Abstract
During C4 photosynthesis, CO2 is concentrated around the enzyme RuBisCO. The net effect is to reduce photorespiration while increasing water and nitrogen use efficiencies. Species that use C4 photosynthesis have evolved independently from their C3 ancestors on more than 60 occasions. Along with mimicry and the camera-like eye, the C4 pathway therefore represents a remarkable example of the repeated evolution of a highly complex trait. In this review, we provide evidence that the polyphyletic evolution of C4 photosynthesis is built upon pre-existing metabolic and genetic networks. For example, cells around veins of C3 species show similarities to those of the C4 bundle sheath in terms of C4 acid decarboxylase activity and also the photosynthetic electron transport chain. Enzymes of C4 photosynthesis function together in gluconeogenesis during early seedling growth of C3 Arabidopsis thaliana. Furthermore, multiple C4 genes appear to be under control of both light and chloroplast signals in the ancestral C3 state. We, therefore, hypothesize that relatively minor rewiring of pre-existing genetic and metabolic networks has facilitated the recurrent evolution of this trait. Understanding how these changes are likely to have occurred could inform attempts to install C4 traits into C3 crops.
This article is part of the themed issue ‘Enhancing photosynthesis in crop plants: targets for improvement’.
Keywords: evolution, C4 photosynthesis, C3 photosynthesis, C4 protein function, gene regulation
1. Introduction
Photosynthesis has shaped life on the Earth by allowing the energy from sunlight to be harvested and used for the assimilation of carbon dioxide. The process of carbon assimilation via the Calvin–Benson–Bassham cycle [1] requires initial fixation of CO2 by the enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) to form the three-carbon molecule 3-phosphoglycerate (3-PGA). RuBisCO is thought to have evolved in bacteria under anoxic conditions approximately 3.5 billion years ago [2,3]. However, approximately 2.3 billion years ago, the proliferation of oxygenic photosynthetic organisms together with an increase in carbonate deposition due to weathering started to deplete atmospheric CO2 concentrations [3–5]. Today, rather than RuBisCO being saturated by CO2, it is now surrounded by 21% oxygen and only 0.04% CO2. Under these conditions, O2 competitively inhibits the carboxylation reaction of RuBisCO to produce 2-phosphoglycolate (2-PG) [6]. 2-PG is toxic and so is rapidly metabolized to prevent its accumulation [7]. The metabolism of PG is known as photorespiration and is energetically costly, especially at high temperatures when rates of oxygenation increase [8]. It has been proposed that high rates of oxygenation by RuBisCO led to the evolution of increased specificity for CO2, but also that an inescapable trade-off between specificity and the rate of catalysis led to a lower turnover rate [9]. Owing to the relatively low rate of catalysis of RuBisCO, C3 species are associated with significant losses of water via stomata, and large investments in nitrogen are required to produce the amounts of RuBisCO needed to maintain reasonable rates of photosynthesis [10].
It would therefore appear logical for photosynthetic organisms to have been subject to significant selection pressures to decrease rates of oxygenation at the active site of RuBisCO. Although there is considerable natural variation in the activity of RuBisCO [11] in photosynthetic lineages as diverse as the cyanobacteria, algae and land plants, it is thought that low CO2 concentrations before the Anthropocene led to the evolution of carbon concentrating mechanisms. These include the carboxysome in cyanobacteria [12], the pyrenoid in algae and hornworts [13], as well as crassulacean acid metabolism [14] and C4 photosynthesis in angiosperms.
The C4 pathway results from a series of metabolic and structural adjustments to leaves that together concentrate CO2 around RuBisCO. In doing so, photorespiration is reduced, less water is lost per unit of carbon fixed, and considerably lower amounts of RuBisCO and therefore nitrogen are accumulated per unit leaf area [15]. Despite its complexity, the C4 pathway has evolved independently in more than 60 lineages that span 18 plant families [16], making it one of the most remarkable examples of convergent evolution found in biology. It is thought that the evolution of C4 photosynthesis relied on a series of coordinated modifications to leaf anatomy, cell biology and biochemistry [17]. However, the basic components, including enzymes of the C4 pathway, are present in species that use the ancestral C3 pathway [18]. In this review, we summarize our current understanding of the role of C4 proteins in C3 species and the regulation of genes encoding these proteins. From these findings, we propose that rewiring of pre-existing metabolic and genetic networks has facilitated the evolution of this novel metabolic pathway.
2. The biochemistry and evolution of C4 photosynthesis
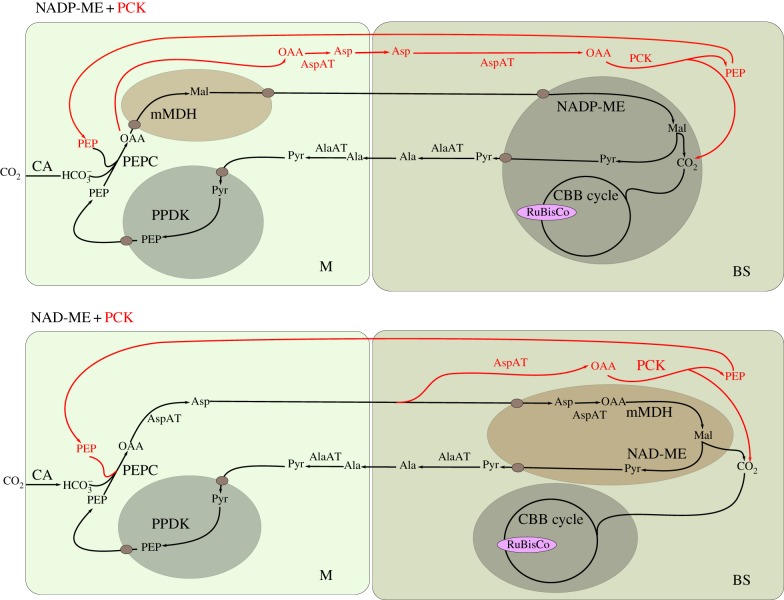
In the majority of C4 plants, CO2 assimilation is divided between mesophyll (M) and bundle sheath (BS) cells [15]. CO2 is first converted to 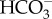 by carbonic anhydrase (CA) and then combined with phosphoenolpyuvate (PEP) by PEP-carboxylase (PEPC) in the M to generate the four-carbon acid oxaloacetate (OAA). Metabolism of OAA to either aspartate or malate is followed by diffusion to the BS where RuBisCO is localized. Decarboxylation of C4 acids typically releases a three-carbon acid and high concentrations of CO2 (figure 1). The three-carbon acid diffuses back to the M where conversion to PEP by pyruvate, orthophosphate dikinase (PPDK) allows the C4 cycle to continue. O2 does not react with PEPC and so CO2 fixation occurs in the absence of oxygenation. Three different C4 acid decarboxylase enzymes are known to operate in the C4 pathway: NADP-dependent malic enzyme (NADP-ME), NAD-dependent malic enzyme (NAD-ME) and PEP-carboxykinase (PCK) (figure 1). Although there is some dominance in the use of individual C4 acid decarboxylases, apparently associated with different C4 lineages, most species use a mixture of the three decarboxylases, the make-up of which varies depending on environmental conditions [19–21].
by carbonic anhydrase (CA) and then combined with phosphoenolpyuvate (PEP) by PEP-carboxylase (PEPC) in the M to generate the four-carbon acid oxaloacetate (OAA). Metabolism of OAA to either aspartate or malate is followed by diffusion to the BS where RuBisCO is localized. Decarboxylation of C4 acids typically releases a three-carbon acid and high concentrations of CO2 (figure 1). The three-carbon acid diffuses back to the M where conversion to PEP by pyruvate, orthophosphate dikinase (PPDK) allows the C4 cycle to continue. O2 does not react with PEPC and so CO2 fixation occurs in the absence of oxygenation. Three different C4 acid decarboxylase enzymes are known to operate in the C4 pathway: NADP-dependent malic enzyme (NADP-ME), NAD-dependent malic enzyme (NAD-ME) and PEP-carboxykinase (PCK) (figure 1). Although there is some dominance in the use of individual C4 acid decarboxylases, apparently associated with different C4 lineages, most species use a mixture of the three decarboxylases, the make-up of which varies depending on environmental conditions [19–21].
Figure 1.
Biochemical subtypes of C4 photosynthesis. Boxes represent the M and BS cells. Chloroplasts are in green and mitochondria brown. NADP-ME, NADP-dependent malic enzyme; PCK, phosphoenolpyruvate carboxykinase; NAD-ME, NAD-dependent malic enzyme; mMDH, mitochondrial malate dehydrogenase; CA, carbonic anhydrase; PEPC, phosphoenolpyuvate-carboxylase; PPDK, pyruvate,orthophosphate dikinase; AspAT, aspartate aminotransferase; RuBisCO, ribulose-1,5 bisphosphate carboxylase/oxygenase; AlaAT, alanine aminotransferase; CBB cycle, Calvin–Benson–Basham cycle; Asp, aspartate; Mal, malate; CO2, carbon dioxide; 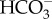 , bicarbonate; PEP, phosphoenolpyruvate; OAA, oxaloacetate; Pyr, pyruvate.
, bicarbonate; PEP, phosphoenolpyruvate; OAA, oxaloacetate; Pyr, pyruvate.
Most estimates suggest that in both monocots and eudicots, the earliest origins of C4 photosynthesis occurred approximately 25–30 Ma during the mid-Oligocene [22,23]. An abrupt reduction in the concentration of atmospheric CO2 during this period is thought to have favoured natural selection for the C4 pathway [2,23]. However, over the next 20–30 Myr, the C4 pathway continued to evolve in other lineages, suggesting that low CO2 concentrations acted as a preconditioning event rather than the sole trigger for C4 evolution [16]. Other factors such as high temperatures, salinity and fire frequency in tropical and subtropical regions have been proposed to contribute to the polyphyletic evolution of C4 photosynthesis [24].
Gene duplication followed by selection or genetic drift are considered important sources for the appearance of new traits [25]. After duplication, most redundant genes tend to be lost as they do not reach sufficient frequencies to become fixed in a population [26,27]. However, those genes that are retained can acquire new functions (neofunctionalization) or mutate to control more than one function (subfunctionalization). Mechanistically, either can occur via changes in cis-regulatory control or through alterations to coding regions resulting in the production of new function [25,28–31]. Gene duplications may therefore have occurred prior to the appearance of the C4 pathway and facilitated its evolution [2,32]. However, until recently the lack of genome sequences for closely related C3 and C4 species precluded accurate assessments of these phenomena, and so evidence for gene duplication followed by neofunctionalization playing a major role in the evolution of core C4 genes was lacking [33–35]. Subsequently, approaches that accurately localized gene duplication events across gene families [36,37] have revealed that in monocotyledons, many C4 cycle genes appear to have duplicated in the last common ancestor of lineages containing C4 plants [38]. There is also evidence that C4 photosynthesis is built on pre-existing components. For example, it makes use of M and BS cells, both of which exist in ancestral C3 leaves. Furthermore, all the enzymes of the C4 pathway identified to date operate in C3 species [18]. Indeed, a number of models depicting evolutionary trajectories from C3 to C4 photosynthesis have been developed in recent years [2,16,39–41]. Although these models take contrasting approaches and focus on slightly different aspects of the C4 system, overall they support the notion that anatomical modifications tended to precede a series of modular changes to metabolic networks that led to evolution of the full C4 pathway. The ancestral role of C4 enzymes in C3 metabolism, from which these evolutionary changes take place, will next be discussed.
3. Characteristics of the C4 pathway in C3 plants
BS cells of C3 species such as rice and barley are capable of carrying out photosynthesis and starch synthesis [42–46]. It is estimated that chloroplasts in BS and M cells of rice contain similar amounts of RuBisCO [47]. Downregulation of chlorophyll synthase in cells associated with the vasculature of C3 Arabidopsis thaliana showed that photosynthetic capacity of these cells makes an important contribution to plant growth and seed production [48]. Thus, although the BS in C3 species is most commonly associated with controlling fluxes of nitrogen, sulfur and water into and out of the leaf [49,50], these results suggest photosynthetic activity contributes significantly to plant fitness. In fact, in a number of species widely distributed from across the land plant phylogeny, cells associated with the vasculature show some characteristics of the C4 pathway. In stems and petioles of celery and tobacco, cells of the mid-vein allow the decarboxylation of organic acids coming from the vasculature and thus release CO2 around RuBisCO for use in photosynthesis [51]. These attributes have also been observed in Arabidopsis and rice leaves [52,53]. In each case, cells associated with veins are photosynthetically active and contain significant activities of C4 acid decarboxylases [51–53]. In the case of rice, just as with the BS of certain C4 species, linear electron transport from photosystem II to photosystem I is reduced in these veinal cells [53]. Thus, BS cells around veins of C3 plants are photosynthetic, but they also contain multiple characteristics more commonly associated with the C4 pathway.
4. The ancestral role of C4 proteins in C3 plants
The fact that core C4 enzymes are present in C3 species meant that they did not need to evolve de novo and so likely facilitated the recurrent evolution of the C4 pathway across land plants. The role of these proteins in C3 species prior to their recruitment into C4 photosynthesis has been addressed recently [18,54]. We therefore next focus on discussing how groups of C4 proteins could have been recruited from pre-existing metabolic networks occurring in C3 species.
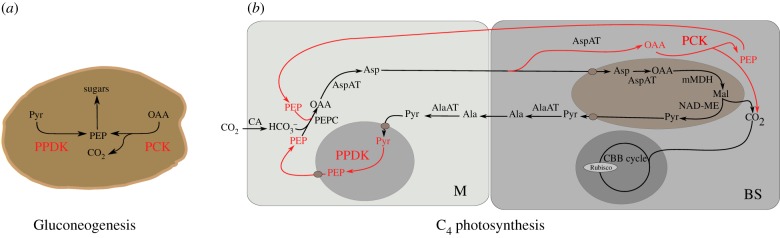
Gluconeogenesis is fundamental to all life, and in plants is particularly important in allowing conversion of storage lipids and proteins into sugars during germination and seedling establishment. Traditionally, it was considered that a single route meditated by PCK allowed the conversion of OAA to PEP, and thus for carbon to enter gluconeogenesis in plants [55–58]. However, disruption of PCK1 function in A. thaliana has only a small effect on early seedling growth [56]. Transcripts derived from the PPDK gene, which encodes the protein catalysing the last committed step of the C4 pathway, are also abundant during seedling establishment [59], and the timing and location of expression within the germinating seed are broadly similar to those derived from PCK [60]. A double ppdk-pck1 mutant showed compromised movement of labelled carbon from storage lipids and proteins into sugars compared with wild-type, and also compared with each single mutant. In addition, seedling establishment was compromised [60]. Based on these findings, it is concluded that two routes into gluconeogenesis operate in C3 plants, both involving proteins associated with C4 photosynthesis (figure 2). It therefore appears that expression of the PCK and PPDK genes is coordinated to ensure proper functioning of gluconeogenesis in C3 plants. We propose that an ancestral gene regulatory system present in C3 species is used to ensure their high and coordinate activity in C4 plants. Clearly, this regulatory system must alter somewhat as C4 evolves. First, it must become operational in mature leaves rather than cotyledons. Second, enhancers of expression must move from the internal promoter that drives expression of cytosolic PPDK in C3 seedlings to the distal promoter driving expression of chloroplastic PPDK in C4 plants. Third, additional regulation must evolve to ensure that expression of the PPDK and PCK genes is restricted to M and BS cells, respectively. If additional genes encoding C4 proteins are co-regulated in the ancestral C3 state to allow the proteins they encode to function together in other metabolic pathways, this may well have further facilitated the evolution of this highly complex state. We next consider our understanding of mechanisms regulating C4 genes in both C3 and C4 plants.
Figure 2.
Genes associated with C4 photosynthesis are coordinated in the ancestral C3 state. (a) The enzymes PPDK and PCK (red) both act during gluconeogenesis in germinating Arabidopsis seedlings [60]. Both genes have been co-opted into the C4 pathway (b) where PPDK regenerates PEP from pyruvate in the M and PCK acts as a C4 acid decarboxylase releasing CO2 around RuBisCO in the BS cells. Abbreviations as for figure 1.
5. Recruitment of pre-existing gene regulatory networks
As with most traits, gene expression associated with the C4 pathway is regulated at multiple levels, including epigenetic, transcriptional, post-transcriptional and post-translational [61,62]. However, it is unclear to what extent these mechanisms are already associated with C4 genes in the ancestral C3 state. It has long been clear that genes encoding proteins of the C4 pathway respond to light [63–66]. Recently, it has become apparent that this key characteristic is found in the ancestral state. In C3 A. thaliana, most genes encoding core C4 proteins are regulated by light [67]. Furthermore, some C4 genes are also subject to control by chloroplast-to-nucleus signalling [67]. Thus, two basic characteristics required for C4 cycle genes to be coordinately expressed with other genes of C3 photosynthesis are already in place in the ancestral C3 state. Again, these networks need to be modified for an efficient C4 system. First, compared with C3 A. thaliana, in C4 Gynadropsis gynandra (formerly designated Cleome gynandra), more C4 cycle genes are controlled by the chloroplast. Second, although an existing system of light-regulation operates in C3 species, this would need to be amplified in order that C4 genes are expressed at sufficiently high levels in leaves undertaking C4 photosynthesis.
In C4 leaves, expression of C4 genes is typically restricted to either M or BS cells [61]. For this to happen, trans-factors must recognize elements in cis in a cell-specific manner. For many years, it appeared that cell-specific expression in C4 leaves was mediated by cis-elements that were not present in orthologous genes from C3 leaves. For example, while the maize PEPC and PPDK genes are expressed in M cells, and RbcS1A expression is limited to BS cells, this was not the case for homologous genes in rice [64,68–71]. In addition, preferential expression of PEPC in the M cells of C4 Flaveria bidentis is associated with two modifications in cis that generate an M-enhancing module (MEM1) [72]. However, it is now clear that multiple genes are expressed preferentially in M or BS cells of C4 G. gynandra because of pre-existing cis-elements located in orthologous genes from A. thaliana. For example, both genes encoding the heterodimeric NAD-ME in G. gynandra contain elements in the coding sequence that determine BS expression, and these elements are found in the orthologues from A. thaliana [73,74]. The genes from A. thaliana are not preferentially expressed in the BS in the ancestral C3 state, but they are when placed into leaves of C4 G. gynandra. A similar situation has been found with PPDK and CA genes. Here, cis-regulatory elements located in untranslated regions generate preferential expression in M cells of C4 G. gynandra [75,76]. Orthologous CA and PPDK genes from C3 A. thaliana contain the same elements, and although they are silent in terms of cell specificity in C3 leaves, when placed into C4 G. gynandra, they lead to expression in the BS. In all these cases, the cis-elements are highly conserved in C3 A. thaliana, suggesting that they carry out an important, but as yet undefined regulatory function. Taken together, these findings indicate that C4 photosynthesis has on multiple occasions made use of cis-regulators found in C3 species, and therefore that its evolution is based on alterations in trans as well as in cis.
Acknowledgements
We thank Steven Burgess for helpful comments.
Data accessibility
This article has no additional data.
Authors' contributions
I.R.-L. and J.M.H. conceived and wrote the manuscript.
Competing interests
We declare we have no competing interests.
Funding
I.R.-L. was funded by CONACyT.
References
- 1.Bassham JA, Benson AA, Kay LD, Harris AZ, Wilson AT, Calvin M. 1954. The path of carbon in photosynthesis. XXI. The cyclic regeneration of carbon dioxide acceptor . J. Am. Chem. Soc. 76, 1760–1770. ( 10.1021/ja01636a012) [DOI] [Google Scholar]
- 2.Sage RF. 2004. Tansley review: the evolution of C4 photosynthesis. New Phytol. 161, 30 ( 10.1111/j.1469-8137.2004.00974.x) [DOI] [PubMed] [Google Scholar]
- 3.Anbar AD, et al. 2007. A whiff of oxygen before the great oxidation event? Science 317, 1903–1906. ( 10.1126/science.1140325) [DOI] [PubMed] [Google Scholar]
- 4.Bekker A, Holland HD, Wang P-L, Rumble D, Stein HJ, Hannah JL, Coetzee LL, Beukes NJ. 2004. Dating the rise of atmospheric oxygen. Nature 427, 117–120. ( 10.1038/nature02260) [DOI] [PubMed] [Google Scholar]
- 5.Canfield DE. 2005. The early history of atmospheric oxygen: homage to Robert M. Garrels. Annu. Rev. Earth Planet. Sci. 33, 1–36. ( 10.1146/annurev.earth.33.092203.122711) [DOI] [Google Scholar]
- 6.Ubierna N, Sun W, Cousins AB. 2011. The efficiency of C4 photosynthesis under low light conditions: assumptions and calculations with CO2 isotope discrimination. J. Exp. Bot. 62, 3119–3134. ( 10.1093/jxb/err073) [DOI] [PubMed] [Google Scholar]
- 7.Bauwe H, Hagemann M, Fernie AR. 2010. Photorespiration: players, partners and origin. Trends Plant Sci. 15, 330–336. ( 10.1016/j.tplants.2010.03.006) [DOI] [PubMed] [Google Scholar]
- 8.Brooks A, Farquhar GD. 1985. Effect of temperature on the CO2/O2 specificity of ribulose-1,5-bisphosphate carboxylase/oxygenase and the rate of respiration in the light. Planta 165, 397–406. ( 10.1007/BF00392238) [DOI] [PubMed] [Google Scholar]
- 9.Tcherkez GGB, Farquhar GD, Andrews TJ. 2006. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl Acad. Sci. USA 103, 7246–7251. ( 10.1073/pnas.0600605103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sage RF, Sharkey TD. 1987. The effect of temperature on the occurrence of O2 and CO2 insensitive photosynthesis in field grown plants. Plant Physiol. 84, 658–664. ( 10.1104/pp.84.3.658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharwood RE, Ghannoum O, Kapralov MV, Gunn LH, Whitney SM. 2016. Temperature responses of Rubisco from Paniceae grasses provide opportunities for improving C3 photosynthesis. Nat. Plants 2, 16186 ( 10.1038/nplants.2016.186) [DOI] [PubMed] [Google Scholar]
- 12.Shively JM, Ball F, Brown DH, Saunders RE. 1973. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182, 584–586. ( 10.1126/science.182.4112.584) [DOI] [PubMed] [Google Scholar]
- 13.Smith EC, Griffiths H. 1996. The occurrence of the chloroplast pyrenoid is correlated with the activity of a CO2-concentrating mechanism and carbon isotope discrimination in lichens and bryophytes. Planta 198, 6–16. ( 10.1007/BF00197580) [DOI] [Google Scholar]
- 14.Ranson SL, Thomas M. 1960. Crassulacean acid metabolism. Annu. Rev. Plant Physiol. 11, 81–110. ( 10.1146/annurev.pp.11.060160.000501) [DOI] [Google Scholar]
- 15.Hatch MD, Slack CR. 1966. Photosynthesis by sugar-cane leaves. A new carboxylation reaction and the pathway of sugar formation. Biochem. J. 101, 103–111. ( 10.1042/bj1010103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sage RF, Sage TL, Kocacinar F. 2012. Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 63, 19–47. ( 10.1146/annurev-arplant-042811-105511) [DOI] [PubMed] [Google Scholar]
- 17.Langdale JA. 2011. C4 cycles: past, present, and future research on C4 photosynthesis. Plant Cell 23, 3879–3892. ( 10.1105/tpc.111.092098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aubry S, Brown NJ, Hibberd JM. 2011. The role of proteins in C3 plants prior to their recruitment into the C4 pathway. J. Exp. Bot. 62, 3049–3059. ( 10.1093/jxb/err012) [DOI] [PubMed] [Google Scholar]
- 19.Furbank RT. 2011. Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types? J. Exp. Bot. 62, 3103–3108. ( 10.1093/jxb/err080) [DOI] [PubMed] [Google Scholar]
- 20.Sommer M, Bräutigam A, Weber APM. 2012. The dicotyledonous NAD malic enzyme C4 plant Cleome gynandra displays age-dependent plasticity of C4 decarboxylation biochemistry. Plant Biol. 14, 621–629. ( 10.1111/j.1438-8677.2011.00539.x) [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Bräutigam A, Weber APM, Zhu X-G. 2014. Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. J. Exp. Bot. 65, 3567–3578. ( 10.1093/jxb/eru058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christin P-A, Osborne CP, Sage RF, Arakaki M, Edwards EJ. 2011. C4 eudicots are not younger than C4 monocots. J. Exp. Bot. 62, 3171–3181. ( 10.1093/jxb/err041) [DOI] [PubMed] [Google Scholar]
- 23.Christin PA, Besnard G, Samaritani E, Duvall MR, Hodkinson TR, Savolainen V, Salamin N. 2008. Oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr. Biol. 18, 37–43. ( 10.1016/j.cub.2007.11.058) [DOI] [PubMed] [Google Scholar]
- 24.Edwards GE, Voznesenskaya EV. 2010. Chapter 4 C4 photosynthesis: Kranz forms and single-cell C4 in terrestrial plants. In C4 photosynthesis and related CO2 concentrating mechanisms (eds A Raghavendra, R Sage), Adv. Photosynthesis Respir. 32, pp. 29–61. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 25.Ohno S. 1970. Evolution by gene duplication, 2013th edn Berlin, Germany: Springer Science & Business Media. [Google Scholar]
- 26.Thomas BC, Pedersen B, Freeling M. 2006. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16, 934–946. ( 10.1101/gr.4708406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodhouse MR, Schnable JC, Pedersen BS, Lyons E, Lisch D, Subramaniam S, Freeling M. 2010. Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homeologs. PLoS Biol. 8, e1000409 ( 10.1371/journal.pbio.1000409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arntz M, Delph L. 2001. Pattern and process: evidence for the evolution of photosynthetic traits in natural populations. Oecologia 127, 455–467. ( 10.1007/s004420100650) [DOI] [PubMed] [Google Scholar]
- 29.Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. ( 10.1126/science.290.5494.1151) [DOI] [PubMed] [Google Scholar]
- 30.Moore RC, Purugganan MD. 2005. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8, 122–128. ( 10.1016/j.pbi.2004.12.001) [DOI] [PubMed] [Google Scholar]
- 31.Freeling M, Lyons E, Pedersen B, Alam M, Ming R, Lisch D. 2008. Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res. 18, 1924–1937. ( 10.1101/gr.081026.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monson RK. 2003. Gene duplication, neofunctionalization, and the evolution of C4 photosynthesis. Int. J. Plant Sci. 164, S43–S54. ( 10.1086/368400) [DOI] [Google Scholar]
- 33.Gowik U, Westhoff P. 2011. The path from C3 to C4 photosynthesis. Plant Physiol. 155, 56–63. ( 10.1104/pp.110.165308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Bergh E, Külahoglu C, Bräutigam A, Hibberd JM, Weber APM, Zhu X-G, Eric Schranz M. 2014. Gene and genome duplications and the origin of C4 photosynthesis: birth of a trait in the Cleomaceae. Curr. Plant Biol. 1, 2–9. ( 10.1016/j.cpb.2014.08.001) [DOI] [Google Scholar]
- 35.Williams BP, Aubry S, Hibberd JM. 2012. Molecular evolution of genes recruited into C4 photosynthesis. Trends Plant Sci. 17, 213–220. ( 10.1016/j.tplants.2012.01.008) [DOI] [PubMed] [Google Scholar]
- 36.Boussau B, Szollosi GJ, Duret L, Gouy M, Tannier E, Daubin V. 2013. Genome-scale coestimation of species and gene trees. Genome Res. 23, 323–330. ( 10.1101/gr.141978.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 ( 10.1186/s13059-015-0721-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emms DM, Covshoff S, Hibberd JM, Kelly S. 2016. Independent and parallel evolution of new genes by gene duplication in two origins of C4 photosynthesis provides new insight into the mechanism of phloem loading in C4 species. Mol. Biol. Evol. 33, 1796–1806. ( 10.1093/molbev/msw057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams BP, Johnston IG, Covshoff S, Hibberd JM. 2013. Phenotypic landscape inference reveals multiple evolutionary paths to C4 photosynthesis. eLife 2, e00961 ( 10.7554/eLife.00961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heckmann D, Schulze S, Denton A, Gowik U, Westhoff P, Weber APM, Lercher MJ. 2013. Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153, 1579–1588. ( 10.1016/j.cell.2013.04.058) [DOI] [PubMed] [Google Scholar]
- 41.Mallmann J, Heckmann D, Bräutigam A, Lercher MJ, Weber APM, Westhoff P, Gowik U. 2014. The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. eLife 3, e02478 ( 10.7554/eLife.02478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyake H, Maeda E. 1976. Development of bundle sheath chloroplasts in rice seedlings. Can. J. Bot. 54, 556–565. ( 10.1139/b76-056) [DOI] [Google Scholar]
- 43.Williams ML, Farrar JF, Pollock CJ. 1989. Cell specialization within the parenchymatous bundle sheath of barley. Plant Cell Environ. 12, 909–918. ( 10.1111/j.1365-3040.1989.tb01970.x) [DOI] [Google Scholar]
- 44.Koroleva OA, Farrar JF, Tomos AD, Pollock CJ. 1998. Carbohydrates in individual cells of epidermis, mesophyll, and bundle sheath in barley leaves with changed export or photosynthetic rate. Plant Physiol. 118, 1525–1532. ( 10.1104/pp.118.4.1525) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsutsumi K, Taniguchi Y, Kawasaki M, Taniguchi M, Miyake H. 2006. Expression of photosynthesis-related genes during the leaf development of a C3 plant rice as visualized by in situ hybridization. Plant Prod. Sci. 9, 232–241. ( 10.1626/pps.9.232) [DOI] [Google Scholar]
- 46.Miyake H. 2016. Starch accumulation in the bundle sheaths of C3 plants: a possible pre-condition for C4 photosynthesis. Plant Cell Physiol. 57, 890–896. ( 10.1093/pcp/pcw046) [DOI] [PubMed] [Google Scholar]
- 47.Yamane K, et al. 2003. Bundle sheath chloroplasts of rice are more sensitive to drought stress than mesophyll chloroplasts. J. Plant Physiol. 160, 1319–1327. ( 10.1078/0176-1617-01180) [DOI] [PubMed] [Google Scholar]
- 48.Janacek SH, et al. 2009. Photosynthesis in cells around veins of the C3 plant Arabidopsis thaliana is important for both the shikimate pathway and leaf senescence as well as contributing to plant fitness. Plant J. 59, 329–343. ( 10.1111/j.1365-313X.2009.03873.x) [DOI] [PubMed] [Google Scholar]
- 49.Leegood RC. 2008. Roles of the bundle sheath cells in leaves of C3 plants. J. Exp. Bot. 59, 1663–1673. ( 10.1093/jxb/erm335) [DOI] [PubMed] [Google Scholar]
- 50.Griffiths H, Weller G, Toy LFM, Dennis RJ. 2013. You're so vein: bundle sheath physiology, phylogeny and evolution in C3 and C4 plants. Plant Cell Environ. 36, 249–261. ( 10.1111/j.1365-3040.2012.02585.x) [DOI] [PubMed] [Google Scholar]
- 51.Hibberd JM, Quick WP. 2002. Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 415, 451–454. ( 10.1038/415451a) [DOI] [PubMed] [Google Scholar]
- 52.Brown NJ, et al. 2010. C4 acid decarboxylases required for C4 photosynthesis are active in the mid-vein of the C3 species Arabidopsis thaliana, and are important in sugar and amino acid metabolism. Plant J. 61, 122–133. ( 10.1111/j.1365-313X.2009.04040.x) [DOI] [PubMed] [Google Scholar]
- 53.Shen W, Ye L, Ma J, Yuan Z, Zheng B, Chuangen LV, Zhu Z, Chen X, Gao Z, Chen G. 2016. The existence of C4-bundle-sheath-like photosynthesis in the mid-vein of C3 rice. Rice 9, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maier A, Zell MB, Maurino VG. 2011. Malate decarboxylases: evolution and roles of NAD(P)-ME isoforms in species performing C4 and C3 photosynthesis. J. Exp. Bot. 62, 3061–3069. ( 10.1093/jxb/err024) [DOI] [PubMed] [Google Scholar]
- 55.Leegood RC, Ap Rees T. 1978. Phosphoenolpyruvate carboxykinase and gluconeogenesis in cotyledons of Cucurbita pepo. Biochim. Biophys. Acta Enzymol. 524, 207–218. ( 10.1016/0005-2744(78)90119-5) [DOI] [PubMed] [Google Scholar]
- 56.Rylott EL, Gilday AD, Graham IA. 2003. The gluconeogenic enzyme phosphoenolpyruvate carboxykinase in Arabidopsis is essential for seedling establishment. Plant Physiol. 131, 1834–1842. ( 10.1104/pp.102.019174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Penfield S, Rylott EL, Gilday AD, Graham S, Larson TR, Graham IA. 2004. Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16, 2705–2718. ( 10.1105/tpc.104.024711) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malone S, Chen Z-H, Bahrami AR, Walker RP, Gray JE, Leegood RC. 2007. Phosphoenolpyruvate carboxykinase in Arabidopsis: changes in gene expression, protein and activity during vegetative and reproductive development. Plant Cell Physiol. 48, 441–450. ( 10.1093/pcp/pcm014) [DOI] [PubMed] [Google Scholar]
- 59.Parsley K, Hibberd JM. 2006. The Arabidopsis PPDK gene is transcribed from two promoters to produce differentially expressed transcripts responsible for cytosolic and plastidic proteins. Plant Mol. Biol. 62, 339–349. ( 10.1007/s11103-006-9023-0) [DOI] [PubMed] [Google Scholar]
- 60.Eastmond PJ, et al. 2015. Arabidopsis uses two gluconeogenic gateways for organic acids to fuel seedling establishment. Nat. Commun. 6, 6659 ( 10.1038/ncomms7659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hibberd JM, Covshoff S. 2010. The regulation of gene expression required for C4 photosynthesis. Annu. Rev. Plant Biol 61, 181–207. ( 10.1146/annurev-arplant-042809-112238) [DOI] [PubMed] [Google Scholar]
- 62.Reeves G, Grangé-Guermente MJ, Hibberd JM. 2016. Regulatory gateways for cell-specific gene expression in C4 leaves with Kranz anatomy. J. Exp. Bot. 68, 107–116. ( 10.1093/jxb/erw438) [DOI] [PubMed] [Google Scholar]
- 63.Collins PD, Hague DR. 1983. Light-stimulated synthesis of NADP malic enzyme in leaves of maize. J. Biol. Chem. 258, 4012–4018. [PubMed] [Google Scholar]
- 64.Sheen J-Y, Bogorad L. 1987. Regulation of levels of nuclear transcripts for C4 photosynthesis in bundle sheath and mesophyll cells of maize leaves. Plant Mol. Biol. 8, 227–238. ( 10.1007/BF00015031) [DOI] [PubMed] [Google Scholar]
- 65.Langdale JA, Zelitch I, Miller E, Nelson T. 1988. Cell position and light influence C4 versus C3 patterns of photosynthetic gene expression in maize. EMBO J. 7, 3643–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Long JJ, Berry JO. 1996. Tissue-specific and light-mediated expression of the C4 photosynthetic NAD-dependent malic enzyme of amaranth mitochondria. Plant Physiol. 112, 473–482. ( 10.1104/pp.112.2.473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burgess SJ, Granero-Moya I, Grangé-Guermente MJ, Boursnell C, Terry MJ, Hibberd JM. 2016. Ancestral light and chloroplast regulation form the foundations for C4 gene expression. Nat. Plants 2, 16161 ( 10.1038/nplants.2016.161) [DOI] [PubMed] [Google Scholar]
- 68.Matsuoka M, Tada Y, Fujimura T, Kano-Murakami Y. 1993. Tissue-specific light-regulated expression directed by the promoter of a C4 gene, maize pyruvate,orthophosphate dikinase, in a C3 plant, rice (C4 photosynthesis/gene evolution/transgenic rice). Proc. Natl Acad. Sci. USA 90, 9586–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nomura M, et al. 2000. The evolution of C4 plants: acquisition of cis-regulatory sequences in the promoter of C4-type pyruvate, orthophosphate dikinase gene. Plant J. 22, 211–221. ( 10.1046/j.1365-313x.2000.00726.x) [DOI] [PubMed] [Google Scholar]
- 70.Nomura M, Katayama K, Nishimura A, Ishida Y, Ohta S, Komari T, Miyao-Tokutomi M, Tajima S, Matsuoka M. 2000. The promoter of rbcS in a C3 plant (rice) directs organ-specific, light-dependent expression in a C4 plant (maize), but does not confer bundle sheath cell-specific expression. Plant Mol. Biol. 44, 99–106. ( 10.1023/A:1006461812053) [DOI] [PubMed] [Google Scholar]
- 71.Taniguchi M, et al. 2000. The promoter for the maize C4 pyruvate,orthophosphate dikinase gene directs cell- and tissue-specific transcription in transgenic maize plants. Plant Cell Physiol. 41, 42–48. ( 10.1093/pcp/41.1.42) [DOI] [PubMed] [Google Scholar]
- 72.Gowik U, Burscheidt J, Akyildiz M, Schlue U, Koczor M, Streubel M, Westhoff P. 2004. cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 16, 1077–1090. ( 10.1105/tpc.019729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, Kajala K, Hibberd JM. 2011. Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331, 1436–1439. ( 10.1126/science.1201248) [DOI] [PubMed] [Google Scholar]
- 74.Reyna-Llorens I, Burgess SJ, Williams BP, Stanley S, Boursnell C, Hibberd JM. 2016. Ancient coding sequences underpin the spatial patterning of gene expression in C4 leaves. bioR xiv, 085795 ( 10.1101/085795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kajala K, Brown NJ, Williams BP, Borrill P, Taylor LE, Hibberd JM. 2012. Multiple Arabidopsis genes primed for recruitment into C4 photosynthesis. Plant J. 69, 47–56. ( 10.1111/j.1365-313X.2011.04769.x) [DOI] [PubMed] [Google Scholar]
- 76.Williams B, Burgess S, Reyna-Llorens I. 2016. An untranslated cis-element regulates the accumulation of multiple C4 enzymes in Gynandropsis gynandra mesophyll cells. Plant Cell 28, 454–465. ( 10.1105/tpc.15.00570) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.