Abstract
Background/Aim: Senescence marker protein-30/gluconolactonase knockout mice (SMP-30/GNL-KO) are a very useful model for clarifying the involvement of vitamin C (VC) in aging-related diseases. In this study, the effects of VC deficiency on skin and hair growth were investigated using SMP-30/GNL-KO mice by RNA sequencing. Materials and Methods: SMP-30/GNL-KO mice were given water containing 1.5 g/l VC until up to 8 weeks after birth to maintain a VC concentration in their organs and plasma equivalent to that in wild-type mice. The mice were then divided into two groups: a VC(+) group, where VC was administered, and a VC(−) group, where VC was not administered. Skin samples were collected at 4 and 8 weeks after the treatment. RNA was extracted from each skin sample, followed by cDNA synthesis and RNA-seq. In addition, hair growth was compared between the VC(−) and VC(+) groups after shaving. Skin samples were collected from the shaved area for histological examination by hematoxylin & eosin (HE) staining. Results: RNA-seq revealed that there were 1,736 (FDR<0.001) differentially expressed genes in the VC(−) and VC(+) groups. From the functional analysis of the differentially expressed genes in the VC(−) and VC(+) groups, predicted functionalities including cell death and cytotoxicity increased in the VC(+) group. Furthermore, it was predicted that the difference in hair growth between the VC(−) and VC(+) groups was caused by the expression of genes including keratin-related genes and the Sonic hedgehog gene. It was confirmed that hair growth was significantly promoted; hair growth from hair papilla cells was also confirmed by HE staining of the shaved backs of SMP-30/GNL-KO mice in the VC(+) group. Conclusion: RNA-seq of the skin from VC-deficient mice showed the effects of VC deficiency on the expression of genes involved in cell growth and the hair cycle. Visual inspection suggested that changes in the expression of the genes are involved in delaying hair growth in the VC(−) group. Further research on the relationship among VC deficiency, the hair cycle, and skin cell growth may contribute to research on hair restoration and skin aging.
Keywords: Vitamin C, ascorbic acid deficiency, RNA sequencing, skin, hair growth
Vitamin C (VC) is involved in collagen synthesis (1) and exhibits anti-oxidative effects in vivo (2). VC is biosynthesized in the liver in most mammals. For example, mice and rats are able to biosynthesize their own VC and thus do not require the intake of VC from their diets. However, some species, including primates and guinea pigs, cannot biosynthesize VC due to mutations in the gulono-gamma-lactone oxidase (GLO) gene (3).
Senescence marker protein-30 (SMP-30) was originally discovered as a biomolecule, the levels of which decreased with age; however, it was later found that SMP 30 was the same gene as gluconolactonase (GNL), which is involved in the penultimate step in the VC biosynthesis pathway (4). SMP-30 is involved one step before GLO, the last step in the VC biosynthesis pathway; therefore, VC-deficient mice can be constructed by knocking out the SMP-30/GNL gene (SMP/GNL-KO).
Arai et al. constructed VC-deficient mice by knocking-out the SMP-30 gene and reported the effects of VC deficiency on the skin (5). The concentration of hydroxyproline, the structural unit of collagen in the skin, markedly decreased after keeping SMP-30/GNL-KO mice in a VC-deficient condition for 60 days. Moreover, skin pigmentation was suppressed when administering VC containing acerola juice followed by UVB irradiation of the skin of VC-deficient hairless mice (6). Other reports regarding the relationship between VC and the skin include a delay in wound healing because of VC deficiency, stimulation of type I collagen gene expression in human skin fibroblasts by VC (7) and VC derivatives, and functional regulation of keratinocytes by the anti-oxidative activity of VC (8).
Furthermore, when new hair growth from dermal papilla cells in skin tissue was investigated, it was found that VC derivatives promoted the growth of cells and hair shafts in cultured human papilla cells; moreover, the growth period during the new hair growth cycle was prolonged by the proliferative and anti-apoptotic effects of VC derivatives. VC derivatives induced the secretion of growth factors, including insulin-like growth factor-1, which, in turn, proliferated and differentiated overlying keratinocytes to promote the elongation of hair shafts (9,10).
Thus, the effect of VC on skin cells and hair growth is becoming clearer; however, changes in gene expression in the skin and regulation of skin function have not been adequately identified following the administration of VC to VC-deficient mice. Therefore, an exploratory investigation was conducted to examine changes in gene expression in the skin by comparing SMP30/GNL-KO mice treated with and without VC administration.
Therefore, various differentially expressed genes (DEGs) were identified between VC(–) and VC(+) when comparing the two groups; moreover, abnormalities at the gene level in skin function and hair growth triggered by VC deficiency were confirmed from the functional analysis of these genes.
Materials and Methods
Animals. SMP30 KO mice were previously generated by gene-targeting techniques (11). Heterozygous female mice (SMP30 +/–) were mated with male KO mice (SMP30 Y/–) to produce SMP30 KO (SMP30 Y/-) and WT (SMP30 Y/+) littermates. Mouse pairs were obtained from the Tokyo Metropolitan Institute of Gerontology, Japan.
SMP30 KO mice were bred at the animal facility of Hokkaido Pharmaceutical University School of Pharmacy. Throughout the experiments, animals were maintained on 12-h light/dark cycles in a controlled environment held at 23˚C±1˚C and 55%±15% relative humidity.
This study conforms to the Guiding Principles for the Care and Use of Experimental Animals of Hokkaido Pharmaceutical University (published 1998, revised in 2001 and 2007). The protocol approval number is H28-010.
Experiment 1: Global analysis of gene expression of skin using RNA-Seq.
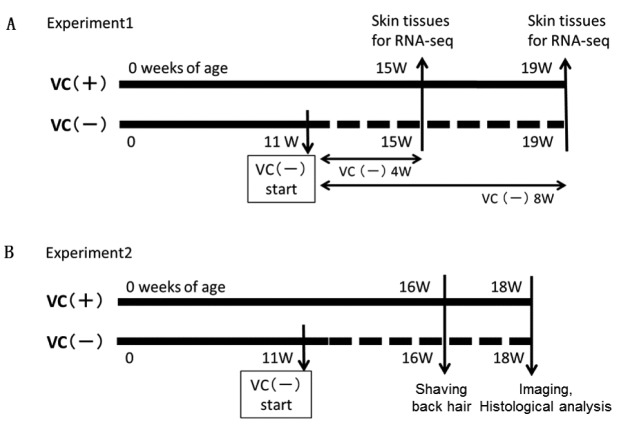
Experimental design. For gene expression analysis of skin by RNA sequencing (RNA-seq), the VC (–) group of male mice (n=5) were fed a diet (CL-2; CLEA Japan, Tokyo) containing VC and distilled water (DW) containing 1.5 g/l L-ascorbic acid and 10 μM EDTA for 11 weeks (11 W) and then fed a VC-deficient diet (CL-2; CLEA Japan, Tokyo) and DW until the end of this experiment. The VC(+) group of male mice (n=5) were fed a diet containing VC and DW containing 1.5 g/l L-ascorbic acid until the end of the experiment. Food and water were available ad libitum.
At 15 W [VC(–) 4 W and VC(+) 4 W, each group n=2] and 19 W [VC(–) 8 W and VC(+) 8 W, each group n=3], the mice were sacrificed under isoflurane anesthesia, and the back skin was collected after removing back hair with an electric shaver. Back skins of 15 W (n=2) and 19 W (n=3) mice were stored in RNAlater® Solution (Thermo Fisher Scientific Inc., Waltham, MA, USA) at –80˚C until isolation of total RNA for RNA-seq analysis (Figure 1A).
Figure 1. Experimental designs on VC(+) or VC(–) SMP30/GNL-KO mice. Experiment 1 was planned for gene expression of skin analysis by RNAseq. Experiment 2 was planned for hair growth observation and dermatological examination.
Total RNA was extracted using an RNeasy Lipid Tissue Mini Kit (Qiagen Co., Ltd., Venlo, Netherlands). The RNA quality and quantity were assessed on a 2100 Bioanalyzer using an RNA 6000 Nano kit (Agilent Technologies Inc., Santa Clara, CA, USA).
In total, 100 ng of total RNA from the skins of the VC (–) and VC (+) groups were used for library preparation. Sequencing libraries were generated using TruSeq RNA Library Preparation Kit v2 (Illumina Inc., San Diego, CA, USA). Library construction procedures were as follows: total RNA was purified to mRNA and mRNA was then fragmented and converted to double-stranded cDNA. Sequencing adapters containing index sequences were ligated to the ends of cDNA. Libraries were sequenced on an Illumina HiSeq 2500 (Illumina Inc.) to generate 100-bp reads.
Sequence reads were trimmed and filtered by quality, and quality reads were mapped to GRCm38 (mm10) with the RNA-Seq mapping algorithm in CLC Genomics Workbench v9.5.3 (Qiagen Co., Ltd.). Mapped reads were approximately 5.5millions for 4W and approximately 20millions for 8W. RNA-seq data analyses were performed using the CLC Genomics Workbench with Advanced RNA-Seq Plugin 1.5.
Principal component analysis. Counts for each individual gene were normalized as log CPM values and projected onto two dimensions for principal component analysis (PCA). Two principal components are specified by the 2D plot. The first principal component specifies the direction with the largest variability in data, while the second component specifies the direction with the second largest variation.
Differential expression analysis. Statistical differential expression tests were performed based on a negative binominal generalized linear model similar to that of edgeR (12). Differentially expressed genes (DEGs) were generated based on a false discovery rate -corrected p-value (FDR) <0.001 using the Wald test for the effect of the two VC treatments (VC(–), VC(+)), while controlling for the weeks of age (4 W, 8 W).
Heat Map with hierarchical clustering of samples and features. Two-dimensional heat maps of expression values were created with each column corresponding to one sample and each row corresponding to one gene. Euclidean distance and complete linkage were set as parameters for the hierarchical clustering of samples and genes. To reduce the number of genes, only DEGs (FDR<000.1) were used for clustering.
Functional annotation analysis of DEGs. Functional analyses of DEGs were performed using the Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Qiagen Co., Ltd.). The activation z-scores of gene networks were predicted using IPA. The activation z-scores represent the activation status (activated or inactivated state) of biofunction and diseases related to gene networks.
Experiment 2: Scoring hair growth and skin tissue observations on hair-shaved SMP30/GNL-KO mice.
Experimental design. For hair growth observations, the VC (–) group of male mice (n=5) were fed a diet containing VC and water for 11 weeks and then fed a VC-deficient diet and DW until the end of the experiment. The VC(+) group of male mice (n=5) were fed a diet containing VC and water (containing 1.5 g/L L-ascorbic acid) until the end of the experiment. Food and water were available ad libitum.
Scoring hair growth and skin tissue observations. Hair was shaved from the backs of 16-W-old mice, and mice were photographed 2 weeks later. Photographs were digitized to score hair growth as follows: hair growth covering 0%-20% of the back was scored as 1 point, 21%-40% as 2 points, 41%-60% as 3 points, 61%-80% as 4 points, and 81%-100% as 5 points.
Each skin sample was fixed in 10% neutral-buffered formalin, embedded in paraffin, and cut in thin sections (5 μm). These sections were stained with hematoxylin & eosin (HE) solution. HE-stained skin tissues were histologically examined using an Olympus AX70 light microscope (Olympus Co., Ltd., Tokyo, Japan), equipped with 40× and 100× objective lenses (Figure 1B).
Statistical analysis. The results are expressed as means±standard deviation. Statistical analyses were performed using the unpaired two-tailed Student’s t-test. All p-values<0.01 were considered statistically significant.
Results
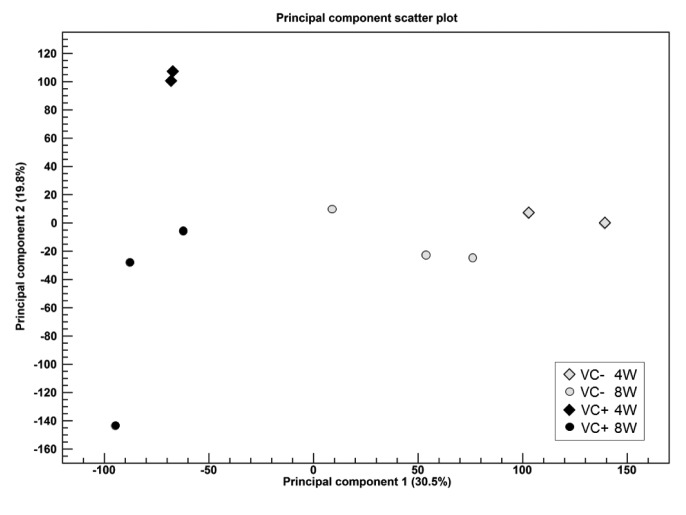
Experiment 1: PCA. To identify outlier samples for quality control and determine the primary causes of variation in the dataset, PCA was conducted (Figure 2). The VC(–) and VC (+) groups could be separated by the first principal component (30.5%, horizontal axis). A difference in the processing period appeared in the first principal component (horizontal axis) in the VC (–) group, while it appeared in the second principal component (19.8%, vertical axis) in the VC(+) group.
Figure 2. Two-dimensional principal component plot of Principal component analysis for RNA-seq. The first principal component is shown for the X axis and the second principal component is shown for the Y axis. The value after the principal component identifier displays the proportion of variance explained by this particular principal component. Colors of symbol are assigned based on VC(–) (gray) or VC(+) (black). Shapes of symbol are assigned based on Week (4W: diamond, 8W: circle).
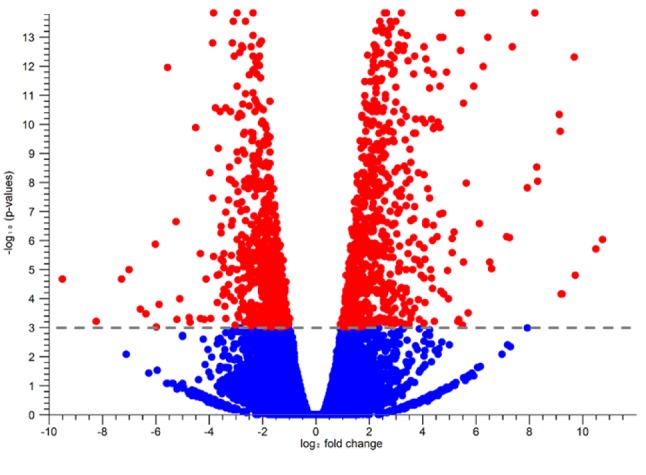
DEG analysis. Expression analysis for comparison between VC(–) and VC(+) identified 1,736 DEGs among 45,706 genes, as FDR < 0.001. Fold change (FC) was calculated using the VC(–) group as the control group. For FC >0, 1,026 genes were identified (minimum FC=1.91), while for FC <0, 710 genes were identified (maximum FC=–1.97). Volcano plots of these genes are indicated in Figure 3.
Figure 3. The volcano plot for statiscal comparison between VC (–)and VC (+). The log2 fold changes which were calculated based on VC (+)/VC (–) were plotted on the x-axis. The -log10 p-values are plotted on the yaxis. Dotted line was drawn on 0.001 FDR value. Each dot represents gene and statistically significant genes were red and others were blue.
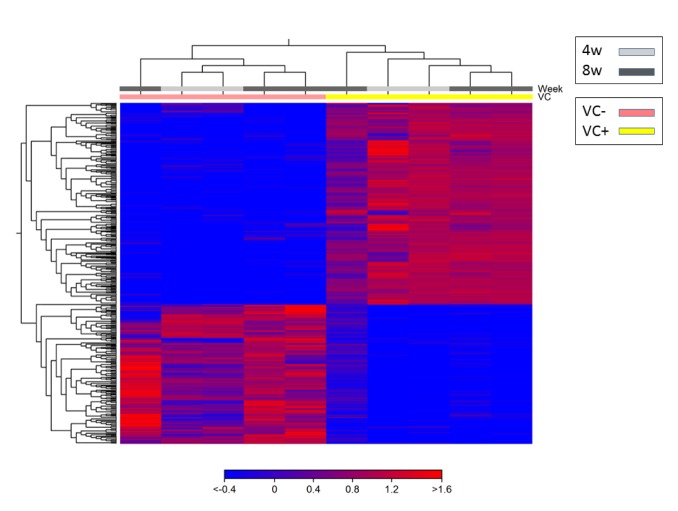
Heat map with hierarchical clustering of samples and features. Using the 1,736 DEGs chosen by expression analysis, clustering of individual mice (horizontal axis) and of information between genes (vertical axis) were conducted to create a heat map (Figure 4). The results indicated that these clusters were divided in the VC(–) and VC(+) groups. Furthermore, any difference was not found between the VC(–) and VC(+) groups caused by a difference in the processing period.
Figure 4. Two dimensional heat map of expression values of differentially expressed genes. Each column corresponds to one sample, and each row corresponds to a gene. The genes are hierarchically clustered by the similarity of their expression profiles over the set of samples and the samples are hierarchically clustered by the similarity of expression patterns over their features. Only differentially expressed genes (1756) were used for creating heat map.
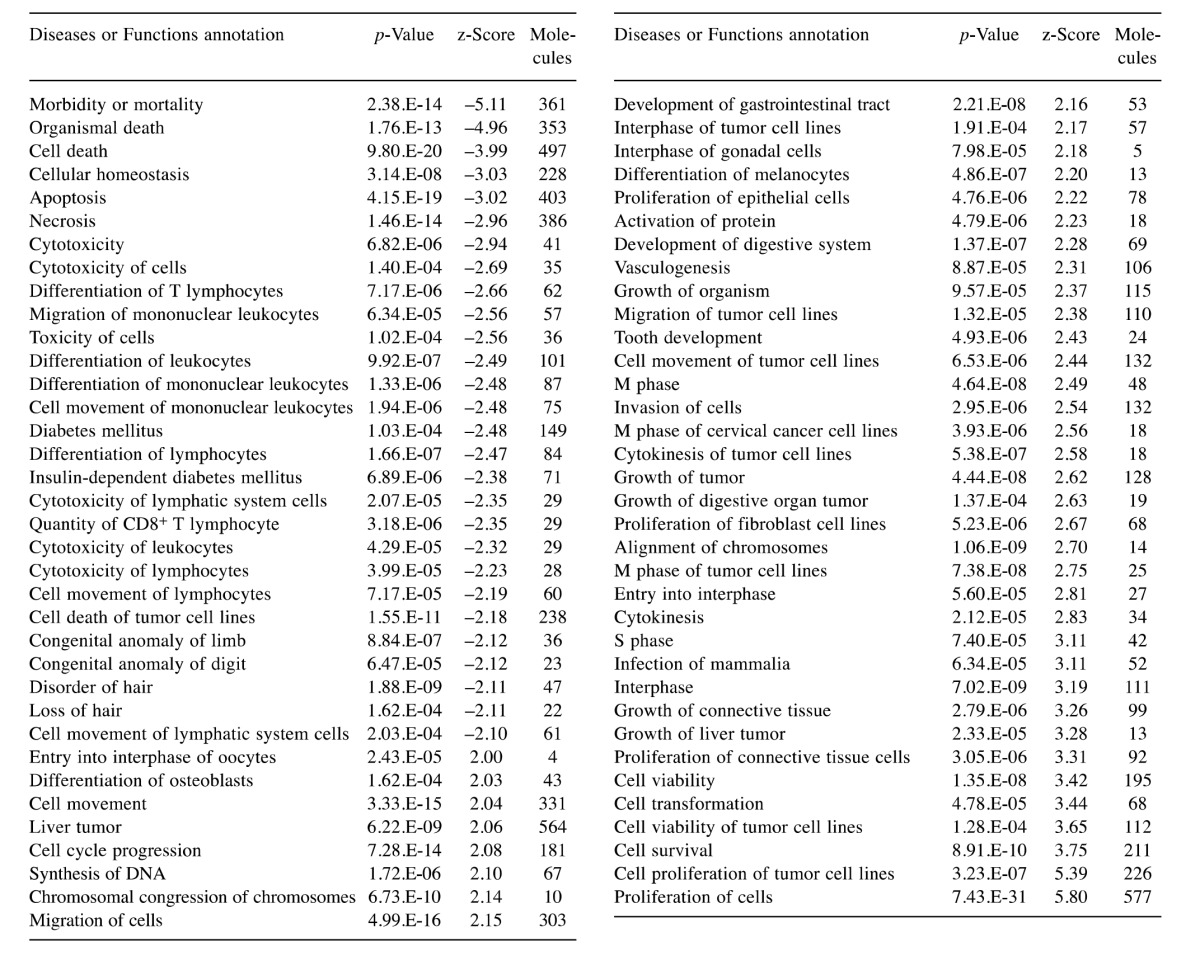
Functional annotation analysis of DEGs. Activation state of diseases and function were predicted according to the functional analysis of the DEGs using IPA (Table I). The results indicated that cell death and cytotoxicity significantly decreased. Moreover, hair disorder and hair loss were decreased. On the other hand, it was indicated that the proliferation, differentiation, and migration of cells increased (Table I). Concerning skin, proliferation of epithelial, fibroblast and connective tissue cells were predicted to be increased. In addition, growth of connective tissue and differentiation of melanocytes were listed.
Table I. Activated Disease or functional annotation (|z-score| >=2) for differentially expressed genes (DEGs) in Ingenuity Pathway Analysis.
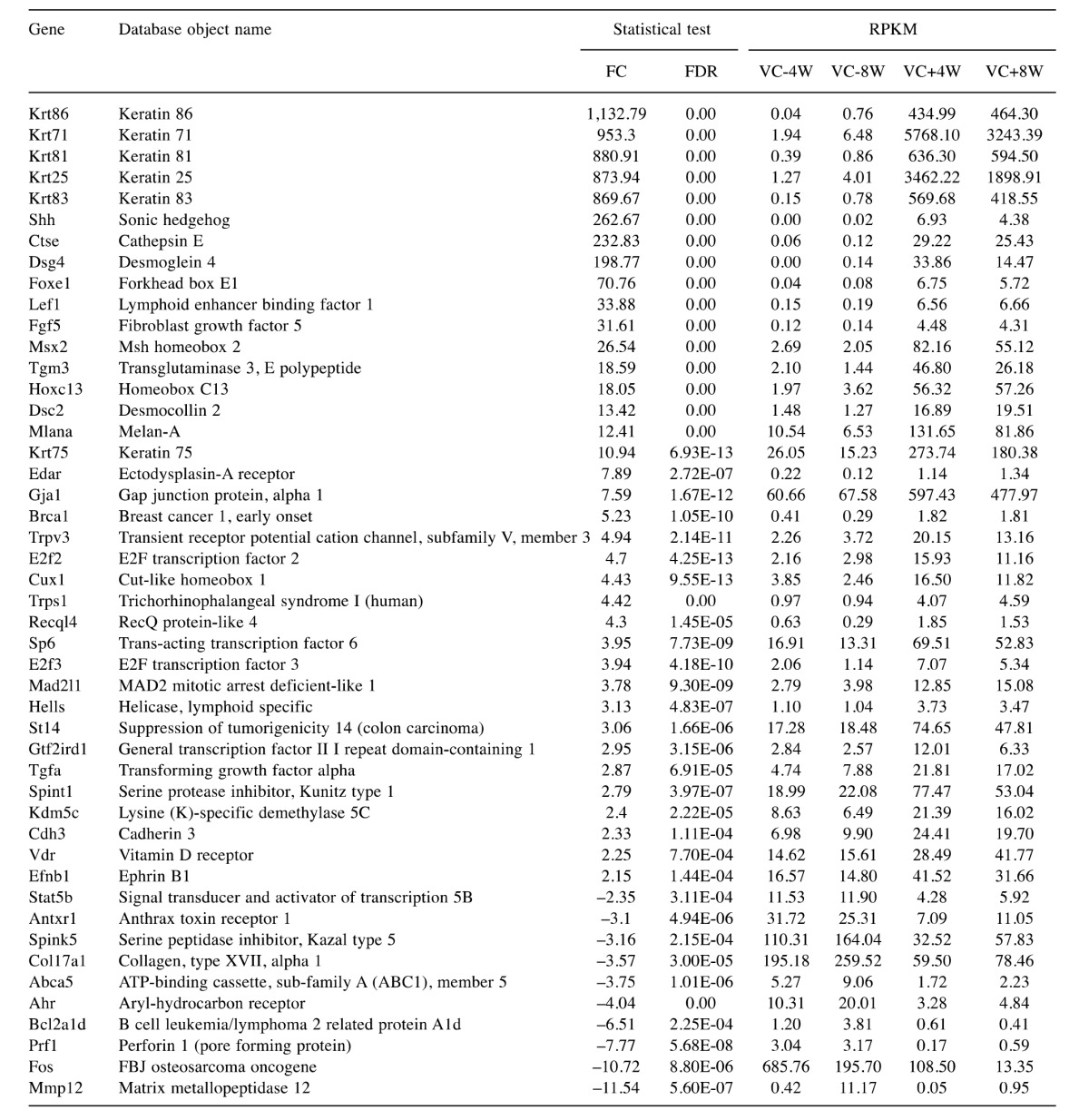
Next, 47 genes, which involved the hair disorder in Table I were selected to examine the expression of genes involved in hair growth. Important genes involved in new hair growth, such as keratin-related genes and Sonic hedgehog (Shh), were included (Table II). In IPA database, 19 genes known to decrease disorder of hair were Ahr, Bcl2a1d, Brca1, Col17a1, Ctse, Cux1, E2f2, Fos, Hells, Lef1, Mad2l1, Mmp12, Msx2, Recql4, Shh, St14, Tgfa, Tgm3 and Vdr in Table II. These genes except Ahr, Bcl2a1d, Col17a1, Fos and Mmp12 were up-regulated in the dataset, therefore predicted to decrease disorder of hair. Other 28 genes are involved in disorder of hair, but do not indicate whether these genes increase or decrease disorder of hair.
Table II. Statistical value of DEG and expression value (RPKM) on each group for genes involved in hair disorder.
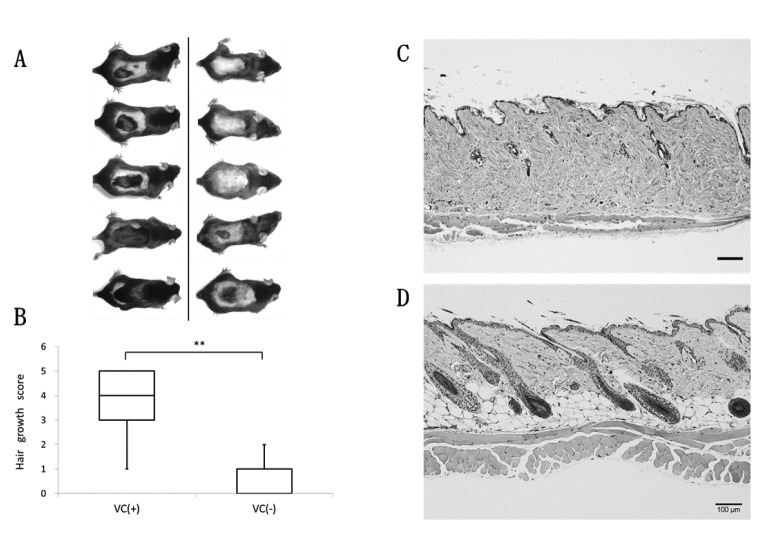
Experiment 2: Scoring hair growth and skin tissue observations. Changes in gene expression involved in hair growth were observed; therefore, the backs of mice in each VC(+) group and VC(–) group were shaved for comparison. Accelerated hair growth was visually observed in the VC(+) group (Figure 5A). The degree of hair growth in each group was scored for comparison, with significantly higher hair growth scores indicated in the VC(+) group than in the VC(–) group (Figure 5B).
Figure 5. Hair growth and skin histological features after shaving back hair in two weeks. (A) Photograph of back hair status in VC(+) group (n=5) (left), VC(–) group (n=5) (right). (B) Changing hair growth score in VC(+) or VC(–) of SMP30/GNL-KO Mice. The back hair of mice was shaved and, after two weeks, hair growth efficiency was evaluated. Data are shown as means±SD. Significant differences (**p<0.01) were obtained between VC(+) group and VC(–) group. (C), (D) Histological features of the back skin lesions were stained with hematoxylin and eosin (HE). C: VC(–); D: VC(+). Scale bars=100 μm.
Moreover, skin tissues were stained with HE for observation, and it was confirmed that the morphology of dermal adipocytes and hair follicles were maintained in the VC(+) group compared with the VC(−) group (Figures 5C, D).
Discussion
VC is involved in anti-oxidative activity and collagen synthesis. It has previously been reported that VC deficiency affects the skin. VC is involved not only in the functional maintenance of the skin, but also in hair growth. Therefore, effects on gene expression caused by decreased VC levels were examined.
According to PCA, expression in each sample was not very high, with a proportion of variance of 30.5%; however, the first principal component in the VC(−) group was different from that in the VC(+) group. A difference in 4W and 8W was observed in the VC(+) group in the second principal component (proportion of variance of 19.8%). Using the 1,736 DEGs chosen by expression analysis, clustering of individual mice and of information between genes were conducted to create a heat map. When clustering samples in the heat map, clustering was not observed with respect to the administration period (4W or 8W) in the VC(+) group; therefore, DEGs identified were thought to be genes that fluctuated within the VC(–) or VC(+) groups, rather than due to the administration of VC for 4W or 8W.
Various DEGs that were significantly different in the VC(–) and VC(+) groups were identified. Furthermore, by analyzing the DEGs, physiological phenomena occurring inside skin tissues could be predicted using IPA. In other words, fluctuations of genes involved in connective tissues and epithelial tissues in skin as well as of genes involved in hair growth were observed as the VC concentration decreased. Moreover, by observing hair growth and considering the observations by Arai et al. (5), significant differences were shown in the VC(–) and VC(+) groups. These results indicated delayed conditions, particularly of hair growth, in skin tissues in the VC(−) group, while physiological conditions were maintained in the VC(+) group. They also reported that VC was completely absent from skin tissues and that the hydroxyproline concentration simultaneously markedly decreased 60 days after the cessation of VC administration (5).
The expression of keratin and keratin-related proteins, which are components of skin tissues, are thought to be important for hair formation. This study found that the expression of genes involved in keratin synthesis (Krt25, 71, 75, 81, 83 and 86) was significantly increased in the VC(+) group compared with the VC(−) group and that their expression was almost non-existent in the VC(−) group.
Regarding changes in gene expression not involved in hair growth and hair formation, decreased apoptosis was predicted in the VC(+) group; therefore, it is hypothesized that oxidative stress increases due to VC deficiency and induces apoptosis. Additionally, it has been predicted that a decrease in the formation of tissues such as the dermal layer of skin (connective tissues and epithelial tissues) is observed due to the inhibition of collagen synthesis in the VC(−) group.
With regard to the maintenance of skin functions and hair growth, collagen is synthesized in osteoblasts in the presence of VC and causes up-regulation of Sonic hedgehog (Shh). Shh is an important gene for cell proliferation and new hair growth not only in osteoblasts but also in various tissues. It has also been confirmed in the present report that expression of Shh was almost non-existent in the VC(−) group, while high expression of the gene was observed in the VC(+) group. Furthermore, IPA analyses indicated that the Shh signaling pathway is significantly up-regulated in the VC(+) group (p-value=1.51E-04, z-score=2.65). Developmental coordination between hair follicles and dermal adipocytes via Shh has been reported to occur during follicle differentiation (13). Furthermore, it has been found that the Wnt/b catenin signaling pathway is significantly up-regulated (p-value= 3.78E-03, z-score=2.07) in addition to the Shh signaling pathway. Shh and Wnt (14) are involved in the differentiation and formation of follicles; therefore, it has been predicted that the differentiation and formation of hair follicles are prevented by VC deficiency. BMP and Notch signaling pathways are involved in hair growth (15); however, changes in these pathways were not identified in the present study.
Function and disease involved in cancers was predicted to be enhanced in the VC(+) group (Table I). Cell proliferation markedly decreased due to VC deficiency but was recovered by VC intake. The predictions would be due to common genes in process of cell proliferation in skin cells of VC (+) and cancer cells. As an example, Shh expression enhances the progression of cancer such as kidney cancer (16), lung cancer (17), and leukemia (18).
When the backs of mice were shaved for observation, differences in hair growth as well as differences in hair growth from hair papilla in skin tissues were visually observed in the VC(+) and VC(−) groups. These data phenomenologically support the results of gene expression analyses in this report.
In the present study, DEGs were revealed by comparing the skin of VC-deficient mice and VC-administered mice. This information is very useful in predicting how VC affects the regeneration of hair and skin. RNA-seq of entire skin tissues was performed in this study; however, it is necessary to perform experiments so that changes in the expression of genes and proteins over time can be examined as well as by the finer dissection of skin tissues and hair papilla cells to understand chronological changes in each intercellular network.
Acknowledgements
This study was supported, in part, by the MEXT-Supported Program for the Strategic Research Foundation at Private Universities 2013-2017 (S1311017). The Authors received generous support from Dr. Akihito Ishigami at the Department of Aging Regulation, Tokyo Metropolitan Institute of Gerontology, Japan. The Authors would also like to thank Rumi Ohtake and Eri Kubota for providing technical support with respect to RNA-Seq and Yuka Saito for his contribution to creating figures.
References
- 1.Murad S, Grove D, Lindberg K, Reynolds G, Sivarajah A, Pinnell S. Regulation of collagen synthesis by ascorbic acid. Proceedings of the National Academy of Sciences. 1981;78:2879–2882. doi: 10.1073/pnas.78.5.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kondo Y, Sakuma R, Ichisawa M, Ishihara K, Kubo M, Handa S, Mugita H, Maruyama N, Koga H, Ishigami A. Potato chip intake increases ascorbic acid levels and decreases reactive oxygen species in SMP30/GNL knockout mouse tissues. J Agric Food Chem. 2014;62:9286–9295. doi: 10.1021/jf502587j. [DOI] [PubMed] [Google Scholar]
- 3.Nishikimi M, Koshizaka T, Ozawa T, Yagi K. Occurrence in humans and guinea pigs of the gene related to their missing enzyme L-gulono-gamma-lactone oxidase. Arch Biochem Biophys. 1988;267:842–846. doi: 10.1016/0003-9861(88)90093-8. [DOI] [PubMed] [Google Scholar]
- 4.Kondo Y, Inai Y, Sato Y, Handa S, Kubo S, Shimokado K, Goto S, Nishikimi M, Maruyama N, Ishigami A. Senescence marker protein 30 functions as gluconolactonase in L-ascorbic acid biosynthesis, and its knockout mice are prone to scurvy. Proc Natl Acad Sci. 2006;103:5723–5728. doi: 10.1073/pnas.0511225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arai KY, Sato Y, Kondo Y, Kudo C, Tsuchiya H, Nomura Y, Ishigami A, Nishiyama T. Effects of vitamin C deficiency on the skin of the senescence marker protein-30 (SMP30) knockout mouse. Biochem Biophys Res Commun. 2009;385:478–483. doi: 10.1016/j.bbrc.2009.05.104. [DOI] [PubMed] [Google Scholar]
- 6.Sato Y, Uchida E, Aoki H, Hanamura T, Nagamine K, Kato H, Koizumi T, Ishigami A. Acerola (Malpighia emarginata DC.) Juice Intake Suppresses UVB-Induced Skin Pigmentation in SMP30/GNL Knockout Hairless Mice. PLoS One. 2017;12:e0170438. doi: 10.1371/journal.pone.0170438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kishimoto Y, Saito N, Kurita K, Shimokado K, Maruyama N, Ishigami A. Ascorbic acid enhances the expression of type 1 and type 4 collagen and SVCT2 in cultured human skin fibroblasts. Biochem Biophys Res Commun. 2013;430:579–584. doi: 10.1016/j.bbrc.2012.11.110. [DOI] [PubMed] [Google Scholar]
- 8.Catani MV, Savini I, Rossi A, Melino G, Avigliano L. Biological Role of Vitamin C in Keratinocytes. Nutr Rev. 2005;63:81–90. doi: 10.1111/j.1753-4887.2005.tb00125.x. [DOI] [PubMed] [Google Scholar]
- 9.Sung YK, Hwang SY, Cha SY, Kim SR, Park SY, Kim MK, Kim JC. The hair growth promoting effect of ascorbic acid 2-phosphate, a long-acting Vitamin C derivative. J Dermatol Sci. 2006;41:150–152. doi: 10.1016/j.jdermsci.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Kim SR, Cha SY, Kim MK, Kim JC, Sung YK. Induction of versican by ascorbic acid 2-phosphate in dermal papilla cells. J Dermatol Sci. 2006;43:60–62. doi: 10.1016/j.jdermsci.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 11.Ishigami A, Kondo Y, Nanba R, Ohsawa T, Handa S, Kubo S, Akita M, Maruyama N. SMP30 deficiency in mice causes an accumulation of neutral lipids and phospholipids in the liver and shortens the life span. Biochem Biophys Res Commun. 2004;315:575–580. doi: 10.1016/j.bbrc.2004.01.091. [DOI] [PubMed] [Google Scholar]
- 12.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, Tsai P-C, Gonzalez-Celeiro M, Chung O, Boumard B, Perdigoto CN, Ezhkova E, Hsu Y-C. Hair follicles’ transit-amplifying cells govern concurrent dermal adipocyte production through Sonic Hedgehog. Genes Dev. 2016;30:2325–2338. doi: 10.1101/gad.285429.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim X, Nusse R. Wnt signaling in skin development, homeostasis, and disease. Cold Spring Harb Perspect Biol. 2013;5:a008029. doi: 10.1101/cshperspect.a008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Tumbar T. Hairy tale of signaling in hair follicle development and cycling. Semin Cell Dev Biol. 2012;23:906–916. doi: 10.1016/j.semcdb.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dormoy V, Jacqmin D, Lang H, Massfelder T. From development to cancer: lessons from the kidney to uncover new therapeutic targets. Anticancer Res. 2012;32:3609–3617. [PubMed] [Google Scholar]
- 17.Jiang WG, Ye L, Ruge F, Sun PH, Sanders AJ, Ji K, Lane J, Zhang L, Satherley L, Weeks HP, Zhi X, Gao Y, Wei C, Wu Y, Mason MD. Expression of Sonic Hedgehog (SHH) in human lung cancer and the impact of YangZheng XiaoJi on SHH-mediated biological function of lung cancer cells and tumor growth. Anticancer Res. 2015;35:1321–1331. [PubMed] [Google Scholar]
- 18.Kawaguchi-Ihara N, Okuhashi Y, Itoh M, Murohashi I, Nara N, Tohda S. Promotion of the self-renewal capacity of human leukemia cells by sonic hedgehog protein. Anticancer Res. 2011;31:781–784. [PubMed] [Google Scholar]