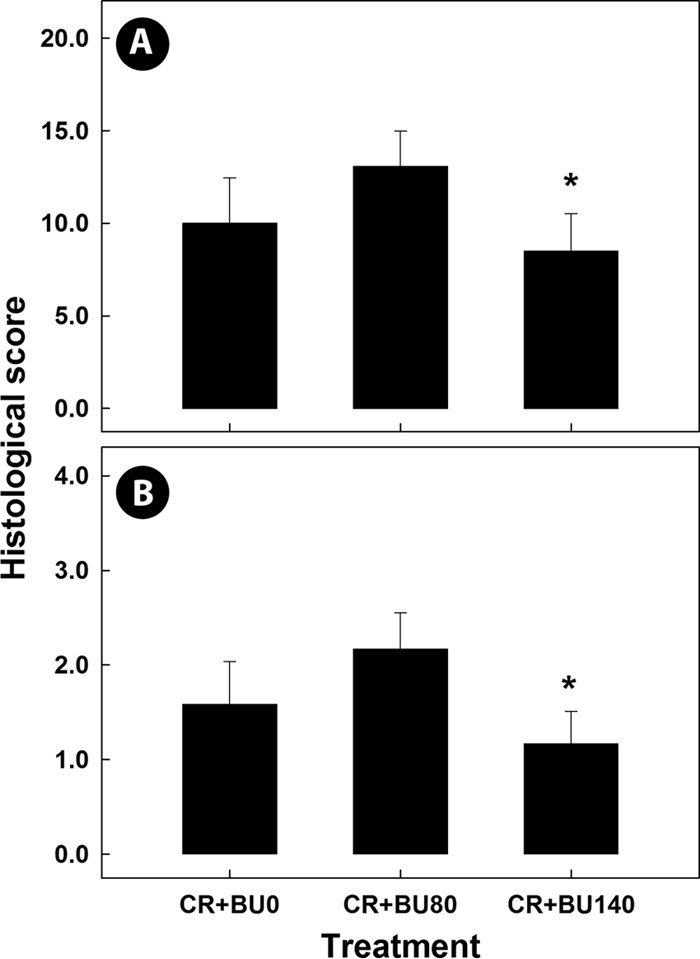
Histopathologic changes in colonic tissue from mice inoculated with C. rodentium (CR) and rectally administered PBS (BU0) or butyrate at concentrations of 80 mM (BU80) and 140 mM (BU140) averaged over peak and late infection. (A) Total histopathologic scores. (B) Epithelial cell hyperplasia. Vertical lines associated with histogram bars represent standard errors of the means (n = 4). *, P < 0.050 (relative to the BU0 treatment). For all treatments, histopathologic changes were reduced over time (P ≤ 0.005) in mice infected with C. rodentium.
Densities of C. rodentium in feces from mice inoculated with the bacterium and rectally administered PBS (BU0), or butyrate at a concentration of 80 mM (BU80), or 140 mM (BU140) from the point of inoculation to day 28 postinoculation. No C. rodentium was isolated from feces of mice that were not inoculated with the bacterium. Download FIG S1, PDF file, 0.1 MB (107.5KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fecal butyrate concentrations were higher in infected mice administered butyrate rectally.
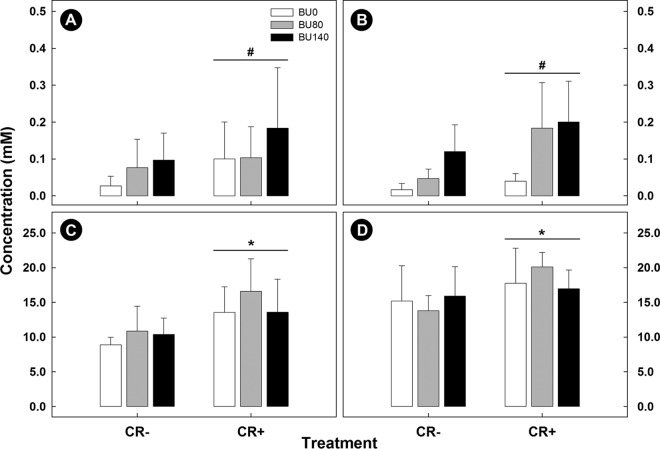
To determine whether rectal butyrate administration altered fecal SCFA concentrations, fresh fecal samples were collected and analyzed for individual SCFAs. There was no effect (P = 0.56) of time p.i. on the overall concentration of butyrate measured in feces (Fig. 3A and B). However, mice with enteritis exhibited a trend for higher (P ≤ 0.073) quantities of butyrate in their feces than mice without enteritis, and infected mice that were administered butyrate had higher (P < 0.050) concentrations of butyrate in their feces than mice administered PBS rectally. Total SCFA concentrations measured in feces were also higher (P ≤ 0.020) in mice inoculated with C. rodentium than in mice administered PBS via oral gavage (Fig. 3C and D).
FIG 3 .
Concentrations of butyrate and total short-chain fatty acids (SCFA) measured in feces from mice inoculated with PBS (CR−) or C. rodentium (CR+) via gavage and rectally administered PBS (BU0) or butyrate at concentrations of 80 mM (BU80) and 140 mM (BU140). Samples were collected on days 14 and 21 postinoculation (p.i.). (A) Butyrate concentrations in feces collected on day 14 p.i. (B) Butyrate concentrations in feces collected on day 21 p.i. (C) Total SCFA concentrations in feces collected on day 14 p.i. (D) Total SCFA concentrations in feces collected on day 21 p.i. Vertical lines associated with histogram bars represent standard errors of the means (n = 4). #, P ≤ 0.073 (between infected and noninfected mice). *, P ≤ 0.020 (between infected and noninfected mice).
Butyrate supplementation increased weight gain and feed consumption in mice with enteritis during peak infection.
In order to measure whether the rectal administration of butyrate had a physiological effect on the mouse during C. rodentium infection, average feed consumption and weight gain values were analyzed. Butyrate administration did not affect feed consumption (P > 0.95) or weight gain (P > 0.44) in mice without enteritis at any of the three sample times (data not presented). At peak infection, mice with enteritis that had been administered butyrate consumed more (P = 0.003) food than mice with enteritis that had not been administered butyrate (Fig. 4). During late infection (P = 0.38) and clearance (P = 0.34), there was no difference in food consumption between control mice and those supplemented with butyrate. Mice with C. rodentium-induced enteritis that were administered butyrate gained weight faster (P ≤ 0.002) than mice not administered butyrate at peak infection (Fig. 5) but not at late infection or clearance (P ≥ 0.53) (data not presented).
FIG 4 .
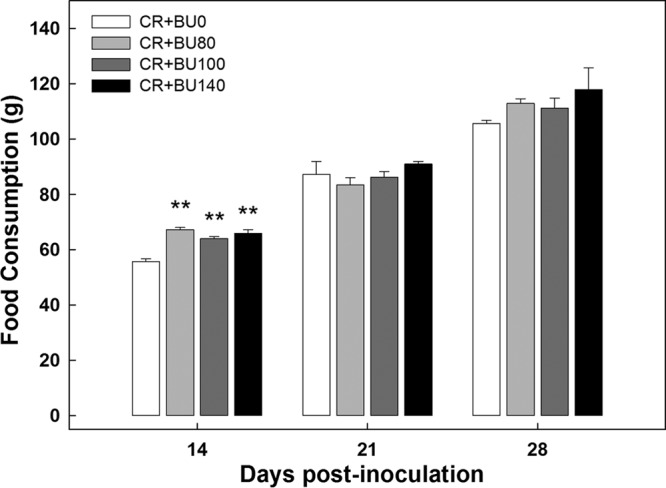
Daily food consumption in mice inoculated with C. rodentium (CR+) and rectally administered PBS (BU0) or butyrate at concentrations of 80 mM (BU80), 100 mM (BU100), and 140 mM (BU140). Vertical lines associated with markers represent standard errors of the means (n = 4). **, P ≤ 0.010 (relative to the BU0 treatment on day 14 postinoculation).
FIG 5 .
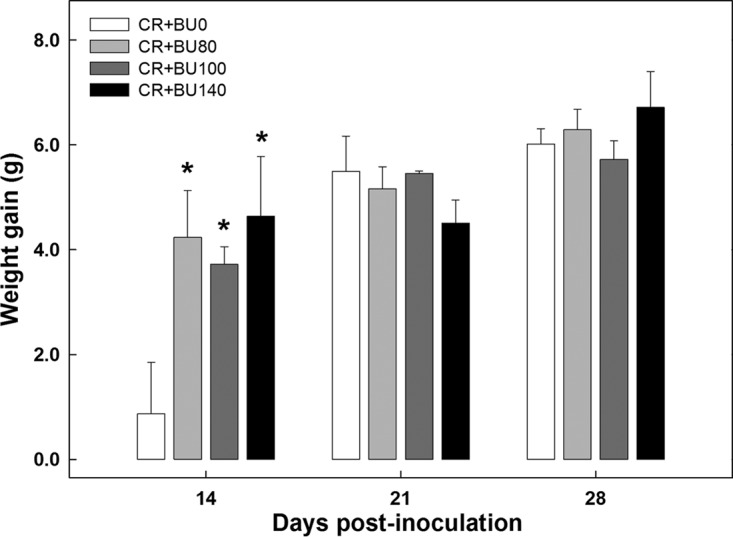
Weight gain in mice inoculated with C. rodentium (CR+) and rectally administered PBS (BU0) or butyrate at concentrations of 80 mM (BU80), 100 mM (BU100), and 140 mM (BU140). Vertical lines associated with histogram bars represent standard errors of the means (n = 4). *, P ≤ 0.050 (relative to the BU0 treatment).
High butyrate concentrations decreased epithelial cell hyperplasia in mice with enteritis.
To evaluate a potential decrease in distal colonic inflammation due to butyrate supplementation, representative samples of distal colonic tissue were collected and analyzed for histopathologic changes. Mice with enteritis exhibited total histopathologic scores of ≥14.0 ± 2.7 at peak infection, ≥9.3 ± 3.5 at late infection, and ≤6.5 ± 0.1 at clearance (Fig. 2A). Scores of total histopathologic changes did not differ (P = 0.19) between peak and late infection but were lower (P ≤ 0.009) at clearance. Averaged over time, total pathological changes (P = 0.020) and epithelial cell hyperplasia levels (P = 0.023) were lower in mice with enteritis that had been administered butyrate at a concentration of 140 mM (Fig. 2) than in mice not administered butyrate. Increased (P = 0.054) mitotic activity was also observed in mice with enteritis that had been administered butyrate at 80 mM at peak infection compared to mice with enteritis not administered butyrate (Fig. S2).
Mitotic cell activity in mice inoculated with C. rodentium (CR+) and rectally administered PBS (BU0), or butyrate at a concentration of 80 mM (BU80), or 140 mM (BU140). Vertical lines associated with histogram bars represent standard errors of the means (n = 4). *, P ≤ 0.050 (relative to the BU0 treatment averaged over the indicated time points). In all instances, infection with C. rodentium affected mitotic cell activity over time (P ≤ 0.005). Download FIG S2, PDF file, 0.1 MB (80.8KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mucus accumulated in the lumen after butyrate supplementation at peak infection.
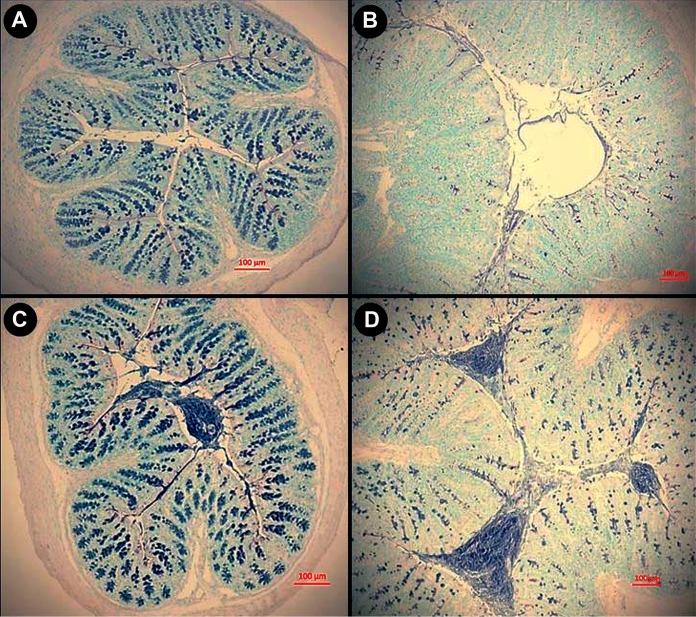
To confirm the hypothesis that butyrate supplementation affects mucus accumulation in the colon, representative tissues from the distal colon were stained for mucus presence. At peak and late infection, mice with enteritis that had been administered butyrate at a concentration of 140 mM exhibited increased accumulation of mucus in the colonic lumen and within goblet cells relative to mice administered PBS enemas (Fig. 6; Fig. S3).
FIG 6 .
Mucus localization in alcian blue–periodic acid-Schiff-stained sections from the distal colons of mice inoculated with PBS (CR−) or C. rodentium (CR+) and rectally administered PBS (BU0) or butyrate at a concentration of 140 mM (BU140) at day 14 p.i. (A) CR–, BU0. (B) CR+, BU0. (C) CR–, BU140. (D) CR+, BU140. Tissue from infected mice administered butyrate exhibited a consistent increase in the density of mucus (blue stain) within goblet cells and the colonic lumen. Bars, 100 µm.
Mucus localization in alcian blue–periodic acid-Schiff-stained sections of the distal colons of mice inoculated with PBS (CR−) or C. rodentium (CR+), and rectally administered PBS (BU0) or butyrate at a concentration of 140 mM (BU140) at day 21 p.i. (A) CR−, BU0. (B) CR+, BU0. (C) CR−, BU140. (D) CR+, BU+. Tissue from infected mice supplemented with butyrate exhibited an increase in the density of mucus (blue stain) within the colonic lumen. Overall, more mucus was observed in the lumen on day 21 p.i. than on day 14 p.i. Bar, 100 μm. Download FIG S3, PDF file, 0.6 MB (680.3KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Butyrate supplementation affected enteric gene expression in mice with enteritis.
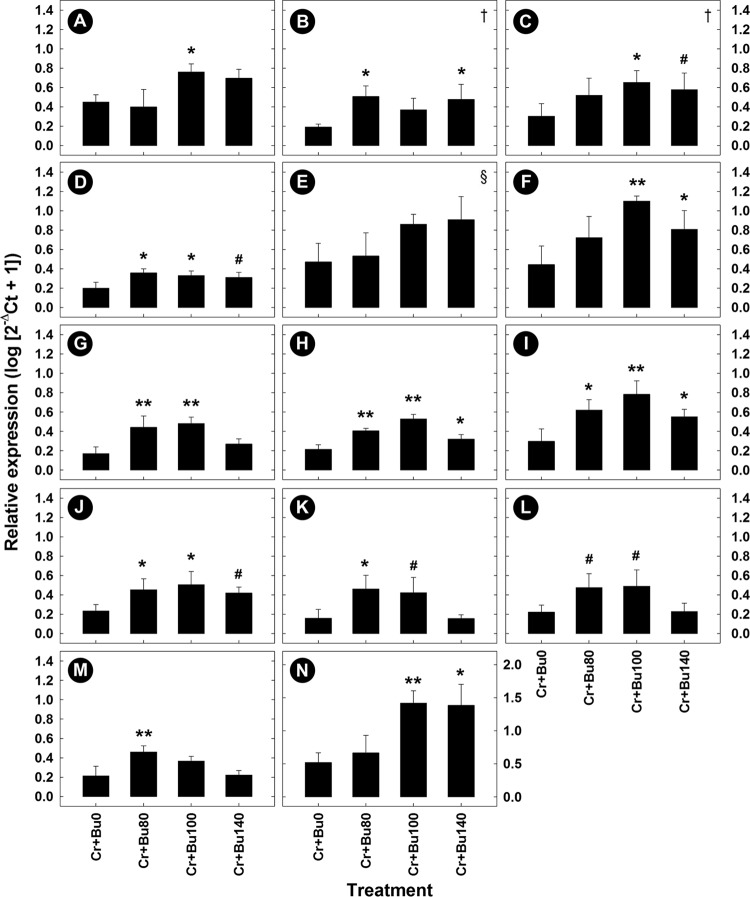
Representative samples from the distal colon were collected and RNA was extracted from them to determine the degree to which butyrate administration influenced mRNA expression of genes related to infection and inflammation. The administration of butyrate had no effect (P ≥ 0.16) on expression of Th1 (Ifnγ, Tnfα)-, Th2 (Il4)-, and Th17 (Il17A, Il22)-associated cytokines or on expression of Myd88, Prg3, RegIIIγ, Tff3, and Tlr9 in mice without enteritis relative to mice that were not administered butyrate (data not presented). In contrast, mice administered butyrate at a concentration of 80 mM exhibited a trend of increased (P = 0.062) expression of Th2 (Il4) cytokines and of significantly increased (P ≤ 0.050) expression of Th1 (Ifnγ, Tnfα) and Th17 (Il22) cytokines in mice with enteritis (Fig. 7); mice administered butyrate at concentrations of 100 mM and higher also exhibited increased (P ≤ 0.050) expression of Il1β. The administration of butyrate at 80, 100, and 140 mM to mice with enteritis increased (P ≤ 0.036) the expression of genes involved in bacterial recognition and defense, including Myd88, Tlr2, and Ltb4r1, but only the mice given butyrate at 80 mM (P = 0.005) and 100 mM (P = 0.004) exhibited increased expression of Tlr9. Mice given butyrate at 80 mM (P = 0.044) and 100 mM (P = 0.076) but not 140 mM (P = 0.990) exhibited increased expression of Prg3. Butyrate given at 100 mM also increased (P = 0.028) expression of RegIIIγ, and administration of 100 mM (P = 0.007) and 140 mM (P = 0.012) butyrate increased expression of Relmβ, while butyrate provided at 80 mM (P = 0.002) and 100 mM increased (P = 0.045) expression of Tff3 in mice with enteritis. There was no difference (P ≥ 0.36) between the PBS and C. rodentium treatments in the expression of the Treg response cytokines Il10 and Tgfβ in mice not administered butyrate (Fig. 8A to D). In mice administered butyrate at 80 and 100 mM, expression of Il10 was increased (P ≤ 0.028), and all concentrations of butyrate stimulated an increase (P = 0.049) in Tgfβ expression in mice with and without enteritis (Fig. 8A to D). Regardless of butyrate administration, mice with enteritis displayed a trend of decreased (P = 0.053) Muc2 expression relative to mice without enteritis. Administration of butyrate at concentrations of 80 and 100 mM increased Muc2 expression in mice without enteritis (P < 0.023) and demonstrated a trend for increased (P < 0.088) Muc2 expression in mice with enteritis (Fig. 8E and F).
FIG 7 .
Relative mRNA gene expression profiles of cytokine-related innate barrier function and host pathogen recognition genes measured in colonic tissue harvested from mice inoculated with C. rodentium (CR) and with rectally administered PBS (BU0) or butyrate at concentrations of 80 mM (BU80), 100 mM (BU100), and 140 mM (BU140) averaged over time. (A) Il17A. (B) Il22. (C) Il1β. (D) MyD88. (E) RegIIIγ. (F) Ifnγ. (G) Tlr9. (H) Tlr2. (I) Tnfα. (J) Ltb4R1. (K) Prg3. (L) Il4. (M) Tff3. (N) Relmβ. Vertical lines associated with histogram bars represent standard errors of the means (n = 4). #, P ≤ 0.100; *, P ≤ 0.050; **, P ≤ 0.010 (relative to the BU0 treatment). †, the statistical value represents a difference determined by excluding a butyrate treatment and comparing CR+ BU0 treatment to only two of the three other treatments; §, the statistical value represents comparison between CR+ BU0 treatment and CR+ BU100 treatment only. Ct, threshold cycle.
FIG 8 .
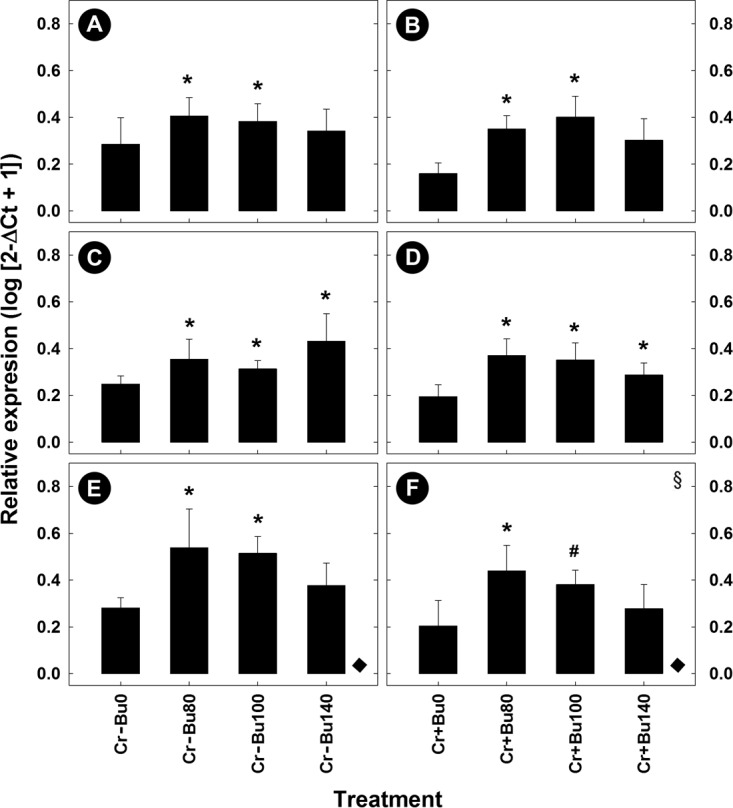
Relative mRNA gene expression profiles of regulatory cytokines and mucus-producing genes in colonic tissue harvested from mice inoculated with PBS (CR−) or C. rodentium (CR+) and rectally administered PBS (BU0) or butyrate at concentrations of 80 mM (BU80), 100 mM (BU100), and 140 mM (BU140). (A and B) Il10. (C and D) Tgfβ. (E and F) Muc2. Vertical lines associated with histogram bars represent standard errors of the means (n = 4). #, P ≤ 0.100; *, P ≤ 0.050 (relative to the BU0 treatment). ♦, P = 0.053 (representing the difference in overall Muc2 expression due to C. rodentium infection). §, the statistical value represents a difference determined by excluding a butyrate treatment and comparing CR+ BU0 treatment to only two of the three other treatments.
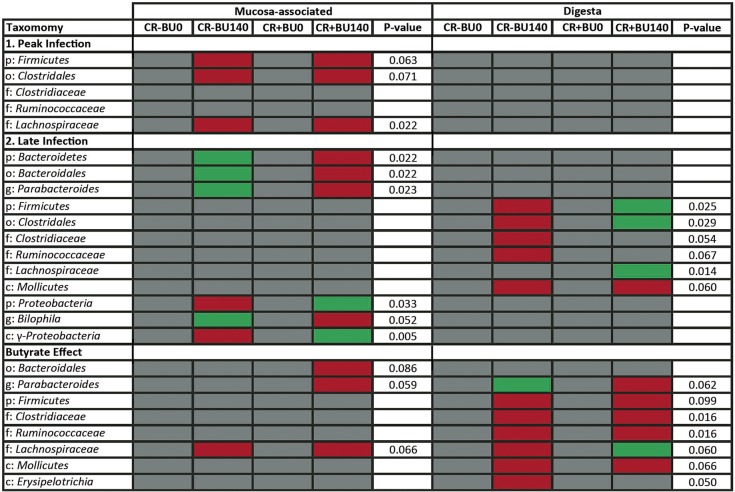
Butyrate supplementation reduced the abundance of Firmicutes and butyrate-producing bacteria in the distal colon.
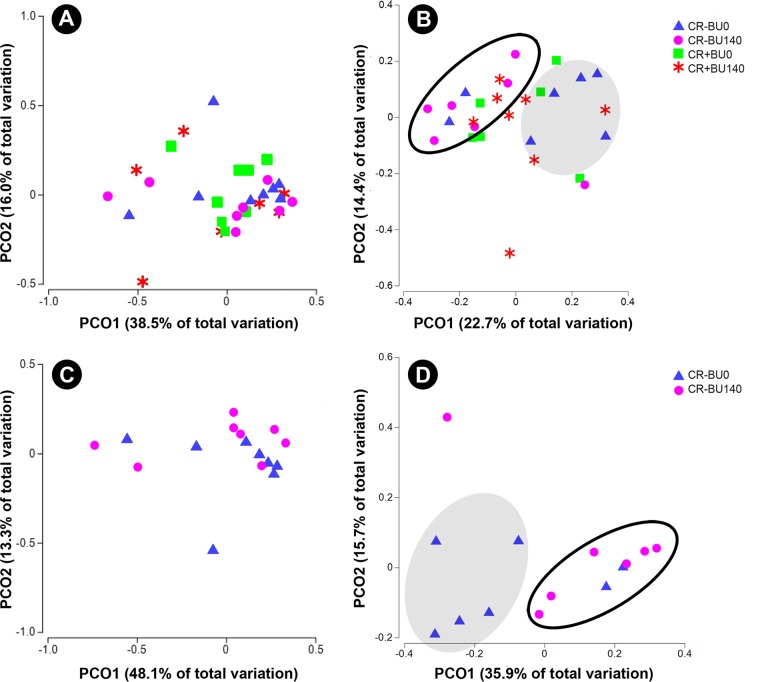
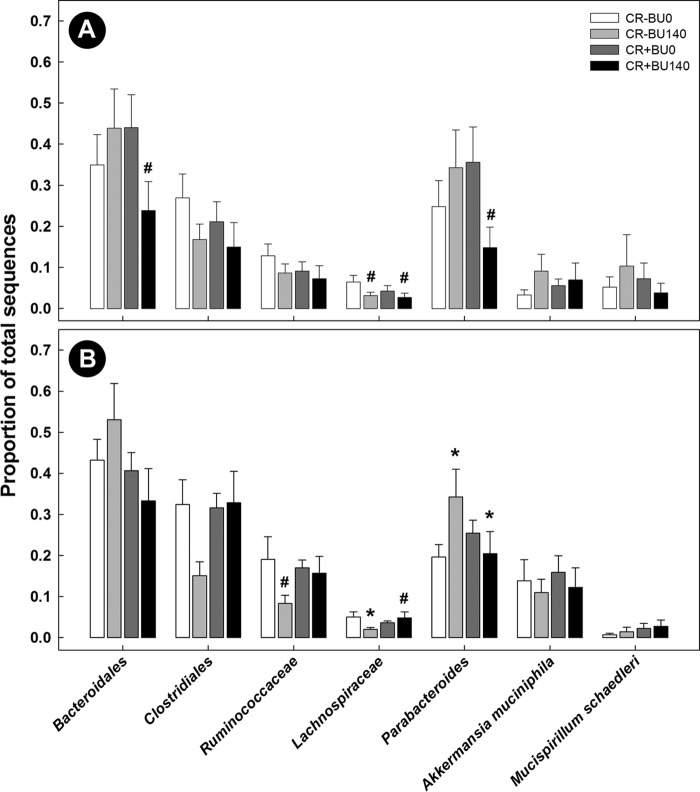
To determine the changes in bacterial community structure due to butyrate supplementation, representative samples from the distal colon (mucosal and digesta) of all mice were collected and community DNA was sequenced for individual animals (i.e., to obtain a measure of variability among replicate mice). Based on UniFrac analyses of community similarities, the structure of the mucosa-associated bacterial community was more variable and differed (P = 0.087) from the community structure of the digesta in the distal colon (Fig. S4). The administration of 140 mM butyrate altered (P = 0.071) the community structure within the colonic digesta but not within the mucosa-associated community in mice without enteritis (Fig. 9). Butyrate affected (P ≤ 0.050) the abundance of bacteria within the Bacteroidetes (i.e., Paraprevotelaceae), Firmicutes (i.e., Bacilli; Clostridiaceae; Erysipelotrichales; Peptostreptococcaceae) and Tenericutes (RF39) phyla (Fig. 10). During peak infection, the administration of butyrate showed a trend for reduced (P = 0.063) abundance of Firmicutes, and, averaged over the two time points, the densities of mucosa-associated Lachnospiraceae were reduced (P = 0.022) in mice with and without enteritis (Fig. 11 and 12). At late infection, mice inoculated with C. rodentium exhibited a lower (P = 0.022) abundance of species within the Bacteroidetes phylum; however, in mice without enteritis, butyrate supplementation increased (P = 0.022) the levels of these bacteria, especially Parabacteroides sp. (P = 0.023), associated with mucosa averaged over time (Fig. 11 and 12A). An increase (P = 0.005) in the abundance of Gammaproteobacteria associated with mucosa was also observed during late infection in mice with enteritis that were administered 140 mM butyrate, and in mice without enteritis, butyrate administration increased (P = 0.052) the presence of Bilophila sp. in association with the mucosa (Fig. 11). Within digesta, butyrate administration altered bacterial communities during late infection (Fig. 11). For example, mice without enteritis that were administered 140 mM butyrate displayed a depletion (P = 0.025) of the abundance of Firmicutes species, namely, Clostridiales (P = 0.029) species, and showed a trend of reduced (P = 0.067) Ruminococcaceae species abundance during late infection. In contrast, mice with enteritis exhibited a higher abundance of Firmicutes (P = 0.025), Clostridiales (P = 0.029), and Lachnospiraceae species (P = 0.014) than mice with enteritis not administered butyrate at late infection (Fig. 11). In mice without enteritis, Parabacteroides spp. increased (P = 0.023) in average abundance over time with butyrate supplementation (Fig. 12). Although significant differences between the treatments were not evident regarding the abundance of mucus-associated species, butyrate administration caused a trend of reduced Akkermansia muciniphila abundance within the digesta collected from the distal colon in mice with and without enteritis (Fig. 12B). In contrast, butyrate administration was associated with a general trend of increased abundance of A. muciniphila associated with mucosa (Fig. 12A). Similarly, in mice without enteritis, we observed a trend of increased abundance of the mucus-dwelling bacterium Mucispirillum schaedleri in the mucosa-associated community (Fig. 12A). However, a trend of lowered abundance was observed in the digesta within the distal colon, and M. schaedleri abundance steadily increased with butyrate administration in mice with enteritis (Fig. 12B). Overall, butyrate administration decreased the abundance of bacteria within digesta and associated with mucosa. However, in digesta collected from the distal colon, butyrate effectively increased the abundance of members of the Lachnospiraceae family during infection.
FIG 9 .
Effects of rectal administration of PBS (BU0) or 140 mM butyrate (BU140) on bacterial community structures in the distal colon of mice (mucosa-associated and within digesta) inoculated with PBS (CR−) or C. rodentium (CR+) as determined by weighted UniFrac analysis. Axes identify percent variation among treatments, and ellipsoids are used to highlight clustering of communities by treatment. (A and B) Mice with (CR+) and without (CR−) enteritis. (C and D) Mice without enteritis (CR−). (A) Mucosa-associated. (B) Digesta. The shaded ellipsoid highlights clustering of communities from CR− BU0 treatment mice, and the open ellipsoid highlights clustering of communities from CR− BU140 treatment mice; a butyrate effect was observed for the CR− treatments (P = 0.071 with 753 random permutations). (C) Mucosa-associated. (D) Digesta. The shaded ellipsoid highlights clustering of communities from CR− BU0 treatment mice, and the open ellipsoid highlights clustering of communities from CR− BU140 treatment mice; a butyrate effect was observed (P = 0.071 with 762 random permutations).
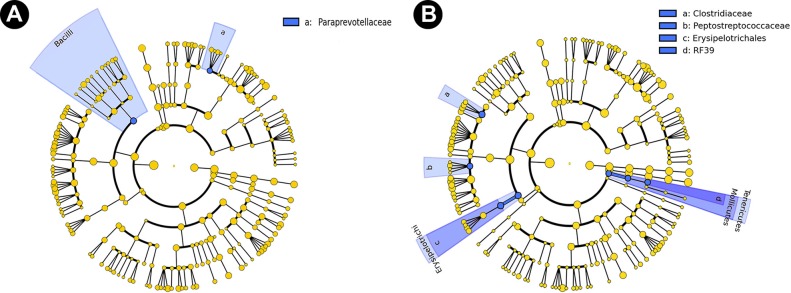
FIG 10 .
Phylogenetic trees of bacteria associated with mucosa and within digesta in the distal colons of mice with (CR+) and without (CR−) enteritis and rectally administered PBS (BU0) or 140 mM butyrate (BU140) as determined using the linear discriminant analysis effect size (LEfSe) method (LDA value, >2.0). The abundances of bacterial taxa highlighted in blue differed (P ≤ 0.050) between the BU0 and BU140 treatments. (A) Mucosa-associated. (B) Digesta. Data to construct the phylogenetic trees used summarized taxonomic values per treatment averaged over four replications.
FIG 11 .
Changes observed in mucosa- and digesta-associated bacterial communities within the distal colon at peak infection and late infection, including the overall butyrate effect. Green boxes indicate a significant (P ≤ 0.100) increase in the abundance of specified taxa supplemented with butyrate (BU140) compared to the PBS control (BU0) within each enteritis treatment group. Red boxes indicate a significant (P ≤ 0.100) decrease in the abundance of specified taxa supplemented with butyrate (BU140) compared to butyrate control taxa (BU0) within each enteritis treatment group. p, phylum; c, class; o, order; f, family; g, genus. A butyrate effect was defined as a significant increase or decrease in bacterial abundance in response to butyrate administration averaged over time.
FIG 12 .
Abundance of bacteria within the distal colon of mice inoculated with PBS (CR−) or C. rodentium (CR+) and rectally administered PBS (BU0) or butyrate at a concentration of 140 mM (BU140). (A) Mucosa-associated. (B) Digesta. Vertical lines associated with histogram bars represent standard errors of the means (n = 4). #, P ≤ 0.100; *, P ≤ 0.050 (comparing butyrate treatment effects within CR− mice [BU0 to BU140] and CR+ mice [BU0 to BU140]).
Principal-coordinate analysis of bacterial communities within digesta and associated with mucosa in the distal colon from mice inoculated with PBS via gavage, and rectally administered PBS (BU0) or 140 mM butyrate (BU140). Ellipsoids show clustering of communities by sample type. Download FIG S4, PDF file, 0.1 MB (138.6KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
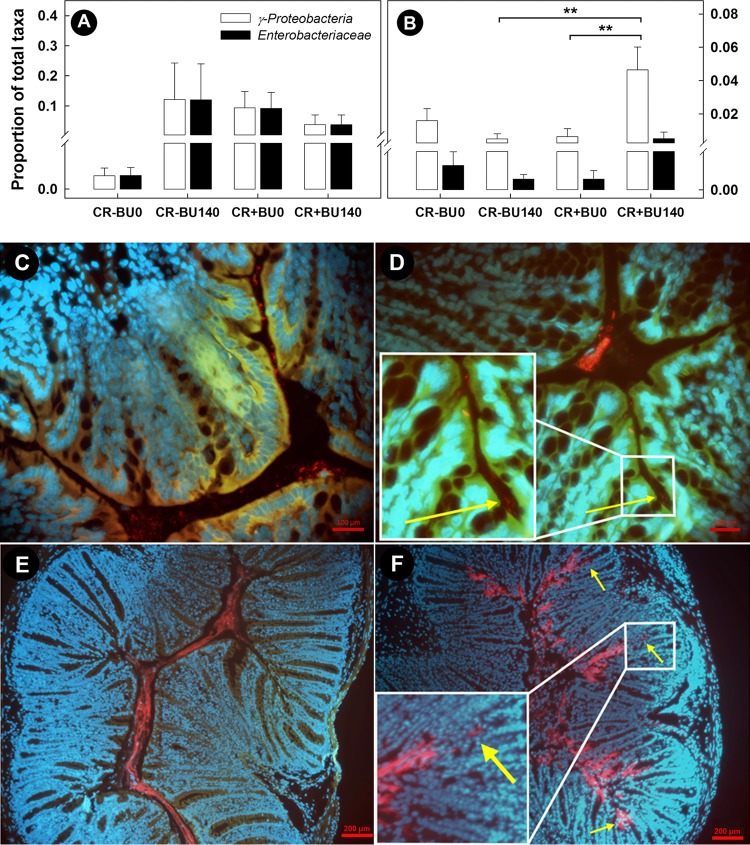
Fluorescent in situ hybridization (FISH) visualization displayed a high abundance of Proteobacteria in butyrate-supplemented tissue.
To confirm the bacterial community sequencing results, we collected representative tissue and stained for specific groups of bacteria. High densities of Gammaproteobacteria and Enterobacteriaceae were associated with the mucosa in the distal colon of mice infected with C. rodentium compared to mice without enteritis, and high densities of Gammaproteobacteria were observed during late infection in mice with enteritis and administered 140 mM butyrate compared to those administered 0 mM butyrate (Fig. 13A and B). It is noteworthy that among the mice with enteritis, higher (P = 0.036) densities of mucosa-associated Pseudomonas spp. were also observed in digesta from the distal colon of mice administered butyrate than in those not given butyrate (Fig. S5). Proteobacteria were more often observed within intestinal colonic crypts in mice rectally administered butyrate at a concentration of 140 mM (Fig. 13D and F) than in mice not administered butyrate (Fig. 13C and E). Butyrate supplementation at a concentration of 140 mM increased the total abundance of Gammaproteobacteria, especially in mice infected with C. rodentium.
FIG 13 .
Localization of Gammaproteobacteria and evidence of Proteobacteria migration into the crypts within the distal colon of mice inoculated with C. rodentium (CR+) and rectally administered PBS (BU0) or butyrate at a concentration of 140 mM (BU140). (A) Abundance of Enterobacteriaceae and Gammaproteobacteria associated with the mucosa on day 14 postinoculation (p.i.) in mice inoculated with PBS and C. rodentium (B) Abundance of Enterobacteriaceae and Gammaproteobacteria associated with mucosa on day 21 p.i. in mice inoculated with PBS and C. rodentium. Vertical lines associated with histogram bars represent standard errors of the means (n = 4). **, P ≤ 0.010 (for treatments linked by the horizontal lines). (C to F) Fluorescence micrographs of distal colonic tissue hybridized with a Gammaproteobacteria probe (red). (C) CR+ BU− at day 14 p.i. (bar, 200 µm). (D) CR+ BU+ at day 14 p.i. (bar, 200 µm). (E) CR+ BU− at day 21 p.i. (bar, 100 µm). (F) CR+ BU+ at day 21 p.i. (bar, 200 µm). The arrows indicate subluminal Proteobacteria.
Proportion of total sequences within the Pseudomonadales order within digesta collected from the distal colon. Vertical lines associated with histogram bars represent standard errors of the means (n = 8). *, P ≤ 0.050 (for treatments linked by the horizontal lines). Download FIG S5, PDF file, 0.1 MB (96.5KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Food consumption and weight gain are characteristics of normal intestinal function and adequate host health, and we observed that infection with C. rodentium caused a reduction in food consumption and weight gain, which has been previously reported (34, 35). We also observed that rectal administration of high concentrations of butyrate resulted in an increase in food consumption and weight gain in infected mice, indicating that butyrate plays an important role in improving feeding behavior and enhancing growth during periods of active enteritis. The concentrations of butyrate administered via enema in the current study were higher than would be expected to be generated by bacterial fermentation in both healthy and inflamed murine intestines (36). In rodents, butyrate within the intestine stimulates gluconeogenesis, and high glucose levels within blood activate brain stimuli that promote the metabolism of glucose, improve feed intake, and subsequently promote growth (37, 38). Thus, it is possible that butyrate administered to mice challenged with C. rodentium ameliorates bacterially induced colitis by enhancing intestinal gluconeogenesis.
Infection with C. rodentium resulted in a strong activation of Th1- and Th17-associated cytokines. These responses are necessary for pathogen clearance (34, 39). Similarly to other studies, we observed elevated levels of epithelial cell hyperplasia in addition to increased expression of Tnfα, Il1β, Il17A, and Il22 in the distal colon of C. rodentium-infected mice (34, 40). In the current study, butyrate treatment further increased expression of cytokines involved in the Th17 and Th1 immune response, which potentially attenuated the infection and promoted epithelial cell restoration (34, 41). This is underscored by the prominent increase in the expression of Ifnγ-, Tnfα-, Il1β-, and Th17-associated cytokines, which are generally used as indicators of active intestinal inflammation (39, 42). Collectively, coordinated Th17 and Th1 immune responses are critical for the clearance of C. rodentium (34), and butyrate further stimulated the expression of cytokines involved in resolving the infection (43). It is noteworthy that although the hallmarks of intestinal inflammation following C. rodentium challenge were observed, the progression of disease was delayed (44). The strain of C. rodentium that was used in the current study was modified to include a chromosomal green fluorescent protein (GFP) insertion, and it is likely that the presence of the insertion altered the bacterial pathogenicity (45).
Expression of the cytokines Il17A and Il17F is important in the clearance of C. rodentium infection, and it has been proposed that these cytokines facilitate the induction of antimicrobial peptides to reduce inflammation (46). Specifically, Th17 cytokines, including IL17 and IL22, have been shown to regulate anti- and proinflammatory and anti- and proapoptotic activities in epithelium cells within inflamed tissue (47–49). These cytokines can also increase production of regulatory proteins such as REGIIIγ during periods of intestinal inflammation (50). We observed that gene expression associated with the intestinal antimicrobial C-type lectin REGIIIγ, an important protein involved in interactions between intestinal bacteria and the epithelium and subsequent tissue repair (51, 52), was increased in mice with enteritis supplemented with butyrate compared to mice with enteritis that were not administered butyrate.
Regulatory T-cell responses are important for modulating and resolving inflammatory responses within the intestine. These responses in association with the stimulation of mucus secretion at the epithelial surface provide an effective strategy to reduce tissue injury following challenges with bacterial pathogens (53, 54). Previous studies have shown that the presence of butyrate increases CD4+ Foxp3 expression from T cells within the colonic mucosa, leading to the mitigation of intestinal injury and promoting inflammatory quiescence (55, 56). Similarly, our findings demonstrated that administration of butyrate at low and high concentrations increased expression of Tgfβ and Il10 in mice with and without enteritis. The maintenance of the physical mucus barrier is important in providing protection to the intestinal epithelium and maintaining intestinal homeostasis (44, 57), and we observed that butyrate administration to mice increased Muc2 gene expression and deposition of mucus at the epithelial layer. The presence of SCFAs within the intestine has the potential to upregulate mucin secretion (58), and butyrate in particular has been linked to increases in Muc2 gene expression in cell lines and within the murine colon (6, 10, 59). In contrast, data in support of the ability of butyrate to increase mucus secretion in vivo are inconsistent (12, 54, 60). Although our research indicates a relationship between increased mucus production and the rectal administration of butyrate, the amount of butyrate measured in fecal samples was not conspicuously altered in mice administered butyrate relative to those administered buffer rectally. A possible explanation is that butyrate was rapidly absorbed by colonocytes but was less rapidly absorbed in mice with enteritis (3, 11), which was exemplified in our alcian blue–periodic acid-Schiff (AB-PAS)-stained sections.
The maintenance of a protective epithelial barrier is a complex process and includes other proteins involved in the production and secretion of mucus and in enterocyte development and turnover. As an example, accessory proteins such as TFF3 and RELMβ are involved in maintaining epithelial cell homeostasis and regeneration (61), and we observed increased expression of genes encoding both proteins following treatment with butyrate. TFF3 is a trefoil peptide secreted from intestinal goblet cells; it increases mucin viscosity to provide further protection to the mucosa and can enhance epithelial repair in the small and large intestines (62, 63). RELMβ is primarily secreted from goblet cells and is mainly found in localized foci of inflammation in mice and humans (64). The functions of RELMβ are poorly understood; however, it has been associated with colonic immunomodulatory function and contributes to intestinal mucus secretion in mice (65–67). We showed that butyrate administration in mice with enteritis contributed to increased expression of Relmβ, Tff3, and Muc2. Thus, through the modulation of proteins associated with mucus secretion and integrity and epithelial cell repair and restitution, butyrate affects the regulation of mucus-associated proteins during periods of intestinal inflammation.
We observed a general decrease in Firmicutes abundance in inflamed intestines in the presence and absence of 140 mM butyrate. Shifts from Bacteroidetes-dominant communities to Firmicutes-dominant communities in inflamed intestines have been reported previously; however, the evidence is conflicting on whether higher densities of either Bacteroidetes or Firmicutes correlate with negative or beneficial effects on intestinal health. Our results are in line with those reported by Schwab et al. (31), who observed that chemically induced injury to the colon increased the abundance of bacteria belonging to the Bacteroidales order and to Enterobacteriales and Deferribacteres. Similarly, we found that mice challenged with C. rodentium exhibited increases in the abundance of Deferribacteres and Proteobacteria species, and these changes were similar to the observations of Hoffmann et al. (68). Finally, we also observed a butyrate-dependent increase in levels of Lachnospiraceae in digesta within the distal colon, suggesting that butyrate was associated with an increase in the abundance of this group of bacteria. Importantly, Lachnospiraceae bacteria produce butyrate in the mammalian intestine (69) and our findings suggest that under conditions of inflammation, these bacteria may increase the amount of butyrate within the colon and subsequently mitigate inflammation (11). Furthermore, as our results suggest that butyrate is a putative inducer of mucus-associated bacteria, our data may signify an increase in the availability of mucus for microbial metabolism. Our data also showed trends of increased abundance of mucolytic A. muciniphila bacteria at the mucosal surface in response to butyrate treatment (70). Increases in the abundance of A. muciniphila during periods of dietary change and intestinal inflammation have also been linked to increased expression of host mucus and immune modulating genes (71, 72), suggesting that this bacterium induces both production and utilization of mucin during episodes of colitis. Collectively, our findings provide evidence that during infection, butyrate modulates the bacterial community in the inflamed distal colon to promote the growth of butyrogenic bacterial species, which also stimulate the butyrate-driven production of mucus by the host.
Although butyrate is an SCFA that has been implicated in improved barrier function, promotion of intestinal epithelial cell growth and repair, and enhanced host mucin secretion (6, 10, 23), we observed that administration of butyrate directly to the colon mildly exacerbated infection in mice with enteritis. In this regard, mice with enteritis that were treated with a low concentration of butyrate tended to have modestly higher histological scores during peak and late infection that had significantly subsided by the time of infection clearance. Histological scores also tended to be the highest in mice treated with butyrate at 80 mM compared to those treated with butyrate at a concentration of 140 mM, suggesting that butyrate administration works at a particular threshold and is most effective at reducing inflammation under conditions of administration to mice at high concentrations. As indicated above, we observed that butyrate-treated mice challenged with C. rodentium showed differing increases in gene expression of cytokines involved in the clearance of infection (Il17A, Tnfα), the reduction of mucosal inflammation (Tgfβ, Il10), and the improvement of host barrier function (Muc2, Relmβ, Tff3). These findings suggest that other processes that stimulate various cytokine pathways may also contribute to modestly heightened levels of tissue inflammation. As an example, in the presence of C. rodentium, butyrate can cause activation of the promoter of the locus of enterocyte effacement (LEE) operon and expression of ler genes necessary for the expression of virulence factors that can exacerbate tissue inflammation (73, 74). Moreover, morphological transformations in the mucosa following C. rodentium infection have been associated with a hyperreactive epithelial reparative response that induces changes such as marked epithelium hyperplasia with increased production of undifferentiated enterocytes (43). This could also potentially increase inflammation scores. In the current study, enhanced expression of epithelial cell-regenerating proteins such as MYD88, TFF3, and RELMβ in butyrate-treated mice likely further elevated the inflammation scores (i.e., as a consequence of increased epithelial hyperplasia, crypt height, and mitotic activity) for C. rodentium-challenged mice, masking some of the anti-inflammatory effects and accentuating proinflammatory tissue responses. Finally, we observed an increase in the abundance of the mucus-degrading bacterium A. muciniphila at the mucosal surface in butyrate-treated mice that was not observed in digesta in the distal colon. As indicated previously, A. muciniphila is known to colonize the intestinal mucus layer, and its presence has been associated with increased epithelial cell turnover, increased biosynthesis of epithelial cell components, and enhanced mucin production (70). Therefore, it is likely that the presence of A. muciniphila increased epithelial growth (i.e., hyperplasia turnover of colonocytes to increase total histological inflammation scores), and this warrants further investigation.
In conclusion, our findings provide evidence that, in a dose-dependent manner, butyrate administration can improve the weight gain of infected mice, enhance clearance of the infection, reduce inflammation through altered cytokine expression, and enhance tissue repair and mucus secretion. Moreover, butyrate treatment also affected the abundance of bacterial populations in both noninflamed and inflamed intestines. Notably, this investigation provided foundational information that can be used to determine the effects of prebiotics and other functional foods on the production of butyrate by enteric bacteria and their impact on intestinal health and host well-being.
MATERIALS AND METHODS
Experimental design.
The experiment was arranged as a completely randomized design with four levels of butyrate concentration (0 mM, 80 mM, 100 mM, and 140 mM), two levels of immunological stress (with C. rodentium and without C. rodentium), and three levels of time postinoculation (p.i.) (14, 21, and 28 days p.i.). Each replicate included 24 mice, and four replicates were performed on separate occasions (96 animals in total).
Ethics statement.
The study was carried out in strict accordance with the recommendations specified in the Canadian Council on Animal Care Guidelines. The project was reviewed and approved by the Lethbridge Research and Development Center (LRDC) Animal Care Committee (Animal Use Protocol Review 1322) and the LRDC Biosafety and Biosecurity Committee before commencement of the research.
Mouse maintenance.
Specific pathogen-free (SPF) C57BL/6J female mice were obtained from Charles River Laboratories, Inc. (Montreal, Quebec, Canada) at 3 weeks of age. For each replicate, mice were housed in groups with six mice per cage upon arrival and were given 10 days to adapt to the animal facility environment under conditions of a 10-h/14-h dark/light cycle. After the adaptation period, mice were transferred to individually ventilated cages (one mouse per cage) operated in containment mode. Mice were provided a low-fiber diet (AIN-93G 103455GI; Dyets Inc., Bethlehem, PA) and were permitted to eat and drink ad libitum. Sterile shredded paper was provided for bedding. The health status of each mouse was monitored daily using a quantitative scoring system (75). Cages (including bedding, food, and water) were replaced weekly. Initial body weights were taken a day before the initial enema and gavage inoculations and again at the time of euthanization. Overall weight gain and feed consumption were measured.
Butyrate administration.
A stock solution of butyric acid (Sigma-Aldrich, Oakville, Ontario, Canada) (>99%; molecular mass, 88.11 g/mol; 100 ml) was diluted with 1× phosphate-buffered saline (PBS) (Sigma-Aldrich) (0.01 M NaH2PO4, 0.04 M Na2HPO4, 0.07 M NaCl; pH 7.4) to attain final concentrations of 80 mM, 100 mM, and 140 mM butyrate; the pH was adjusted to 7.4 ± 0.2 with 10 M sodium hydroxide. The butyrate solution was prepared the day prior to administration and was stored at 4°C until use. Solutions were warmed to room temperature (RT) for 30 min before administration. Phosphate-buffered saline (PBS) served as the butyrate control treatment. Butyrate treatments were administered via enemas (300 µl) at 2-day intervals throughout the experimental period. For administration of enemas, mice were inverted at a 45° angle, and a 22-gauge-by-2.5-cm-long gavage needle with a 1.25-mm ball tip was gently inserted into the colon, the liquid was slowly injected, and mice were maintained in an inverted position for 30 s after administration of the enema. Animals were monitored for discomfort/pain for 4 h after the enemas were administered.
Citrobacter rodentium inoculation.
Green fluorescent protein-labeled C. rodentium DBS100 (ATCC 51459) was used to incite acute inflammation. The bacterium was grown aerobically on lysogeny broth agar (LA) with 30 µg/ml chloramphenicol at 37°C for 24 h. To differentiate GFP-labeled C. rodentium from nonlabeled C. rodentium, a chloramphenicol resistance gene was incorporated into the genome, which required chloramphenicol to be used for growth and isolation techniques. Biomass was removed from the surface of the agar and transferred into sterile lysogeny broth (LB) containing 15 µg/ml chloramphenicol (Sigma-Aldrich). Cultures were maintained for 2 h at 37°C at 100 rpm until an optical density at 600 nm (OD600) of >0.1 was obtained. Cultures were centrifuged at 2,256 × g for 15 min, supernatants were removed, and C. rodentium cells were resuspended in 3.0 ml PBS. To confirm densities of viable cells, inoculum was diluted in a 10-fold dilution series and 100 µl of each dilution was spread in duplicate onto LA. Cultures were incubated at 37°C, and the number of C. rodentium colonies was counted at the dilution yielding 30 to 300 CFU after 24 h. Cell densities were adjusted to 3 × 109 CFU/ml with PBS. For each replicate, 12 mice were administered C. rodentium cells and PBS (100 µl) or PBS alone (100 µl) via gavage on two consecutive days using a 22-gauge-by-2.5-cm-long gavage needle with a 1.25-mm ball tip.
Quantification of C. rodentium in feces.
Fecal samples from mice were collected at ∼7-day intervals and homogenized in 1.0 ml LB, the homogenate was diluted in a 10-fold dilution series, and 100-µl aliquots of each dilution were spread in duplicate on MacConkey agar (Becton, Dickinson and Company, Mississauga, Ontario, Canada) containing 15 µg/ml chloramphenicol (Sigma-Aldrich). Cultures were incubated at 37°C for 24 h, and colonies of C. rodentium were enumerated at the dilution yielding 30 to 300 CFU. To confirm the identity of C. rodentium, colony PCR was performed on arbitrarily selected colonies. The EspB protein is responsible for C. rodentium attachment to host membranes to cause infection, and this effector molecule was chosen as a target gene for identification (76). Primers specific for the espB gene (CrodF [5′-GCTTCTGCGAAGTCTGTCAA-3′] and CrodR [5′-CAGTAAAGCGACTTAACAGATT-3′]) were used to confirm the identity of C. rodentium isolates (36). PCR conditions commenced with one cycle of 15 min at 95°C, followed by 35 cycles of 45 s at 95°C, 1 min at 57°C, and 1 min at 72°C and a final cycle of 5 min at 72°C. The amplicon was 270 bp in size.
Densities of C. rodentium were also enumerated in feces on days 14, 21, and 28 p.i. using SYBR green quantitative PCR (qPCR). Genomic DNA from feces (200 mg ± 5 mg) was extracted using a QIAamp DNA stool minikit (Qiagen Inc.) according to the manufacturer’s recommendation. The primers targeting the espB gene described above were used. PCR was conducted using an Mx 3005p thermocycler (Agilent Technologies Canada Inc., Mississauga, Ontario, Canada). The conditions for amplification were 1 cycle at 95°C for 15 min followed by 40 cycles of 15 s at 94°C, 30 s at 57°C, and 30 s at 72°C for data acquisition. Melt curve analysis was conducted over a range of 55 to 95°C, with increments set at 0.5°C (80 cycles). A linear equation established from genomic DNA extracted from C. rodentium bacteria of known densities was used to interpolate the numbers of copies present in the unknown samples. In all instances, each sample was run twice (i.e., two subsamples), and the mean value was used.
Animal euthanization and intestinal sample collection.
On days 14, 21, and 28 p.i., one randomly selected mouse from each treatment was anesthetized with isoflurane followed by euthanasia by cervical dislocation under anesthesia. Immediately after death, a mid-line laparotomy was used to exteriorize the intestine, and a gross pathological assessment of the intestine was completed. The colon was longitudinally incised, and digesta was collected and stored at −20°C for analysis of SCFAs. Sections of distal colon (∼4 mm in length) were weighed and placed at −20°C for DNA analyses and in RNAlater (Qiagen, Toronto, Ontario, Canada) at −20°C for mRNA extraction. Tissue from the distal colon was also collected for histopathologic and mucus analyses, as well as for fluorescent in situ hybridization (FISH).
Histopathology.
Harvested colonic tissue was fixed in Surgipath 10% neutral buffered formalin (Leica Biosystems, Concord, Ontario, Canada) for 24 h. Formalin-fixed tissues were dehydrated in ethanol and placed in Histo-Clear (Diamed Lab Supplies, Mississauga, Ontario, Canada) prior to embedding in paraffin at 60°C. Sections (5 µm) were deparaffinized with xylene and stained with hematoxylin and eosin (H&E). Tissues were scored for mucosal damage by an experienced veterinary pathologist blind to the treatments using an established scoring guide (34) that ranked common characteristics of mucosal damage at scores from 0 to 4, with 4 representing pronounced damage and 0 representing minimal to no damage (see Table S1 in the supplemental material). Sections were also scored (0 to 3 or 4) for epithelial cell wall hyperplasia based on a mild to severe increase in the number of cells found within crypt columns; for crypt height based on minimal to maximal increases in height; for epithelium cell injury (noting the degree of focal erosions and cell shedding); for the degree of inflammation based on the number of neutrophils and mononuclear cells present in the lamina propria; for goblet cell depletion based on the number of goblet cells and mucin droplet size; and for the degree of mitotic activity based on how much of the epithelial cell displayed increased activity (Table S1).
Description of the histological scoring criteria applied to cross sections from the murine distal colon. Download TABLE S1, PDF file, 0.1 MB (114.4KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Short-chain fatty acid analysis.
To quantify SCFA levels, fecal pellets were collected and stored on ice until they were weighed (within 30 min of collection). After the weight was calculated, samples were homogenized in PBS at a 1:9 (wt/vol) ratio. Meta-phosphoric acid (Sigma-Aldrich) was added to the homogenate at a 1:4 (vol/vol) ratio, and the reaction mixture was incubated at RT for 30 min. Samples were then centrifuged for 75 min at 16,100 × g, and the supernatants were collected and stored at −20°C. Acetate, butyrate, and propionate concentrations were quantified with a gas chromatograph (model 6890 N with 7683 series injector; Agilent Technologies, Mississauga, Ontario, Canada) according to an established protocol (77, 78).
Characterization of digesta- and mucosa-associated bacterial communities.
Mucosa-associated bacterial genomic DNA was extracted from distal colonic samples using a DNeasy blood and tissue extraction kit (Qiagen Inc.). Genomic DNA was also extracted from the distal colonic digesta using a QIAamp Fast DNA stool extraction kit (Qiagen Inc.). Extracted DNA was processed using an Illumina protocol for creating 16S rRNA gene metagenomic sequencing libraries (79). Extracted DNA was normalized to 5 ng/µl in 10 mM Tris (pH 8.5). Following this, 2.5 µl of purified DNA was PCR amplified with 5 µl of each amplicon primer, spanning the V3 and V4 regions of the 16S rRNA gene (F [5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′] and R [5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′]), and 12.5 µl of 2× Kapa HotStart Ready mix (Kapa Biosystems, Inc., Wilmington, MA) for a final volume of 25 µl. The resulting 550-bp product underwent a PCR cleanup using AMPure XP beads (Beckman Coulter Canada Inc., Mississauga, Ontario, Canada) on a magnetic stand to isolate the DNA, and the product was washed with 80% ethanol and eluted with 10 mM Tris (pH 8.5). An indexing PCR was used to add forward and reverse indices to each sample. Conditions included 5 µl of DNA, 5 µl of each index primer (specific nonrepeating pair per sample), and 25 µl of 2× Kapa HiFi HotStart Ready mix and nuclease-free water (Qiagen Inc.) added to reach a final volume of 50 µl per sample. A final PCR cleanup was performed on the 630-bp product. Indexed DNA libraries were quantified and normalized to 4 nM with 10 mM Tris (pH 8.5), and 5 µl of each normalized library was pooled into one sample for sequencing using a MiSeq system (Illumina, San Diego, CA). A PhiX control was run in parallel with the normalized DNA libraries, and both were denatured and diluted to 4 pM prior to loading onto the MiSeq cartridge.
Forward reads were assembled using the Quantitative Insights Into Microbial Ecology (QIIME; version 1.8.0) software package (80), resulting in a total of 13,125,994 sequences. Barcodes were extracted from each sample FASTQ file, and each was joined with its corresponding forward read. Libraries were split according to barcode, and sequences were filtered to include only those sequences with a base-calling-accuracy Phred value (Q) of 20 or greater, indicating the probability of 1 in 100 base calls being incorrect. Sequence reads were filtered to exclude those with more than three consecutive low-quality base calls, those with less than 75% of the read length containing consecutive high-quality base calls, and/or those corresponding to barcode having more than 1.5 errors present. These sequences (n = 5,390,077) were then subjected to chimera checking using USEARCH 6.1 software, and the resulting chimeras were filtered out prior to picking of operational taxonomic units (OTU) from the Greengene reference database. In total, 371,065 OTUs were identified using a 97% similarity parameter, and the most common sequence was used to define the groups of similar OTUs. OTUs were then aligned to the Ribosomal Database project (RDP) with a classifier value of 0.5 (sequences having at least 50% similarity to reference database sequences) using the NAST algorithm (81). Taxonomies were assigned to each sequence cluster using UCLUST (82) and classified using the Greengenes reference database (83). An OTU table was produced, and data from all samples were rarified such that 3,450 OTUs were randomly chosen and compared between samples for analysis (the number of OTUs per biological sample ranged from 3,450 to 100,000). Diversity among species (β-diversity) was examined using Bray-Curtis analysis and weighted and unweighted UniFrac analyses (84).
Visualization of intestinal bacteria.
The presence and localization of bacteria within the colon were determined using FISH. Preparation of colonic samples for FISH was performed using the method described below for AB-PAS staining. Tissue sections were circled with a hydrophobic pen, and sections were incubated in the dark overnight at 37°C with either the Alexa Fluor 555-conjugated total bacterial probe EUB338 (Life Technologies, Inc., Burlington, Ontario, Canada) (5′-GCTGCCTCCCGTAGGAGT-3′) or the Alexa Fluor 555-conjugated Gammaproteobacteria probe Gam42a (Life Technologies, Inc.) (5′-GCCTTCCCACATCGTTT-3′). Probes were stored in 0.25 µg/µl stocks and diluted (1:100) with hybridization buffer (0.9 M NaCl, 0.1 M Tris [pH 7.2], 30% formamide, 0.1% SDS) prior to addition of the solutions to the sections. The sections were stored in a dark humidifying box. After incubation with the fluorescent probe, sections were washed with hybridization buffer in the dark for 15 min and then with wash buffer (0.9 M NaCl, 0.1 M Tris [pH 7.2]). Sections were mounted with ProLong Gold Antifade solution with DAPI (4′,6-diamidino-2-phenylindole) (44) and analyzed with a Zeiss Axioskop II Plus microscope (Carl Zeiss Canada, Ltd., North York, Ontario, Canada) using Zen2 (Blue edition) core imaging software.
Quantification of gene expression.
Gene expression (Table S2) was analyzed for cytokine profiles from total RNA that was extracted using an RNeasy minikit (Qiagen Inc.). The concentration and quality of the total RNA extracted were analyzed using an RNA 600 Nano Chip and a 2100 Bioanalyzer (Agilent Technologies). Using 1,000 ng of total RNA, reverse transcription was performed using a QuantiTect reverse transcription kit (Qiagen Inc.). The reference genes used to normalize the measured threshold cycle (CT) values were Ppia, Hprt, and GusB. Quantitech SYBR green Mastermix (Qiagen Inc.) was used as an indicator of double-stranded DNA and product amplification. Individual PCRs consisted of 1 µl of cDNA; 3 µl of nuclease-free water (Qiagen Inc.); 0.5 µl of 10 µM forward primer; 0.5 µl of 10 µM reverse primer; and 5 µl SYBR green (Table S2). Reactions were run in triplicate per cDNA sample. Quantitative PCRs were run on a 384-well ABI 7900HT qPCR thermocycler (Life Technologies, Inc.), with an activation step of 95°C for 15 min and 40 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s, followed by melt curve analysis. Normalized gene expression data were calculated using qbasePLUS (Biogazelle, Zwijnaarde, Belgium) on the basis of geNorm and qBase quantification models (85, 86).
Targets and primer sequences used to analyze the gene expression of cDNA harvested from murine colonic tissue. Download TABLE S2, PDF file, 1 MB (1,000.9KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of mucus.
Colonic tissue samples were collected from the same region of the distal colon from each sampled mouse and were fixed overnight in methacarn (60% methanol, 30% chloroform, 10% glacial acetic acid) (87) prior to dehydration with ethanol and Histo-Clear (Diamed Lab Supplies). Sections (5 µm) were deparaffinized for 5 min on a 60°C heating bed, cleared with xylene, and rehydrated in a decreasing ethanol gradient (100%, 90%, 70%, and 50%) according to a standard protocol (Abcam, Inc., Toronto, Ontario, Canada). To visualize mucus, sections were stained with alcian blue (American MasterTech, Lodi, CA) (pH 2.5) for 30 min, 0.5% periodic acid (American MasterTech, Lodi, CA) for 5 min, and Schiff’s solution (American MasterTech, Lodi, CA) and were stored at 4°C for 15 min (AB-PAS).
Statistical analyses.
Most statistical analyses were performed using SAS (SAS Institute, Cary, NC). Continuous data were checked for normality and analyzed using the MIXED procedure of SAS (SAS Institute, Cary, NC). Where applicable (i.e., for samples were not independent), collection time was treated as a repeated measure; the appropriate covariance structure was utilized according to the lowest Akaike’s information criterion. In the event of a significant main effect, the least-squares method (LSM) was used to compare treatments within factors. Categorical data (i.e., histopathology results) were analyzed using the GLIMMIX procedure. For bacterial community analyses, both SAS and Primer 7 were used. In Primer 7, PERMANOVA (permutational multivariate analysis of variance) and principal-coordinate analyses (PCoA) were used to determine levels of β-diversity whereas analysis of variance (MIXED procedure) was used with a protected LSM test to determine levels of α-diversity. Differences in the abundances of microbial OTUs were also analyzed using the MIXED procedure with a protected LSM test. For both parametric and nonparametric statistical analyses, P values of ≤0.050 were considered to represent statistical significance, whereas P values of >0.050 and ≤0.100 were considered to represent a statistically significant trend.
ACKNOWLEDGMENTS
We thank the following individuals at Agriculture and Agri-Food Canada Lethbridge Research and Development Centre: Tara Shelton for overseeing and assisting with all aspects of the animal component of the study; Jasmin Teske for her assistance with sample collection and animal husbandry; Darrell Vedres for quantifying SCFAs; Dallas Thomas, Kirsty Brown, and Devin Holman for their assistance with the analysis of bacterial community data; and Toby Entz for experimental design and statistical advice. We also thank Martin Kalmokoff (Agriculture and Agri-Food Canada, Kentville, Nova Scotia, Canada) and Stephen Brooks (Health Canada, Ottawa, Ontario, Canada) for their advice on various aspects of the project and Bruce Vallance (University of British Columbia, Vancouver, British Columbia, Canada) for providing the GFP-labeled C. rodentium strain used in the study.
This work was supported by Alberta Innovates Bio Solutions and General Mills (QFH-11-013 to G.D.I., R.R.E.U., and D.W.A.).
REFERENCES
- 1.Lin HV, Frassetto A, Kowalik EJ Jr, Nawrocki AR, Lu MM, Kosinski JR, Hubert JA, Szeto D, Yao X, Forrest G, Marsh DJ. 2012. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One 7:e35240. doi: 10.1371/journal.pone.0035240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cummings JH. 1984. Colonic absorption: the importance of short chain fatty acids in man. Scand J Gastroenterol Suppl 93:89–99. [PubMed] [Google Scholar]
- 4.Leonel AJ, Alvarez-Leite JI. 2012. Butyrate: implications for intestinal function. Curr Opin Clin Nutr Metab Care 15:474–479. doi: 10.1097/MCO.0b013e32835665fa. [DOI] [PubMed] [Google Scholar]
- 5.Barcelo A, Claustre J, Moro F, Chayvialle JA, Cuber JC, Plaisancié P. 2000. Mucin secretion is modulated by luminal factors in the isolated vascularly perfused rat colon. Gut 46:218–224. doi: 10.1136/gut.46.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaudier E, Rival M, Buisine MP, Robineau I, Hoebler C. 2009. Butyrate enemas upregulate Muc genes expression but decrease adherent mucus thickness in mice colon. Physiol Res 58:111–119. [DOI] [PubMed] [Google Scholar]
- 7.Pacheco RG, Esposito CC, Müller LC, Castelo-Branco MT, Quintella LP, Chagas VL, de Souza HS, Schanaider A. 2012. Use of butyrate or glutamine in enema solution reduces inflammation and fibrosis in experimental diversion colitis. World J Gastroenterol 18:4278–4287. doi: 10.3748/wjg.v18.i32.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimotoyodome A, Meguro S, Hase T, Tokimitsu I, Sakata T. 2000. Short chain fatty acids but not lactate or succinate stimulate mucus release in the rat colon. Comp Biochem Physiol A Mol Integr Physiol 125:525–531. doi: 10.1016/S1095-6433(00)00183-5. [DOI] [PubMed] [Google Scholar]
- 9.D’Argenio G, Cosenza V, Delle Cave M, Iovino P, Delle Valle N, Lombardi G, Mazzacca G. 1996. Butyrate enemas in experimental colitis and protection against large bowel cancer in a rat model. Gastroenterology 110:1727–1734. doi: 10.1053/gast.1996.v110.pm8964397. [DOI] [PubMed] [Google Scholar]
- 10.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. 2008. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 11.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 12.Hamer HM, Jonkers DM, Renes IB, Vanhoutvin SA, Kodde A, Troost FJ, Venema K, Brummer RJ. 2010. Butyrate enemas do not affect human colonic MUC2 and TFF3 expression. Eur J Gastroenterol Hepatol 22:1134–1140. doi: 10.1097/MEG.0b013e32833a6ca0. [DOI] [PubMed] [Google Scholar]
- 13.Russo I, Luciani A, De Cicco P, Troncone E, Ciacci C. 2012. Butyrate attenuates lipopolysaccharide-induced inflammation in intestinal cells and Crohn’s mucosa through modulation of antioxidant defense machinery. PLoS One 7:e32841. doi: 10.1371/journal.pone.0032841. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Fung KY, Cosgrove L, Lockett T, Head R, Topping DL. 2012. A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrate. Br J Nutr 108:820–831. doi: 10.1017/S0007114512001948. [DOI] [PubMed] [Google Scholar]
- 15.Plöger S, Stumpff F, Penner GB, Schulzke JD, Gäbel G, Martens H, Shen Z, Günzel D, Aschenbach JR. 2012. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci 1258:52–59. doi: 10.1111/j.1749-6632.2012.06553.x. [DOI] [PubMed] [Google Scholar]
- 16.Zimmerman MA, Singh N, Martin PM, Thangaraju M, Ganapathy V, Waller JL, Shi H, Robertson KD, Munn DH, Liu K. 2012. Butyrate suppresses colonic inflammation through HDAC1-dependent Fas upregulation and Fas-mediated apoptosis of T cells. Am J Physiol Gastrointest Liver Physiol 302:G1405–G1415. doi: 10.1152/ajpgi.00543.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, Calignano A. 2011. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol 17:1519–1528. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hýzd’alová M, Hofmanová J, Pacherník J, Vaculová A, Kozubík A. 2008. The interaction of butyrate with TNF-alpha during differentiation and apoptosis of colon epithelial cells: role of NF-kappaB activation. Cytokine 44:33–43. doi: 10.1016/j.cyto.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Usami M, Kishimoto K, Ohata A, Miyoshi M, Aoyama M, Fueda Y, Kotani J. 2008. Butyrate and trichostatin A attenuate nuclear factor kappaB activation and tumor necrosis factor alpha secretion and increase prostaglandin E2 secretion in human peripheral blood mononuclear cells. Nutr Res 28:321–328. doi: 10.1016/j.nutres.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Hofmanová J, Straková N, Vaculová AH, Tylichová Z, Safaříková B, Skender B, Kozubík A. 2014. Interaction of dietary fatty acids with tumour necrosis factor family cytokines during colon inflammation and cancer. Mediators Inflamm 2014:848632. doi: 10.1155/2014/848632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Wang F, Luo H, Liu A, Li K, Li C, Jiang Y. 2016. Protective effect of butyrate against ethanol-induced gastric ulcers in mice by promoting the anti-inflammatory, anti-oxidant and mucosal defense mechanisms. Int Immunopharmacol 30:179–187. doi: 10.1016/j.intimp.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 22.Song M, Xia B, Li J. 2006. Effects of topical treatment of sodium butyrate and 5-aminosalicylic acid on expression of trefoil factor 3, interleukin 1beta, and nuclear factor kappaB in trinitrobenzene sulphonic acid induced colitis in rats. Postgrad Med J 82:130–135. doi: 10.1136/pgmj.2005.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guilloteau P, Martin L, Eeckhaut V, Ducatelle R, Zabielski R, Van Immerseel F. 2010. From the gut to the peripheral tissues: the multiple effects of butyrate. Nutr Res Rev 23:366–384. doi: 10.1017/S0954422410000247. [DOI] [PubMed] [Google Scholar]
- 24.Atuma C, Strugala V, Allen A, Holm L. 2001. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol 280:G922–G929. [DOI] [PubMed] [Google Scholar]
- 25.Pelaseyed T, Bergström JH, Gustafsson JK, Ermund A, Birchenough GM, Schütte A, van der Post S, Svensson F, Rodríguez-Piñeiro AM, Nyström EE, Wising C, Johansson ME, Hansson GC. 2014. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev 260:8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson ME, Larsson JM, Hansson GC. 2011. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A 108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belzer C, de Vos WM. 2012. Microbes inside--from diversity to function: the case of Akkermansia. ISME J 6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson BR, O’Rourke JL, Neilan BA, Vandamme P, On SL, Fox JG, Lee A. 2005. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol 55:1199–1204. doi: 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]
- 29.Wrzosek L, Miquel S, Noordine ML, Bouet S, Joncquel Chevalier-Curt M, Robert V, Philippe C, Bridonneau C, Cherbuy C, Robbe-Masselot C, Langella P, Thomas M. 2013. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol 11:61. doi: 10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez I, Kim J, Duffy PR, Schlegel VL, Walter J. 2010. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One 5:e15046. doi: 10.1371/journal.pone.0015046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwab C, Berry D, Rauch I, Rennisch I, Ramesmayer J, Hainzl E, Heider S, Decker T, Kenner L, Müller M, Strobl B, Wagner M, Schleper C, Loy A, Urich T. 2014. Longitudinal study of murine microbiota activity and interactions with the host during acute inflammation and recovery. ISME J 8:1101–1114. doi: 10.1038/ismej.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen TS, Lærke HN, Theil PK, Sørensen JF, Saarinen M, Forssten S, Knudsen KE. 2014. Diets high in resistant starch and arabinoxylan modulate digestion processes and SCFA pool size in the large intestine and faecal microbial composition in pigs. Br J Nutr 112:1837–1849. doi: 10.1017/S000711451400302X. [DOI] [PubMed] [Google Scholar]
- 33.Kumar A, Alrefai WA, Borthakur A, Dudeja PK. 2015. Lactobacillus acidophilus counteracts enteropathogenic E. coli-induced inhibition of butyrate uptake in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 309:G602–G607. doi: 10.1152/ajpgi.00186.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa E, Uwiera RR, Kastelic JP, Selinger LB, Inglis GD. 2011. Non-therapeutic administration of a model antimicrobial growth promoter modulates intestinal immune responses. Gut Pathog 3:14. doi: 10.1186/1757-4749-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhinder G, Sham HP, Chan JM, Morampudi V, Jacobson K, Vallance BA. 2013. The Citrobacter rodentium mouse model: studying pathogen and host contributions to infectious colitis. J Vis Exp 2013:e50222. doi: 10.3791/50222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiminez JA, Uwiera TC, Abbott DW, Uwiera RRE, Inglis GD. 2016. Impacts of resistant starch and wheat bran consumption on enteric inflammation in relation to colonic bacterial community structures and short-chain fatty acid concentrations in mice. Gut Pathog 8:67. doi: 10.1186/s13099-016-0149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boets E, Deroover L, Houben E, Vermeulen K, Gomand SV, Delcour JA, Verbeke K. 2015. Quantification of in vivo colonic short chain fatty acid production from inulin. Nutrients 7:8916–8929. doi: 10.3390/nu7115440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. 2014. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 39.Weaver CT, Elson CO, Fouser LA, Kolls JK. 2013. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol 8:477–512. doi: 10.1146/annurev-pathol-011110-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luperchio SA, Schauer DB. 2001. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect 3:333–340. doi: 10.1016/S1286-4579(01)01387-9. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Friedrich C, Hagemann SC, Korte WH, Goharani N, Cording S, Eberl G, Sparwasser T, Lochner M. 2014. Regulatory T cells promote a protective Th17-associated immune response to intestinal bacterial infection with C. rodentium. Mucosal Immunol 7:1290–1301. doi: 10.1038/mi.2014.17. [DOI] [PubMed] [Google Scholar]
- 42.Kojouharoff G, Hans W, Obermeier F, Männel DN, Andus T, Schölmerich J, Gross V, Falk W. 1997. Neutralization of tumour necrosis factor (TNF) but not of IL-1 reduces inflammation in chronic dextran sulphate sodium-induced colitis in mice. Clin Exp Immunol 107:353–358. doi: 10.1111/j.1365-2249.1997.291-ce1184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins JW, Keeney KM, Crepin VF, Rathinam VA, Fitzgerald KA, Finlay BB, Frankel G. 2014. Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol 12:612–623. doi: 10.1038/nrmicro3315. [DOI] [PubMed] [Google Scholar]
- 44.Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, Chadee K, Vallance BA. 2010. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog 6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knodler LA, Bestor A, Ma C, Hansen-Wester I, Hensel M, Vallance BA, Steele-Mortimer O. 2005. Cloning vectors and fluorescent proteins can significantly inhibit Salmonella enterica virulence in both epithelial cells and macrophages: implications for bacterial pathogenesis studies. Infect Immun 73:7027–7031. doi: 10.1128/IAI.73.10.7027-7031.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanoue T, Honda K. 2015. Regulation of intestinal Th17 and Treg cells by gut microbiota. Inflamm Regen 35:99–105. doi: 10.2492/inflammregen.35.099. [DOI] [Google Scholar]
- 47.Neufert C, Pickert G, Zheng Y, Wittkopf N, Warntjen M, Nikolaev A, Ouyang W, Neurath MF, Becker C. 2010. Activation of epithelial STAT3 regulates intestinal homeostasis. Cell Cycle 9:652–655. doi: 10.4161/cc.9.4.10615. [DOI] [PubMed] [Google Scholar]
- 48.Mizoguchi A. 2012. Healing of intestinal inflammation by IL-22. Inflamm Bowel Dis 18:1777–1784. doi: 10.1002/ibd.22929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang M, Zhou Q, Dorfman RG, Huang X, Fan T, Zhang H, Zhang J, Yu C. 2016. Butyrate inhibits interleukin-17 and generates Tregs to ameliorate colorectal colitis in rats. BMC Gastroenterol 16:84. doi: 10.1186/s12876-016-0500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 51.Saleh M, Trinchieri G. 2011. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat Rev Immunol 11:9–20. doi: 10.1038/nri2891. [DOI] [PubMed] [Google Scholar]
- 52.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. 2011. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Büller HA, Dekker J, Van Seuningen I, Renes IB, Einerhand AW. 2006. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 55.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 57.Bergstrom KS, Xia L. 2013. Mucin-type O-glycans and their roles in intestinal homeostasis. Glycobiology 23:1026–1037. doi: 10.1093/glycob/cwt045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kles KA, Chang EB. 2006. Short-chain fatty acids impact on intestinal adaptation, inflammation, carcinoma, and failure. Gastroenterology 130:S100–S105. doi: 10.1053/j.gastro.2005.11.048. [DOI] [PubMed] [Google Scholar]
- 59.Hatayama H, Iwashita J, Kuwajima A, Abe T. 2007. The short chain fatty acid, butyrate, stimulates MUC2 mucin production in the human colon cancer cell line, LS174T. Biochem Biophys Res Commun 356:599–603. doi: 10.1016/j.bbrc.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 60.Gaudier E, Jarry A, Blottière HM, de Coppet P, Buisine MP, Aubert JP, Laboisse C, Cherbut C, Hoebler C. 2004. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am J Physiol Gastrointest Liver Physiol 287:G1168–GG1174. doi: 10.1152/ajpgi.00219.2004. [DOI] [PubMed] [Google Scholar]
- 61.Nair MG, Guild KJ, Du Y, Zaph C, Yancopoulos GD, Valenzuela DM, Murphy A, Stevens S, Karow M, Artis D. 2008. Goblet cell-derived resistin-like molecule beta augments CD4+ T cell production of IFN-gamma and infection-induced intestinal inflammation. J Immunol 181:4709–4715. doi: 10.4049/jimmunol.181.7.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim YS, Ho SB. 2010. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep 12:319–330. doi: 10.1007/s11894-010-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kindon H, Pothoulakis C, Thim L, Lynch-Devaney K, Podolsky DK. 1995. Trefoil peptide protection of intestinal epithelial barrier function: cooperative interaction with mucin glycoprotein. Gastroenterology 109:516–523. doi: 10.1016/0016-5085(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 64.Artis D, Wang ML, Keilbaugh SA, He W, Brenes M, Swain GP, Knight PA, Donaldson DD, Lazar MA, Miller HR, Schad GA, Scott P, Wu GD. 2004. RELMbeta/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proc Natl Acad Sci U S A 101:13596–13600. doi: 10.1073/pnas.0404034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krimi RB, Kotelevets L, Dubuquoy L, Plaisancié P, Walker F, Lehy T, Desreumaux P, Van Seuningen I, Chastre E, Forgue-Lafitte ME, Marie JC. 2008. Resistin-like molecule beta regulates intestinal mucous secretion and curtails TNBS-induced colitis in mice. Inflamm Bowel Dis 14:931–941. doi: 10.1002/ibd.20420. [DOI] [PubMed] [Google Scholar]
- 66.Hogan SP, Seidu L, Blanchard C, Groschwitz K, Mishra A, Karow ML, Ahrens R, Artis D, Murphy AJ, Valenzuela DM, Yancopoulos GD, Rothenberg ME. 2006. Resistin-like molecule β regulates innate colonic function: barrier integrity and inflammation susceptibility. J Allergy Clin Immunol 118:257–268. doi: 10.1016/j.jaci.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bergstrom KSB, Morampudi V, Chan JM, Bhinder G, Lau J, Yang H, Ma C, Huang T, Ryz N, Sham HP, Zarepour M, Zaph C, Artis D, Nair M, Vallance BA. 2015. Goblet cell derived RELM-β recruits CD4+ T cells during infectious colitis to promote protective intestinal epithelial cell proliferation. PLoS Pathog 11:e1005108. doi: 10.1371/journal.ppat.1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoffmann C, Hill DA, Minkah N, Kirn T, Troy A, Artis D, Bushman F. 2009. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infect Immun 77:4668–4678. doi: 10.1128/IAI.00493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meehan CJ, Beiko RG. 2014. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol 6:703–713. doi: 10.1093/gbe/evu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Derrien M, Van Baarlen P, Hooiveld G, Norin E, Müller M, de Vos WM. 2011. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol 2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Núñez G, Martens EC. 2016. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167:1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takao M, Yen H, Tobe T. 2014. LeuO enhances butyrate-induced virulence expression through a positive regulatory loop in enterohaemorrhagic Escherichia coli. Mol Microbiol 93:1302–1313. doi: 10.1111/mmi.12737. [DOI] [PubMed] [Google Scholar]
- 74.Franzin FM, Sircili MP. 2015. Locus of enterocyte effacement: a pathogenicity island involved in the virulence of enteropathogenic and enterohemorragic Escherichia coli subjected to a complex network of gene regulation. Biomed Res Int 2015:534738. doi: 10.1155/2015/534738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Council NR. 2009. Recognition and alleviation of pain in laboratory animals. The National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 76.Newman JV, Zabel BA, Jha SS, Schauer DB. 1999. Citrobacter rodentium espB is necessary for signal transduction and for infection of laboratory mice. Infect Immun 67:6019–6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Playne MJ. 1985. Determination of ethanol, volatile fatty acids, lactic and succinic acids in fermentation liquids by gas chromatography. J Sci Food Agric 36:638–644. doi: 10.1002/jsfa.2740360803. [DOI] [Google Scholar]
- 78.Cottyn BG, Boucque CV. 1968. Rapid method for the gas-chromatographic determination of volatile fatty acids in rumen fluid. J Agric Food Chem 16:105–107. doi: 10.1021/jf60155a002. [DOI] [Google Scholar]
- 79.Illumina 2013, posting date 16S metagenomic sequencing library preparation. Illumina, San Diego, CA: https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html. [Google Scholar]
- 80.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 83.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. 2011. UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johansson ME, Hansson GC. 2012. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Methods Mol Biol 842:229–235. doi: 10.1007/978-1-61779-513-8_13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Densities of C. rodentium in feces from mice inoculated with the bacterium and rectally administered PBS (BU0), or butyrate at a concentration of 80 mM (BU80), or 140 mM (BU140) from the point of inoculation to day 28 postinoculation. No C. rodentium was isolated from feces of mice that were not inoculated with the bacterium. Download FIG S1, PDF file, 0.1 MB (107.5KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mitotic cell activity in mice inoculated with C. rodentium (CR+) and rectally administered PBS (BU0), or butyrate at a concentration of 80 mM (BU80), or 140 mM (BU140). Vertical lines associated with histogram bars represent standard errors of the means (n = 4). *, P ≤ 0.050 (relative to the BU0 treatment averaged over the indicated time points). In all instances, infection with C. rodentium affected mitotic cell activity over time (P ≤ 0.005). Download FIG S2, PDF file, 0.1 MB (80.8KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mucus localization in alcian blue–periodic acid-Schiff-stained sections of the distal colons of mice inoculated with PBS (CR−) or C. rodentium (CR+), and rectally administered PBS (BU0) or butyrate at a concentration of 140 mM (BU140) at day 21 p.i. (A) CR−, BU0. (B) CR+, BU0. (C) CR−, BU140. (D) CR+, BU+. Tissue from infected mice supplemented with butyrate exhibited an increase in the density of mucus (blue stain) within the colonic lumen. Overall, more mucus was observed in the lumen on day 21 p.i. than on day 14 p.i. Bar, 100 μm. Download FIG S3, PDF file, 0.6 MB (680.3KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Principal-coordinate analysis of bacterial communities within digesta and associated with mucosa in the distal colon from mice inoculated with PBS via gavage, and rectally administered PBS (BU0) or 140 mM butyrate (BU140). Ellipsoids show clustering of communities by sample type. Download FIG S4, PDF file, 0.1 MB (138.6KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proportion of total sequences within the Pseudomonadales order within digesta collected from the distal colon. Vertical lines associated with histogram bars represent standard errors of the means (n = 8). *, P ≤ 0.050 (for treatments linked by the horizontal lines). Download FIG S5, PDF file, 0.1 MB (96.5KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Description of the histological scoring criteria applied to cross sections from the murine distal colon. Download TABLE S1, PDF file, 0.1 MB (114.4KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Targets and primer sequences used to analyze the gene expression of cDNA harvested from murine colonic tissue. Download TABLE S2, PDF file, 1 MB (1,000.9KB, pdf) .
© Crown copyright 2017.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.