Abstract
About 40% of lung cancer cases globally are diagnosed at the advanced stage. Lung cancer has a high mortality and overall survival in stage I disease is only 70%. This study was aimed at finding a candidate of transcription regulator that initiates the mechanism for metastasis by integrating computational and functional studies. The genes involved in lung cancer were retrieved using in silico software. 10 kb promoter sequences upstream were scanned for the master regulator. Transient transfection of shRNA NFIXs were conducted against A549 and NCI-H1299 cell lines. qRT-PCR and functional assays for cell proliferation, migration and invasion were carried out to validate the involvement of NFIX in metastasis. Genome-wide gene expression microarray using a HumanHT-12v4.0 Expression BeadChip Kit was performed to identify differentially expressed genes and construct a new regulatory network. The in silico analysis identified NFIX as a master regulator and is strongly associated with 17 genes involved in the migration and invasion pathways including IL6ST, TIMP1 and ITGB1. Silencing of NFIX showed reduced expression of IL6ST, TIMP1 and ITGB1 as well as the cellular proliferation, migration and invasion processes. The data was integrated with the in silico analyses to find the differentially expressed genes. Microarray analysis showed that 18 genes were expressed differentially in both cell lines after statistical analyses integration between t-test, LIMMA and ANOVA with Benjamini-Hochberg adjustment at p-value < 0.05. A transcriptional regulatory network was created using all 18 genes, the existing regulated genes including the new genes PTCH1, NFAT5 and GGCX that were found highly associated with NFIX, the master regulator of metastasis. This study suggests that NFIX is a promising target for therapeutic intervention that is expected to inhibit metastatic recurrence and improve survival rate.
Keywords: master regulator, transcription factor, transcriptional regulator, lung cancer, NFIX, metastasis
Introduction
The 5-year survival rate of patients with stage 1A and 1B non–small cell lung cancer (NSCLC) is about 49% and 45% respectively (American Cancer Society, 2015). The poor prognosis in lung cancer suggests that some cells in the primary tumours are programmed to metastasize (Quail and Joyce, 2013; Seyfried and Huysentruyt, 2013). Lung cancer is usually asymptomatic in the early stage and typically symptoms arise when the tumor has metastasized (Slatore et al., 2011). Besides drug resistance, metastasis is also a major cause in cancer recurrence and a big concern in treatment effectiveness (Tang et al., 2013). Therefore, it is crucial to determine the regulatory network that controls the metastatic genes in lung cancer to improve prognosis and increase the survival rate of patients. These genes are not well defined because of the gene transcription changes at every stage of cancer (Shibayama et al., 2011). However, identification of the transcription factors (TFs) that regulate the metastasis mechanisms could be a novel approach for reducing metastasis in lung cancer.
Previous studies of colorectal cancer have indicated that distinct gene expression profiles are associated with disease progression and metastatic recurrence (Shibayama et al., 2011). Metastasis is a multi-step process where tumor cells interact with their microenvironment (Chiang and Massagué, 2008). Expression of the metastatic signature has been directly associated with poor prognosis in most cancers, including colorectal and breast cancers (Hanahan and Weinberg, 2000). For example, expression of the ELAC1 gene in colorectal cancer is essential for inducing tumor development, cell growth and disruption of apoptosis pathways (Kleivi et al., 2007). MTA1 gene is involved in the progression and metastasis of breast cancer (Gururaj et al., 2006). Overexpression of MTA1 also promotes the transcription of oncogenes (Gururaj et al., 2006; Pakala et al., 2013).
Patients with metastatic gene expression shows poor prognosis compared to patients with no metastatic gene signatures (Ramaswamy et al., 2003). The authors suggested that rare cells within a primary tumor have the metastatic phenotype, where they can migrate and invade other cells (Ramaswamy et al., 2003). Although more than 90% of metastasis signatures have been discovered, the metastatic mechanisms of these signatures are largely unknown (Sleeman and Steeg, 2010). Gene expression profiling for identification of molecular signatures results in extensive data on differentially expressed genes (Rohrbeck et al., 2008). The gene expression data provides an insight into every step of lung carcinogenesis according to the different tumor morphology (Borczuk et al., 2003). Using the bioinformatics software, a molecular network on metastasis can be constructed and the TFs that control the metastasis process can be identified.
Predicting the specific binding of TFs to a DNA sequence is the main key in constructing a specific transcriptional regulation network which leads to the metastatic process. To facilitate understanding of the metastatic mechanisms, the TFs need to be identified first; hence the regulatory network can be constructed. These TFs could potentially be targeted to control lung cancer metastasis, thus could improve the survival and prognosis of patients with lung cancer.
Materials and Methods
Identification of Candidate Metastatic Lung Cancer Genes
Nine lung cancer datasets were selected from the Oncomine database1: Bhattacharjee, Beer, Bild, Hou, Lee, The Cancer Genome Atlas (TCGA), TCGA2, Tomida and Lung. These datasets are based on the original microarray analyses on lung cancer by various researchers that published and pooled together in data-mining platform of Oncomine for easy discovery. The difference between both TCGAs is that the second dataset (TCGA2) is actually containing the data from new samples profiled since the first published. Genes related to non–small cell primary lung cancer adenocarcinoma, the 5-year survival rate and stage I and II pathology subtypes were retrieved using the given filters. All datasets were compared except for the outliers, and the cut-off point was set at p-value < 0.01. FunDO is a functional disease ontology annotation database (django.nubic.northwestern.edu/fundo/) lists the genes involved in various cancers (Osborne et al., 2009). The common genes involved in cancers and lung cancer obtained were classified according to their biological processes and molecular functions (cell–cell adhesion, cell migration, cell motion, cell death, apoptosis, programmed cell death, regulation of locomotion, cell migration, cell cycle, cell division, localization of cell, cellular differentiation, cell viability, inflammation, regulation of transcription, angiogenesis and oncogenic signaling pathway) were using DAVID Bioinformatics Database Version 6.7 (Huang et al., 2008). Integrated genes from DAVID analysis underwent further filtration by Pathway Studio analysis with the cut-off point at p-value < 0.01. Overlapping genes were selected using GeneVenn (Pirooznia et al., 2007).
Identification of the Master Regulator of Metastatic Lung Cancer
10kb promoter sequences upstream from the transcription start site (TSS) of the final candidate genes were retrieved from the Eukaryotic Promoter Database (EPD2) and the National Centre for Biotechnology Information (NCBI3) for TF recognition in FASTA format. MATCH, a TRANSFAC® gene regulation database program, was used for predicting putative transcription factor binding sites (TFBS) in DNA sequences. TFBS library in the TRANSFAC® database was used to construct specific binding site weight matrices in TFBS prediction. A maximum core dissimilarity cut-off value of 15% (85% core similarity) was chosen for each matrix. This parameter shows the similarity of a sequence and the weight matrix used by the system to report the TF as a true binding site. PROMO version 3.0.24 scans and identifies TFBS in promoter regions by weight matrix search (Farré et al., 2003; Messeguer et al., 2012). Factors predicted within a dissimilarity margin less or equal to 15%.
Identification of Specific Genes and Their Master Regulator
Transcription factor binding sites derived from both MATCH and PROMO were integrated to identify the common TFs of the regulated genes. To obtain novel findings, we constructed a network that includes all regulated genes and the predicted TFs using Pathway Studio and String 2.0. One of the direct networks was selected based on in silico prediction results as shown in Section “Results.”
Lentiviral Vector and Cell Lines Used
The lentiviral vector with NFIX plasmids (shRNA) was purchased from Thermo Scientific Open Biosystems (BD Biosciences, United States). Plasmids were cultured in Luria broth with ampicillin (AMRESCO, United States) and DNA was extracted using Qiagen purification kit (Germany) for transient transfection. Two human lung cancer cell lines, A549 and NCI-H1299 were purchased from American Type Culture Collection (United States). A549 cells, from a human lung carcinoma cell, maintained in Kaighn’s modification of Ham’s F-12 (F-12K) medium (Thermo Fisher Scientific, United States). NCI-H1299 cells, also derived from a human lung carcinoma and metastatic site lymph nodes, maintained in Roswell Park Memorial Institute1640 (RPMI1640) medium (Thermo Fisher Scientific, United States). Both medium were supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, United States).
Transient Transfection of NFIX shRNA
4 × 104 cells were cultured with serum-free medium without antibiotics in a 6-well culture plate until 60–70% confluent. One microgram of shRNA DNA was transiently transfected into the cells using TurboFECT (Thermo Fisher Scientific, United States). Three NFIX shRNAs, GAPDH shRNA (endogenous control) and non-silencing plasmid were used to study the effects of NFIX loss of function. Transfection efficiency was measured using the fluorescent imaging software NIS-Elements (Nikon Instrument Inc., United States) standardized in all materials and methods. The experiment was performed in triplicate.
Gene Expression Analyses Using Quantitative PCR (qPCR)
Total RNA was isolated using a NucleoSpin® RNA II kit (Macherey-Nagel, United States) according to the manufacturer’s protocol and purity was determined using NanoDrop (Thermo Fisher Scientific, United States). Following the High-Capacity RNA-to-cDNATM kit (Thermo Fisher Scientific, United States) protocol, 1 μg RNA was converted to cDNA. Gene expression analyses were conducted using TaqManFast Advanced Master Mix (Thermo Fisher Scientific) and TaqMan® GeneExpression Assay (Thermo Fisher Scientific, United States) using 7500 Fast Real-Time PCR machine (Thermo Fisher Scientific, United States). The experiment was conducted in triplicate.
Cell Proliferation Assay
1 × 104 cells were cultured in a 96-well culture plate and transfected with 1 μg of each NFIX shRNA in the respective wells and incubated for 48 h. CellTiter 96®AQueous One Solution Cell Proliferation (MTS) (20 μl) (Promega, United States) was added to each well and incubated for 4 h before readings were obtained using Varioskan Flash (Thermo Fisher Scientific) at 490 nm. The experiment was conducted in triplicate.
Cell Migration and Cell Invasion Assays
The QCMTM Chemotaxis (3 μm) migration kit (Chemicon, United States) and QCMTM 24-well Fluorimetric Cell Invasion assay kit (Millipore, United States) were used to measure the migration and invasion activities of the transfected cell lines. The membrane of the ECMatrixTM in the invasion kit only allows invasive cells to migrate toward the underside of the insert filter into the bottom well. In total, 1 × 106 cells were harvested in 1ml serum-free medium and 250 μl cells were pipetted into the insert in triplicate for each treatment. About 400 and 500 μl complete culture medium was filled in the bottom of the well before soaking the insert. After 48 h incubation, the migrated and invaded cells were detached using cell detachment solution and dyed with cyQUANT® GR and 4X Lysis Buffer in a ratio of 1:75. The migrated and invaded cells at the bottom of wells were measured using VarioskanFlash (Thermo Fisher Scientific, United States) at 480/520 nm. The experiment was conducted in triplicate.
Gene Expression Microarray
Transfected cells were collected and RNA was extracted using NucleoSpin® RNA II (Macherey-Nagel, Germany) and diluted with RNase-free water to a final concentration of 150 ng/μl. RNA concentration and purity were measured using NanoDrop (Thermo Fisher Scientific, United States) and Bioanalyzer 2100 RNA 6000 kit (Agilent Technologies, United States). The RNA integrity number (RIN) must be >7 for microarray experiments. TargetAmpTM Nano Labelling Kit for Illumina® Expression BeadChip®(Epicentre Biotechnologies, United States) was used for RNA labeling. The cDNA purification step was carried out using a MinElute Reaction Clean-up (50) kit (Qiagen, Germany). A HumanHT-12v4.0 BeadChip expression kit was used to hybridize the purified cDNA samples. A gene expression direct hybridisation assay system from Illumina was used to process the BeadChip. Cy3-streptavidin (Thermo Fisher Scientific, United States) was introduced as biotin to the analytical probes in the hybridized BeadChip. The iScan Image BeadChip software system was used to scan the BeadChip. Genome Studio, Partek and R software were utilized to analyze the microarray data.
Results
In Silico Analyses for Identifying Master Regulator and the Candidate Genes
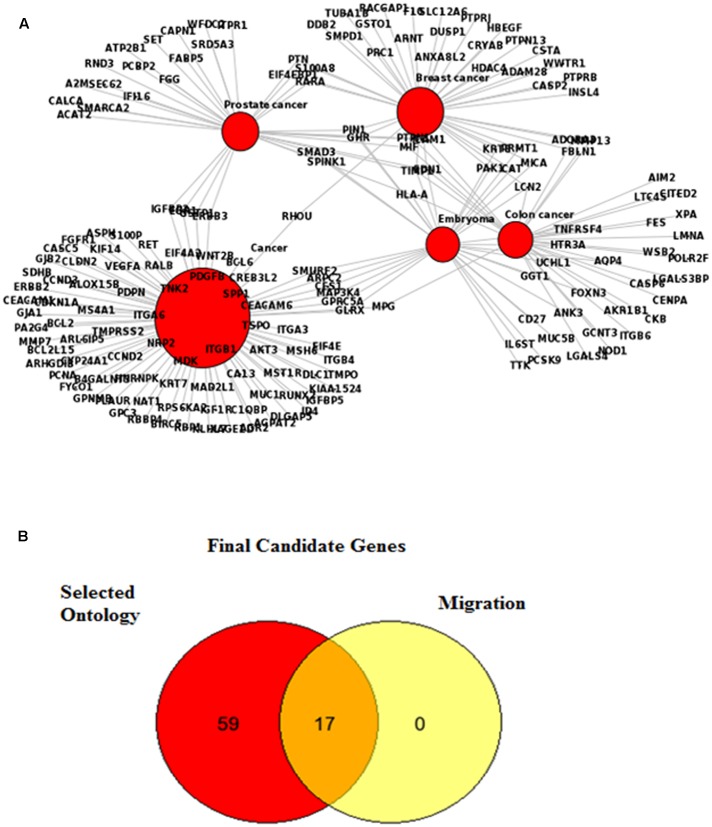
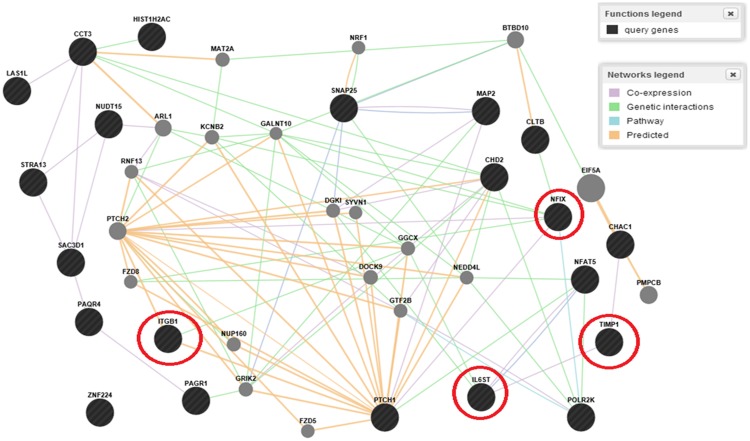
Nine datasets were obtained from the Oncomine database and 3561 genes were selected based on p-value < 0.01 after filtration for the criteria of lung adenocarcinoma, 5-year survival rate and stage I and II pathological subtypes. Early stages of cancer were used to find the genes commonly appear compared to the late stages as we believed they are aiding to metastasis in the micrometastasis stage. The FunDO results were presented in two data sets: one is showing the gene list in lung cancer and the other one is containing the gene list for various cancers such as colorectal, breast, embryoma and prostate. Tables 1, 2 show the gene list involved in lung cancer and various cancers respectively. Figure 1A shows the network involved in various diseases from 549 up-regulated and 674 down-regulated genes. About 18 genes are involved in lung cancer regulation and 88 genes are shared by various cancers based on the gene network that is automatically generated by FunDO analysis using the algorithm set up with Unified Medical Language System (UMLS) MetaMap Transfer tool (MMTx) to discover gene-disease relationship from the GeneRIF database. All 106 genes were enriched and classified by selected ontology using DAVID and Pathway Studio. All genes involved in the migration pathway were selected as final candidate genes. Figure 1B shows all candidate of transcription regulator and Table 3 shows the final gene list commonly involved in lung cancer with their respected direction of regulation in lung cancer including ALOX15B, CALCA, CEACAM1, DLC1, ICAM1, IL6ST, ITGB1, RET, S100P, VEGFA, BCL2, IGF1R, IGFBP5, ITGA6, NRP2, PDGFB and PDPN. Table 4 shows the final candidate of TFs involved in lung cancer metastasis regulation, i.e., AP1, SP1, GR, FOXJ1, GATA6, NF1, GATA5, TTF1(NKX2.1), ATF2, C/EBP, C/EBPG, C-JUN, myogenin/NF1, HNF3B, RXR:RAR, CTF1, FOXL1, TAX/CREB and NF1A/NFIX. We selected nuclear factor I X (NFIX) as the candidate master regulator as it is a family of closely related TFs that constitutively bind as dimers to specific DNA sequences with high affinity. We chose NFIX instead of another TFs since it has higher occurrence. NFIX is also less studied especially in lung cancer. Furthermore, NFIX has a high occurrence in 10 kb upstream of the promoter sequences of the candidate genes. We constructed a network consist of all our known regulated genes and the predicted TFs using Pathway Studio and String 2.0, however, we only pick one direct network based on in silico prediction results as shown in Figure 2. The signaling network of IL6ST was evaluated and TIMP1 was established as the gene that is co-expressed in the migration program. In addition, ITGB1 was selected as the final candidate gene to complete the network as it is involved in the migration and invasion pathways as determined in the previous analyses.
Table 1.
Functional disease ontology annotation database (FunDO) identified gene involved in lung cancer regulation.
| Rank | Gene | Entrez ID |
|---|---|---|
| 1 | GHR | 2690 |
| 2 | CSTA | 1475 |
| 3 | TIMP1 | 7076 |
| 4 | ADAM8 | 101 |
| 5 | P2RY6 | 5031 |
| 6 | FGFR1OP | 11116 |
| 7 | LGALS3BP | 3959 |
| 8 | KLRB1 | 3820 |
| 9 | CCNB2 | 9133 |
| 10 | ADAM28 | 10863 |
| 11 | KRT8 | 3856 |
| 12 | UCHL1 | 7345 |
| 13 | XPA | 7507 |
| 14 | IL6ST | 3572 |
| 15 | ICAM1 | 3383 |
| 16 | MIF | 4282 |
| 17 | CALCA | 796 |
| 18 | FEN1 | 2237 |
Table 2.
Functional disease ontology annotation database (FunDO) identified genes involved in various cancer networks.
| Rank | Gene | Entrez ID | Rank | Gene | Entrez ID | Rank | Gene | Entrez ID |
|---|---|---|---|---|---|---|---|---|
| 1 | CDKN1A | 1026 | 30 | DLC1 | 10395 | 59 | WNT2B | 7482 |
| 2 | NAT1 | 9 | 31 | GPC3 | 2719 | 60 | RBP1 | 5947 |
| 3 | ERBB2 | 2064 | 32 | KLHL7 | 5597 | 61 | ID4 | 3400 |
| 4 | ERBB3 | 2065 | 33 | VEGFA | 7422 | 62 | BIRC5 | 332 |
| 5 | PDGFB | 5155 | 34 | GLRX | 2745 | 63 | CA13 | 377677 |
| 6 | SPP1 | 6696 | 35 | TSPO | 706 | 64 | RET | 5979 |
| 7 | GPRC5A | 9052 | 36 | CIQBP | 708 | 65 | RUNX2 | 860 |
| 8 | CES1 | 1066 | 37 | KIFI4 | 9928 | 66 | AGPAT2 | 10555 |
| 9 | RPS6KA2 | 6196 | 38 | PLAUR | 5329 | 67 | CLDN2 | 9075 |
| 10 | CYP24A1 | 1591 | 39 | FGFR1 | 2260 | 68 | ARPC2 | 10109 |
| 11 | DLGAP5 | 9787 | 40 | GPNMB | 10457 | 69 | CCND2 | 894 |
| 12 | MST1R | 4486 | 41 | GSTP1 | 2950 | 70 | CCND3 | 896 |
| 13 | ITGA6 | 3655 | 42 | MSH6 | 2956 | 71 | B4GALNT3 | 283358 |
| 14 | CEACAM6 | 4680 | 43 | SMURF2 | 64750 | 72 | MMP7 | 4316 |
| 15 | FYCO1 | 79443 | 44 | SDHB | 63905 | 73 | ARHGDIB | 397 |
| 16 | BCL2 | 596 | 45 | ALOX15B | 247 | 74 | IGF1R | 3480 |
| 17 | ITGA3 | 3675 | 46 | CASC5 | 57082 | 75 | IGFBP2 | 3485 |
| 18 | BCL6 | 604 | 47 | CREB3L2 | 64764 | 76 | IGFBP5 | 3488 |
| 19 | MDK | 4192 | 48 | MPG | 4350 | 77 | MS4A1 | 931 |
| 20 | ITGB1 | 3688 | 49 | RALB | 5899 | 78 | EGR1 | 1958 |
| 21 | ITGB4 | 3691 | 50 | KRT7 | 3855 | 79 | PA2G4 | 5036 |
| 22 | RHOU | 58480 | 51 | AKT3 | 10000 | 80 | EIF4A2 | 1974 |
| 23 | HNRNPK | 3190 | 52 | BCL2L15 | 440603 | 81 | EIF4E | 1977 |
| 24 | MAP3K4 | 4216 | 53 | XAGEID | 9503 | 82 | ASPH | 444 |
| 25 | CEACAM1 | 634 | 54 | PDPN | 10630 | 83 | TMPO | 7112 |
| 26 | NRP2 | 8828 | 55 | RBBP4 | 5928 | 84 | TMPRSS2 | 7113 |
| 27 | GJA1 | 2697 | 56 | KIAA1524 | 57650 | 85 | TNK2 | 10188 |
| 28 | S100P | 6286 | 57 | ARL6IP5 | 10550 | 86 | MUC1 | 4582 |
| 29 | GJB2 | 2706 | 58 | AGR2 | 10551 | 87 | MAD2L1 | 4085 |
| 88 | PCNA | 5111 | ||||||
FIGURE 1.
FunDO analysis revealed the molecular networks for various cancers. (A) Genes such as SMAD3, HLA-A, S100A8 and RHOU were common in prostate, breast and colon cancers. (B) Venn diagram showing the 17 candidate genes involved in migration as well as other common ontology in cancers.
Table 3.
Final gene list prior to transcription factors (TFs) scanning by National Centre for Biotechnology Information (NCBI), Eukaryotic Promoter Database (EPD) and PROMO.
| No. | Expression in Lung Cancer | Gene |
|---|---|---|
| 1 | UP | ALOX15B |
| 2 | UP | CALCA |
| 3 | UP | CEACAM1 |
| 4 | UP | DLC1 |
| 5 | UP | ICAM1 |
| 6 | UP | IL6ST |
| 7 | UP | ITGB1 |
| 8 | UP | RET |
| 9 | UP | S100P |
| 10 | UP | VEGFA |
| 11 | DOWN | BCL2 |
| 12 | DOWN | IGF1R |
| 13 | DOWN | IGFBP5 |
| 14 | DOWN | ITGA6 |
| 15 | DOWN | NRP2 |
| 16 | DOWN | PDGFB |
| 17 | DOWN | PDPN |
Table 4.
Transcription factor binding sites (TFBS) of up-regulated and down-regulated genes that occurred in MATCH and PROMO, scanned up to 10 kb upstream of the promoter sequences from the transcription start site (TSS).
| Rank of occurrence | TFBS |
|---|---|
| 1 | AP1 |
| 1 | SP1 |
| 2 | GR |
| 3 | FOXJ1 |
| 4 | GATA5 |
| 5 | Myogenin/NF1 |
| 6 | GATA6 |
| 7 | TTF1 (NKX2.1) |
| 8 | ATF2 |
| 9 | C/EBP |
| 9 | C/EBPG |
| 9 | C-JUN |
| 9 | NF1 |
| 10 | C/EBPA |
| 11 | HNF3B |
| 11 | RXR:RAR |
| 12 | CTF1 |
| 12 | FOXL1 |
| 13 | NF1A/NFIX |
| 13 | TAX/CREB |
FIGURE 2.
Circled are the known regulated genes and predicted master regulator NFIX as a final direct network that underwent the validation based on in-silico prediction results.
Effect of NFIX Silencing on the Regulated Genes IL6ST, TIMP1 and ITGB1
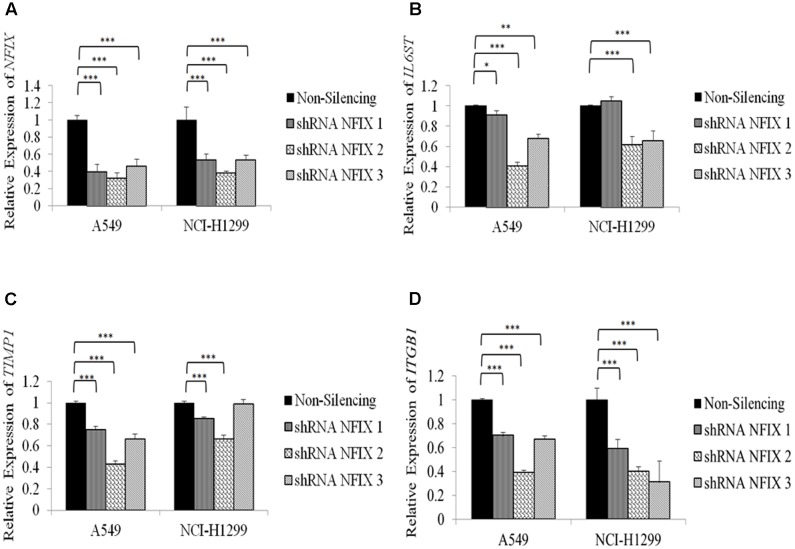
Relative expression of NFIX in A549 cells was decreased significantly with p-value < 5.0 × 10-4 for each construct 48h post-transfection. NCI-H1299 cells transfected with NFIX shRNA also showed significant reduction of NFIX gene expression. NFIX_2 shRNA caused the most significant reduction of IL6ST expression in A549 and NCI-H1299 cells (p-value = 1.2 × 10-4 and 7.99 × 10-5, respectively). NFIX_2 shRNA caused the most significant reduction of TIMP1 expression in A549 cells (p-value = 2.71 × 10-8) and NCI-H1299 cells (p-value = 4.98 × 10-7). All NFIX shRNAs caused significant reduction in ITGB1 expression in both A549 and NCI-H1299 cells. Figure 3 shows NFIX, IL6ST, TIMP1 and ITGB1 expression in both cell lines after 48 h transfection with NFIX shRNAs.
FIGURE 3.
A549 and NCI-H1299 cell lines transfected for 48 h with NFIX shRNAs had reduced NFIX expression. (A) Relative expression of NFIX after 48 h NFIX shRNAs transfection. (B) Transfection of NFIX shRNAs reduced IL6ST gene expression in both cell lines. (C) Transfection with NFIX_2shRNA showed the most significant reduction of TIMP1gene expression. (D) Transfection of NFIX shRNAs significantly reduced ITGB1 gene expression in both cell lines. ∗p < 0.01;∗∗ 0.01 < p < 0.05; ∗∗∗p < 0.05.
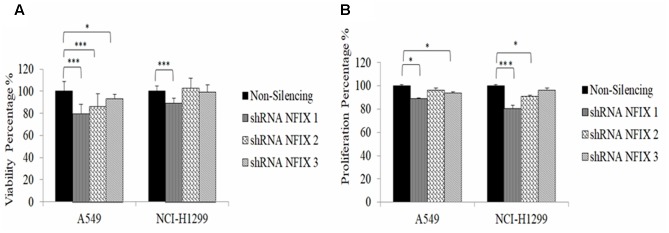
NFIX Reduced Cellular Viability and Proliferation
NFIX_1 shRNA transfection reduced cellular viability significantly in both cell lines (A549: p-value = 5.87 × 10-5; NCI-H1299: p-value = 2.4 × 10-4) 48h post-transfection. Cellular proliferation was reduced significantly at 48 h post-transfection with NFIX_1 shRNA in both A549 and NCI-H1299 (p-value = 0.05 and 0.01, respectively). Figure 4 shows the reduction in cellular viability and proliferation in both cells 48 h after NFIX shRNAs transfection.
FIGURE 4.
Cellular viability and proliferation activities were reduced in both cell lines at 48 h post-transfection with NFIX shRNAs. (A) A549 cell viability was decreased following transfection with all NFIX shRNAs. (B) NCI-H1299 cell viability was reduced only by NFIX_1shRNA. ∗p < 0.01; ∗∗∗p < 0.05.
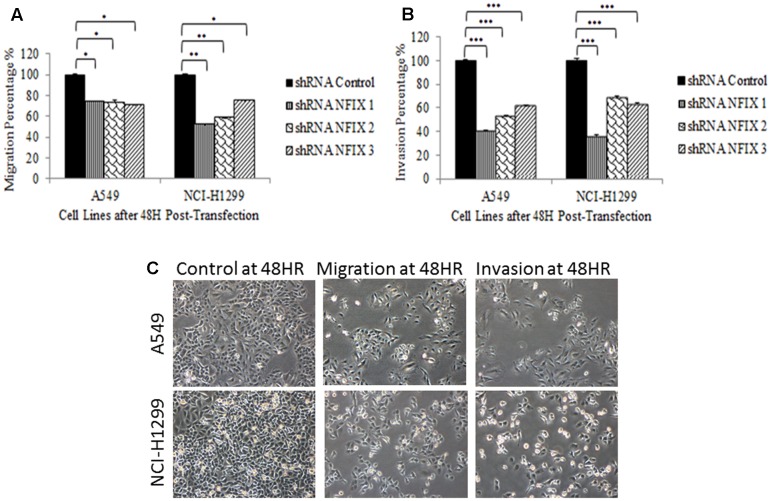
NFIX Decreased Cellular Migration and Invasion
Transfection of all NFIX shRNAs significantly reduced the migration and invasion activities in both cell lines at 48h post-transfection. Transfection of NFIX_1 shRNA showed the most significant reduction in both cell lines after 48h (A549: p-value = 7.9 × 10-3; NCI-H1299: p-value = 7.3 × 10-3). This observation suggests that NFIX may play an important role in lung cancer metastasis. Figure 5 shows that NFIX shRNAs transfection reduced migration and invasion activities in both cell lines.
FIGURE 5.
Reduction in cellular migration and invasion activities at 48 h post-transfection. (A) All NFIX shRNAs decreased migration in both cell lines. (B) All NFIX shRNAs significantly reduced invasion in both cell lines. (C) Transwell migration and invasion assays compared with each control cell lines. (∗p < 0.01; ∗∗0.01 < p < 0.05; ∗∗∗p < 0.05).
Microarray Analysis of Master Regulator and Regulatory Network in Lung Cancer
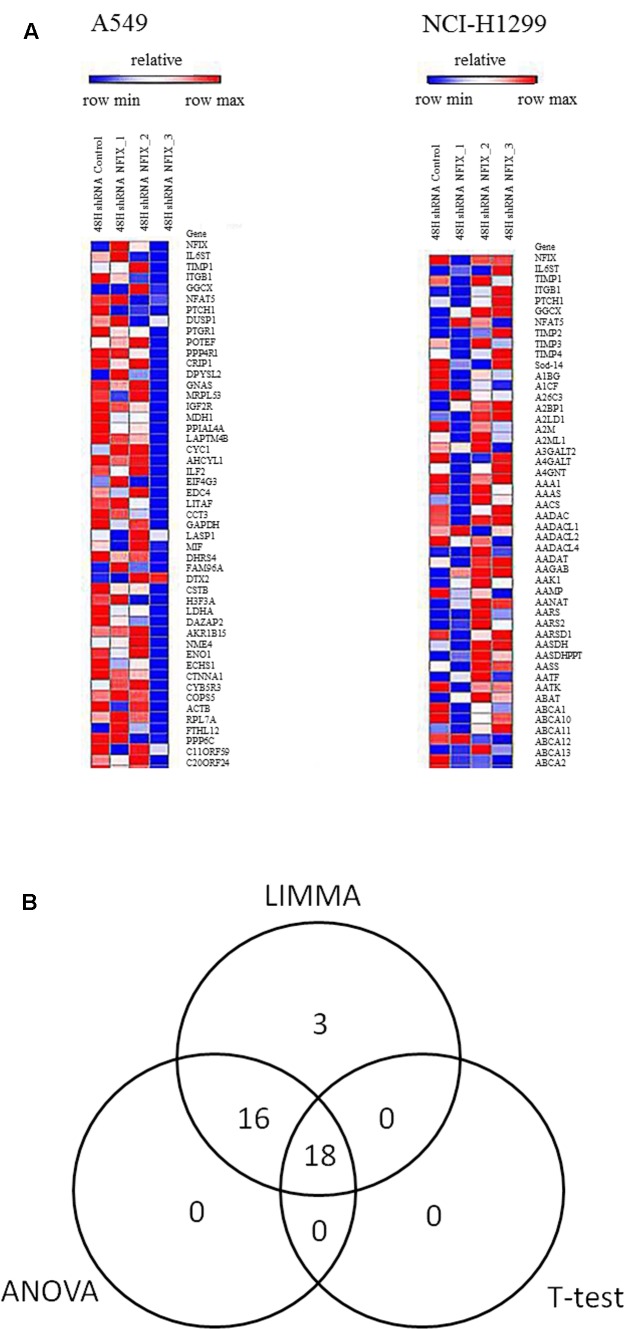
Gene expression microarray analyses were carried out to find differentially expressed genes before and after NFIX knockdown by comparing each cell line with respective controls at p-value < 0.05. Figure 6A depicts the heatmap of the top 50 genes including NFIX, IL6ST, TIMP1 and ITGB1, for both cell lines at 48 h post-transfection. All differentially expressed genes in both cell lines from microarray analyses were integrated before undergo the statistical analyses whereas Figure 6B demonstrates the results of integrated analyses from all statistical software used. Table 5 listed 18 genes identified via integrated analyses with respect to their fold change. All 18 genes were mapped into pathways to determine their involvement in lung cancer metastasis and how they are regulated by NFIX.
FIGURE 6.
Heatmap from microarray gene expression analyses showing the differentially expressed genes in the A549 and NCI-H1299cell lines. (A) Shows differentially expressed genes in both cell lines at48h post–NFIX shRNAs transfection as compared to the non-silenced samples. (B) T-test, LIMMA and ANOVA were used to identify differentially expressed genes between NFIX silencing and non-silencing in the A549 and NCI-H1299cell lines. In total, 18 genes were identified as differentially expressed between both cell lines (∗with Benjamini-Hochberg adjustment with p-value < 0.05).
Table 5.
The 18 differentially expressed genes in A549 and NCI-H1299 cell lines.
| No. | Gene | Fold change (FC) | Log FC |
|---|---|---|---|
| 1 | CHAC1 | 3.26 | 1.70 |
| 2 | HIST1H2AC | 2.34 | 1.22 |
| 3 | MAP2 | 2.13 | 1.09 |
| 4 | NFAT5 | 1.94 | 0.95 |
| 5 | ZNF224 | 1.92 | 0.94 |
| 6 | MAP2 | 1.91 | 0.93 |
| 7 | CHD2 | 1.86 | 0.89 |
| 8 | SNAP25 | 1.78 | 0.83 |
| 9 | PTCH1 | 1.45 | 0.54 |
| 10 | CLTB | 0.77 | -0.38 |
| 11 | NUDT15 | 0.76 | -0.39 |
| 12 | POLR2K | 0.75 | -0.41 |
| 13 | LAS1L | 0.73 | -0.45 |
| 14 | CCT3 | 0.70 | -0.51 |
| 15 | C16orf53 | 0.64 | -0.64 |
| 16 | PAQR4 | 0.63 | -0.67 |
| 17 | SAC3D1 | 0.58 | -0.79 |
| 18 | STRA13 | 0.56 | -0.83 |
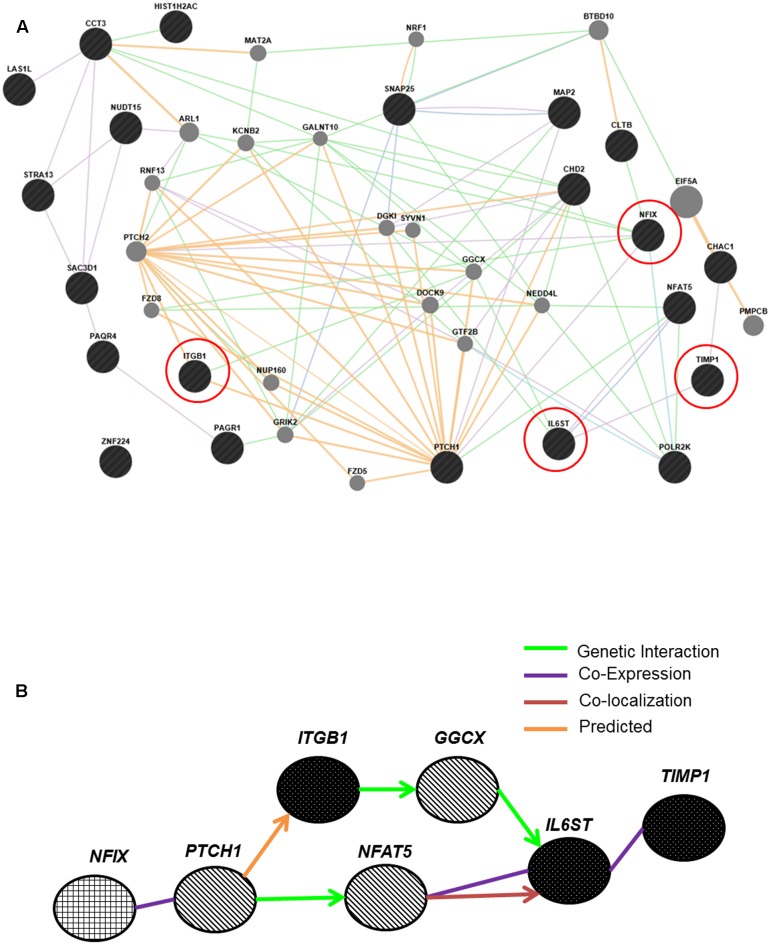
Construction of a New Regulatory Pathway from Genes Identified in Microarray Analyses
The regulated genes ITGB1, TIMP1 and IL6ST, were included in the pathway altogether with 17 genes identified in the gene expression microarray using GeneMANIA. One pathway was selected as a new regulatory pathway network where it contained the most regulated genes in lung cancer metastasis as explained in Figure 2 above. Figure 7A shows several new pathways created from genes identified in the meta-analysis and the microarray analyses. One direct network was selected prior to determination of their functions using GeneCards. NFIX, a master regulator regulates ITGB1, TIMP1 and IL6ST, as well as three new entities, PTCH1, NFAT5 and GGCX, were included in this new transcriptional network as shown in Figure 7B.
FIGURE 7.
Pathways created by GeneMANIA connect the master regulator NFIX and its regulated genes IL6ST, TIMP1 and ITGB1. (A) 17 differentially expressed genes from microarray analyses showed connection with all regulated genes and NFIX. Some genes showed co-expression with NFIX, including PTCH1. (B) Successful construction of a new transcriptional network from both meta-analysis and microarray analyses. NFIX was identified as a master regulator that regulates the ITGB1, TIMP1 and IL6ST genes. Three new entities: the PTCH1, NFAT5 and GGCX, were also included in this metastasis regulatory network for lung cancer. NFIX is the master regulator; IL6ST, TIMP1 and ITGB1 are the NFIX-regulated genes and PTCH1, NFAT5 and GGCX are genes involved in the NFIX transcriptional regulatory network.
Discussion
The understanding of tumor progression and TFs are critical in the efforts of identifying new biomarkers, inventing novel therapeutics, making patients’ prognosis and designing an advance cancer detection tools (Gomez, 2016). The identification of gene signatures for metastasis is crucial for preventing tumor cells from metastasizing, to improve lung cancer prognosis as well as increased survival. The target gene could be used for future therapy to inhibit the metastasis process in lung cancer. The identification of TFs that regulate gene expression is essential for understanding the whole transcription process, particularly the genes involved in metastasis. In this study, we identified 17 genes involved in the migration pathway using DAVID and Pathway Studio. Subsequently, 10 kb upstream of the promoter sequences of these 17 genes were retrieved and finally we postulated that NFIX is a master regulator for lung cancer metastasis. We showed evidence that NFIX regulates IL6ST, TIMP1 and ITGB1 genes. Altogether, these genes promote cell migration and invasion, which lead to lung cancer aggressiveness, poor survival and high mortality rates. However, further study need to be done to understand the molecular mechanism of NFIX in regulating these regulated genes and functional pathways of metastasis.
In addition to NFIX, AP1 was also identified more occurrences at 10 kb upstream of the promoter sequence. The AP1 gene is well studied in the migration pathway of breast cancer via the extracellular signal–regulated kinase pathway (Chen et al., 2009). It plays an important role in regulating endothelin-1 (ET1) gene transcription in endothelial cells (Kawana et al., 1995). The TF SP1 also identified with the highest occurrences; it is highly expressed in the tumor cells progression of gastric cancer (Wang et al., 2003). Blomquist et al. (1999) showed that NFIX acts as a specific dimer to DNA sequences with high affinity which can increase the specificity for molecular recognition and aid targeted therapy in lung cancer (Blomquist et al., 1999).
Silencing of NFIX reduced the expression of its regulated genes IL6ST, TIMP1 and ITGB1. Necula et al. (2012) showed that increased levels of IL6ST in tissues (>758 pg/ml) and plasma (>38.21 pg/ml) are associated with shorter survival in gastric adenocarcinoma. IL6 manipulates the tumorigenesis process by activating the genes involved in differentiation, survival, apoptosis and proliferation (Lin et al., 2007). In addition, IL6 may be involved in promoting inflammation by inducing anti-apoptotic signals which is mediated by STAT3 (Necula et al., 2012). Lissat et al. (2015) showed that high expression of IL6ST aids Ewing sarcoma tumor progression and renders it resistant to apoptosis and promotes metastasis, thus suggesting that IL6ST plays a role in cancer metastasis. TIMP1 promotes melanoma through activation of the AKT pathway independent of the PI3K signaling pathway (Toricelli et al., 2013). In human melanoma cell lines, TIMP interacts with ITGB1 and CD63 and assists melanomagenesis and resistance to the cell death program (Toricelli et al., 2013).
A previous study of ITGB1 demonstrated its association with migration activity, invasion and wound healing in an in vitro experiment of lung adenocarcinoma (Wang et al., 2013). The authors reported that osteopontin, LAMB3 and ITGB1 are pro-metastatic genes for lung cancer. Osteopontin also leads to increased vascular endothelial cell migration, proliferation, angiogenesis and tumor growth in lung cancer (Cui et al., 2007; Fong et al., 2009). Lymphatic metastasis in lung cancer has a high concentration of ITGB1 as compared to non-lymphatic metastasis (Wang et al., 2013). Our data demonstrate similar results in the in vitro experiments, where significant reduction of cell proliferation, migration and invasion was observed post–NFIX knockdown. The in vitro studies were in concordance with the in silico analyses predictions, where NFIX regulates IL6ST, TIMP1 and ITGB1 expression.
Microarray analyses were carried out to discover differentially expressed genes in A549 and NCI-H1299 cell lines post–NFIX silencing as compared to non-silenced cells. Silencing of cell line A549 using NFIX_3 shRNA does not show any difference compared to the other shRNAs. We suggest that the sequence in NFIX_1 shRNA is the most powerful in silencing the master regulator NFIX especially in aggressive cell line NCI-H1299. NFAT5 was identified as a new entity in this network. NFAT5 is a crucial component in tumor development and progression, particularly in regulating inflammation in the carcinogenesis process (Neuhofer, 2010; Yoon et al., 2011). We also observed that PTCH1 is co-expressed with NFIX. PTCH1 serves as a tumor suppressor gene as demonstrated in NSCLC cell lines (Shikata et al., 2011). Li et al. (2012) showed that PTCH1 interacts with the Hedgehog (Hh) signaling pathway via the effect of miRNA-212 on cell proliferation. However, our microarray data showed that PTCH1 is co-expressed with NFIX. Further studies on the co-expression between these two genes need to be performed in the future. The GGCX gene, encoding the enzyme responsible for the post-translational modification of vitamin K–dependent proteins in haemostasis (Stafford, 2005; Weston and Monahan, 2008), was also found in our newly constructed transcriptional regulatory network. Our findings suggest that NFIX may be a potential biomarker with high potential for inhibiting the metastatic process in NSCLC as demonstrated by the in-silico and in vitro experiments.
Conclusion
In silico analyses identified NFIX as a predicted master regulator. Based on the in silico findings, functional assays and microarray, a new transcriptional network involving the inflammation, migration and invasion pathways specific to metastatic lung cancer was created. We acknowledged the in-silico results are considered the limitation of our study as the results of the same analyses will always be different with the new additions from new discoveries as the programmer might update and upgrade the software and algorithms. We used several in silico analyses to strengthen and support our findings. We also ran all NFIX shRNAs lentiviral with different sequences in our study to check the best shRNA needed to significantly reduce the expression of the master regulator and gene functions that lead to metastasis phenotypic outcome. We suggest that NFIX is a master regulator that regulates metastasis in lung cancer by transcribing the genes responsible in activating the pathways leading to micrometastasis and finally cause the cancer cells to metastasize such as inflammation, proliferation, migration and invasion. Where its’ silencing can reduce cellular proliferation, migration and invasion in vitro especially as observed in aggressive cell line. Hence, NFIX with IL6ST, TIMP1, ITGB1, PTCH1, GGCX and NFAT5 genes may regulate the migration and invasion process and they could serve as potential therapeutic targets in patients with lung cancer to predict survival and improve prognosis. We couldn’t carry the in vivo and also protein study experiments due to lack of funding. However, more detailed functional studies, in vivo and clinical validation need to be carried out in the future to prove this concept and make the study more transparent and reliable.
Author Contributions
This study was planned by RH. All experiments were carried out by NIAR as described in the manuscript including interpretation of data and statistical analyses. MMM participated in microarray data analysis. RH and NAAM supervised the student, assisted in study and manuscript writing. The manuscript was written by NIAR with comments from all co-authors and mostly from NAAM, RH, and RJ. All co-authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Nor Adzimah Johdi, Mrs. Nurmi Nasir, Mrs. Sri Noraima Othman and Mrs. Intan Fairuz Ramli for their skilful assistance in cell culture and bioinformatics. We also would like to thank the staff and students of UKM Medical Molecular Biology Institute (UMBI).
Funding. This research was supported by UKM Research University Grant (GUP): UKM-GUP-2011-287.
References
- American Cancer Society (2015). Cancer Facts & Figures 2015. Atlanta, GA: American Cancer Society. [Google Scholar]
- Blomquist P., Belikov S., Wrange O. (1999). Increased nuclear factor 1 binding to its nucleosomal site mediated by sequence-dependent DNA structure. Nucleic Acids Res. 27 517–525. 10.1093/nar/27.2.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borczuk A. C., Gorenstein L., Walter K. L., Assaad A. A., Wang L., Powell C. A. (2003). Non-small-cell lung cancer molecular signatures recapitulate lung developmental pathways. Am. J. Pathol. 163 1949–1960. 10.1016/S0002-9440(10)63553-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhu G., Li Y., Padia R. N., Dong Z., Pan Z. K. (2009). Extracellular signal-regulated kinase signaling pathway regulates breast cancer cell migration by maintaining slug expression. Cancer Res. 69 9228–9235. 10.1158/0008-5472.CAN-09-1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang A. C., Massagué J. (2008). Molecular basis of metastasis. N. Engl. J. Med. 359 2814–2823. 10.1056/NEJMra0805239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R., Takahashi F., Ohashi R., Gu T., Yoshioka M., Nishio K. (2007). Abrogation of the interaction between osteopontin and alphavbeta3 integrin reduces tumor growth of human lung cancer cells in mice. Lung Cancer 5 302–310. 10.1016/j.lungcan.2007.03.019 [DOI] [PubMed] [Google Scholar]
- Farré D., Roset R., Huerta M., Adsuara J. E., Rosellõ L., Albà M. M. (2003). Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 31 3651–3653. 10.1093/nar/gkg605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y. C., Liu S. C., Huang C. Y., Li T. M., Hsu S. F., Kao S. T. (2009). Osteopontin increases lung cancer cells migration via activation of the alphavbeta3 integrin/FAK/Akt and NF-kappaB-dependent pathway. Lung Cancer 64 263–270. 10.1016/j.lungcan.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Gomez C. R. (2016). Editorial: tumor hypoxia: impact in tumorigenesis, diagnosis, prognosis and therapeutics. Front. Oncol. 6:229 10.3389/fonc.2016.00229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururaj A. E., Singh R. R., Rayala S. K., Holm C., den Hollander P., Zhang H. (2006). MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proc. Natl. Acad. Sci. U.S.A. 103 6670–6675. 10.1073/pnas.0601989103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2000). The hallmarks of cancer. Cell Press Review. 100 57–70. 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2008). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawana M., Lee M. E., Quertermous E. E., Quertermous T. (1995). Cooperative interaction of GATA-2 and AP1 regulates transcription of the endothelin-1 gene. Mol. Cell Biol. 15 4225–4231. 10.1016/S0092-8674(00)81683-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleivi K., Lind G. E., Diep C. B., Meling G. I., Brandal L. T., Nesland J. M. (2007). Gene expression profiles of primary colorectal carcinomas, liver metastases, and carcinomatoses. Mol. Cancer 6 2–20. 10.1186/1476-4598-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang D. Q., Chen C. W., Ruan Z. C., Li Y., Huang Y. (2012). MicroRNA-212 displays tumor-promoting properties in non-small cell lung cancer cells and targets the hedgehog pathway receptor PTCH1. Mol. Biol. Cell 23 1423–1423. 10.1091/mbc.E11-09-0777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. T., Lin B. R., Chang C. C., Chu C. Y., Su H. J., Chen S. T. (2007). IL-6 induces AGS gastric cancer cell invasion via activation of the c-Src/RhoA/ROCK signaling pathway. Int. J. Cancer 120 2600–2608. 10.1002/ijc.22599 [DOI] [PubMed] [Google Scholar]
- Lissat A., Joerschke M., Shinde D. A., Braunschweig T., Meier A., Makowska A. (2015). IL6 secreted by the Ewing sarcoma tumor microenvironment confers anti-apoptosis and cell-disseminating paracrine responses in Ewing sarcoma cells. BMC Cancer 15:552 10.1186/s12885-015-1564-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer X., Escudero R., Farré D., Núňez O., Martínez J., Albà M. M. (2012). PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18 333–334. 10.1093/bioinformatics/18.2.333 [DOI] [PubMed] [Google Scholar]
- Necula L. G., Chivu-Economescu M., Stanciulescu E. L., Bleotu C., Dima S. O., Alexiu I. (2012). IL-6 and IL-11 as markers for tumor aggressiveness and prognosis in gastric adenocarcinoma patients without mutations in Gp130 subunits. J. Gastrointestin. Liver Dis. 21 23–29. [PubMed] [Google Scholar]
- Neuhofer W. (2010). Role of NFAT5 in inflammatory disorders associated with osmotic stress. Curr. Genomics 11 584–590. 10.2174/138920210793360961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne J. D., Flatow J., Holko M., Lin S. M., Kibbe W. A., Zhu L. H. (2009). Annotating the human genome with Disease Ontology. BMC Genomics 10(Suppl. 1): S6 10.1186/1471-2164-10-S1-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakala S. B., Rayala S. K., Wang R. A., Ohshiro K., Mudvari P., Reddy S. D. N. (2013). MTA1 promotes STAT3 transcription and pulmonary metastasis in breast cancer. Cancer Res. 73 3761–3770. 10.1158/0008-5472.CAN-12-3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia M., Nagarajan V., Deng Y. (2007). GeneVenn - a web application for comparing gene lists using Venn diagrams. Bioinformation 1 420–422. 10.6026/97320630001420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail D. F., Joyce J. A. (2013). Microenvironment regulation of tumour progression and metastasis. Nat. Med. 19 1423–1437. 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S., Ross K. N., Lander E. S., Golub T. R. (2003). A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33 49–54. [DOI] [PubMed] [Google Scholar]
- Rohrbeck A., Neukirchen J., Rosskopf M., Pardillos G. G., Geddert H., Schwalen A. (2008). Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers. J. Transl. Med. 6 69–86. 10.1186/1479-5876-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried T. N., Huysentruyt L. C. (2013). On the origin of cancer metastasis. Crit. Rev. Oncog. 18 43–73. 10.1615/CritRevOncog.v18.i1-2.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibayama M., Maak M., Nitsche U., Gotoh K., Rosenberg R., Janssen K. P. (2011). Prediction of metastasis and recurrence in colorectal cancer based on gene expression analysis: ready for the clinic? Cancers 3 2858–2869. 10.3390/cancers3032858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata Y., Okada T., Hashimoto M., Ellis T., Matsumaru D., Shiroishi T. (2011). Ptch1-mediated dosage-dependent action of Shh signaling regulates neural progenitor development at late gestational stages. Dev. Biol. 349 147–159. 10.1016/j.ydbio.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Slatore C. G., Gould M. K., Au D. H., Deffebach M. E., White E. (2011). Lung cancer stage at diagnosis; individual associations in the prospective VITamins and lifestyle (VITAL) cohort. BMC Cancer 11:228 10.1186/1471-2407-11-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J., Steeg P. S. (2010). Cancer metastasis as a therapeutic target. Eur. J. Cancer 46 1177–1180. 10.1016/j.ejca.2010.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford D. W. (2005). The vitamin K cycle. J. Thromb. Haemost. 3 1873–1878. 10.1111/j.1538-7836.2005.01419 [DOI] [PubMed] [Google Scholar]
- Tang T., Salama R., Gadgeel S. M., Sarkar F. H., Ahmad A. (2013). Erlotinib resistance in lung cancer: current progress and future perspectives. Front. Pharmacol. 4:15 10.3389/fphar.2013.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toricelli M., Melo F. H. M., Peres G. B., Silva D. C. P., Jasiulionis M. G. (2013). Timp1 interacts with beta-1 integrin and CD63 along melanoma genesis and confers anoikis resistance by activating PI3-K signaling pathway independently of Akt phosphorylation. Mol. Cancer 12 22–37. 10.1186/1476-4598-12-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wei D., Huang S., Peng Z., Le X., Wu T. T. (2003). Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin. Cancer Res. 9 6371–6380. [PubMed] [Google Scholar]
- Wang X. M., Li J., Yan M. X., Liu L., Jia D. S., Geng Q. (2013). Integrative analyses identify osteopontin, LAMB3 and ITGB1 as critical pro-metastatic genes for lung cancer. PLoS ONE 8:e55714 10.1371/journal.pone.0055714 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Weston B. W., Monahan P. E. (2008). Familial deficiency of vitamin K-dependent clotting factors. Haemophilia 14 1209–1213. 10.1111/j.1365-2516.2008.01853.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. J., You S., Yoo S. A., Kim N. H., Kwon H. M., Yoon C. H. (2011). NFAT5 is a critical regulator of inflammatory arthritis. Arthritis Rheum. 63 1843–1852. 10.1002/art.30229 [DOI] [PMC free article] [PubMed] [Google Scholar]