Abstract
In worldwide conditions of increasingly antibiotic-resistant hospital infections, it is important to research alternative therapies. Bdellovibrio bacteriovorus bacteria naturally prey on Gram-negative pathogens, including antibiotic-resistant strains and so B. bacteriovorus have been proposed as “living antibiotics” to combat antimicrobially-resistant pathogens. Predator-prey interactions are complex and can be altered by environmental components. To be effective B. bacteriovorus predation needs to work in human body fluids such as serum where predation dynamics may differ to that studied in laboratory media. Here we combine mathematical modelling and lab experimentation to investigate the predation of an important carbapenem-resistant human pathogen, Klebsiella pneumoniae, by B. bacteriovorus in human serum versus buffer. We show experimentally that B. bacteriovorus is able to reduce prey numbers in each environment, on different timescales. Our mathematical model captures the underlying dynamics of the experimentation, including an initial predation-delay at the predator-prey-serum interface. Our research shows differences between predation in buffer and serum and highlights both the potential and limitations of B. bacteriovorus acting therapeutically against K. pneumoniae in serum, informing future research into the medicinal behaviours and dosing of this living antibacterial.
Introduction
Infections caused by antibiotic-resistant Gram-negative bacteria have steadily risen over the past decade and pose a serious risk to public health. This is illustrated by the fact that Gram-negative bacteria are currently responsible for more than 30% of all hospital-acquired infections1 and are associated with high levels of morbidity and mortality in the Intensive Care Unit setting2–5. Consequently, new therapies effective against drug resistant isolates are urgently needed. One option mooted is the use of predatory bacteria, such as Bdellovibrio bacteriovorus, as therapeutic agents6, 7. B. bacteriovorus HD100 is a small (0.3 µm × 1 µm) Gram-negative bacterium that preys on other Gram-negative bacteria as part of its developmental lifecycle, which has a number of well-defined stages in liquid conditions8. During this lifecycle, the prey act as both food source and housing for the predator9. Firstly, highly motile attack phase predator- cells are propelled by a single flagellum until they encounter a prey cell, at which point they attach to the outer membrane and invade the prey periplasm, forming a predator+dead-prey structure called a bdelloplast. Once established within the periplasm, B. bacteriovorus begin filamentous replication by virtue of sequestration of prey cell nutrients. Depending on the prey and external environment, the typical lifecycle of B. bacteriovorus lasts 3–4 hours and when complete results in lysis of the prey and release of new predatory progeny. The number of predators released is dependent on the size of the prey cell10.
Understanding the dynamics of B. bacteriovorus predation under the in vivo conditions found in a treatment setting, is critical to developing this predatory bacterium as a “living antibiotic”. Formulating a dosage regime for any future administration of a living organism inside a complex system, such as the human body, requires a comprehensive understanding of the factors affecting predation. Previously published in vitro predation studies in buffer and bacteriological media have been informative7, 11 but need expanding towards physiologically relevant conditions. In order to address this issue, we employ an interdisciplinary approach to investigate experimentally and mathematically model the predation by B. bacteriovorus HD100 of an antibiotic-resistant, carbapenemase producing (KPC) clinical isolate of Klebsiella pneumoniae in whole human serum at 37 °C; conditions that better represent those found in vivo. K. pneumoniae KPC is an important human pathogen and member of the ESKAPE group12 of antibiotic resistant human pathogens with carbapenem resistance, which further limits the treatment options for patients. It is a major cause of nosocomial infections globally and has an associated high mortality rate13.
Early mathematical modelling of B. bacteriovorus predation, in buffers/simple lab media alone, includes the influential work of Varon and Zeigler14 who used a Lotka-Volterra model15, 16 to investigate predation on a Photobacterium species. Experimentally they used low concentrations of both predator and prey in a tri-salt buffer to determine the concentrations necessary for survival of B. bacteriovorus and to induce oscillatory waves of predatory behaviour in prey populations. They then used this data to estimate parameters for their Lotka-Volterra model, which predicted that “the prey density required to give bdellovibrios a 50% chance of survival was calculated to be at least 3.0 × 106 cells per ml”, although this has been later disputed17. In contrast, driven by the potential therapeutic application, our model focuses on prey elimination rather than B. bacteriovorus survival. We take the Lotka-Volterra model as a basis for our model, but adapt it significantly to reflect the unique predation mechanism of B. bacteriovorus. Most significantly, our model allows us to capture temporal aspects of predation not accounted for in Varon and Zeiglers’ model, and also assess the impact of prey resistance and environmental variables on predation outcome.
Other early work includes a model of B. bacteriovorus predation on Escherichia coli which explicitly includes a bdelloplast stage with an incubation period during which the predator B. bacteriovorus grows and replicates inside prey18. In contrast to our model, this model only considered predation in buffer with no consideration of immune system effects, and only looked at short time scales (<10 hours after inoculation), allowing the authors to use a much simpler model than the one we present. We also note that the parameter estimates reported for progeny number (from E. coli) are larger than those that have since been observed experimentally. Later modelling work by Wilkinson19 and Hobley20 examined predation in the presence of a third ‘decoy’ species, either inert or live. These studies are an important step away from the previous tightly controlled two species model systems, and the presence of other bacterial species is important in assessing the clinical application of B. bacteriovorus against mixed bacterial infections. These studies, however, unlike the model presented in this work, consider predation in buffer rather than in a complex growth environment as would be the case in any future clinical application.
Most recently, Dattner21 modelled B. bacteriovorus predation in a habitat of sand suspended in buffer. The authors proposed an alternative approach to the standard (numerical integration based) parameter estimation approach used in analysis of non-linear dynamics. By taking a simplified ‘strategic’ model rather than a mechanistic, model the authors were able to use a statistical direct integral approach to estimate the model parameters and produced a fit to a small sample of experimental data points. Whilst this approach may give insights into predation dynamics in an artificially simplified experimental environment, it is unlikely to be suitable for the more complex environment that would be present in the clinical application of B. bacteriovorus, such as serum. In a whole human environment, several immunological and/or antimicrobial factors (such as antibodies, antimicrobial peptides and leukocytes) will act upon the predatory bacteria, potentially killing the predators before predation can occur. These factors are complex but important to understand and quantify as they will impact efficacy of B. bacteriovorus used as a therapeutic.
In this study, we used standard numerical integration to estimate the model parameters and simulate time course behaviour in human serum at the physiological temperature of 37 °C. This results in a complex parameterisation of the predation process, even for serum (albeit without the leukocytes that would be present in whole blood, an area that requires significant further experimentation). By capturing the mechanistic complexity of the experimental system, we are able to use our model to guide the design of experiments and to suggest mechanisms to further investigate. Furthermore, our model allows rapid prediction of predation outcomes over longer timescales, which would be difficult and labour-intensive to obtain experimentally.
Mathematical modelling of B. bacteriovorus predation cannot be thought of as a variation of more traditional animal predator-prey dynamics22, 23. The B. bacteriovorus intra-prey, life cycle length and uniformity leads to a synchronicity of prey bursting and replicated progeny-predator release, in controlled laboratory media (such as buffer), and when an almost equal number of predators to prey are present at the start of predation. Whilst this will not affect the long term (relative to life cycle length) dynamics of the model, it has a significant impact on the shorter term dynamics (less than the cycle length). Previous modelling work for predation in buffer14 did not include the impact that synchronous prey-bursting may have on the validity of the model at early time points, and the early mathematical models were not able to fully account for the differences between bacterial and animal predation. In this paper, we present a more clinically applicable ordinary differential equation (ODE) population-based model of bacterial predator-prey dynamics which is experimentally validated and informs upon both short- and long-term dynamics in buffer and human serum. We also consider some important aspects of predation that as yet have not been fully investigated. Firstly we consider the baseline conditions of predation in buffer, over longer time scales (up to 96 hours). This allows modelling of the prey outgrowing their predators, linking to reports from previous experimental work14, 17. Secondly, we mathematically model and experimentally determine the dynamics of B. bacteriovorus predation of K. pneumoniae in human serum; not the usual buffer employed for most previous predation studies. We quantify and demonstrate that pathogen predation in human serum does occur under certain conditions, that the dynamics of predation in serum is markedly different to that in buffer and that there is serum to serum variation.
A more complete understanding of the potential and limitations of predation in human serum is essential if ultimately B. bacteriovorus is to be considered for use as a living antibiotic. Both our experimental work and model show a pattern of delayed predation in serum initially lowering the K. pneumoniae load, but this is followed by prey regrowth, which will need to be fully understood in future work if we and other Bdellovibrio researchers are to prevent re-infection. The interdisciplinary approach we take here is crucial to achieving that aim, with the experimental work highlighting the marked differences between predation in serum and buffer, and the mathematical modelling providing insight into potential reasons for these differences. This paper takes important steps towards increasing our understanding of B. bacteriovorus predation in a realistic environment and provides a framework for further developing B. bacteriovorus as a potential therapeutic.
Methods
Bacterial strains, serum, media and growth conditions
Klebsiella pneumoniae EARSSU271 is a carbapenemase producing (KPC) clinical isolate and was provided via Dr Mathew Diggle (EMPATH Nottingham University Hospitals UK). K. pneumoniae was cultured on LB agar and YT broth at 37 °C. B. bacteriovorus HD10024 was cultured as described previously25. Briefly, B. bacteriovorus stocks were prepared by culture on stationary phase E. coli S17-126 prey suspended in Ca/HEPES buffer (5.49 g/l HEPES free acid, 0.284 g/l calcium chloride dehydrate, pH 7.6). Predatory cultures were prepared by culturing of E. coli S17-1 in YT broth (5 g/l sodium chloride, 5 g/l peptone, 8 g/l tryptone, pH 7.5) at 37 °C with shaking for approximately 16 h. Stationary phase prey cells (3 ml) were then added to 1 ml of a previous B. bacteriovorus predatory culture in 50 ml Ca/HEPES buffer and incubated at 29 °C for at least 16 h until the culture had cleared. (Predator and prey cell numbers were enumerated from cultures as described in experimentation below). Human serum (pooled from male AB plasma) was sourced from Sigma (H4522, Sigma-Aldrich Corporation, St. Louis, MO, USA).
Assaying reduction of K. pneumoniae numbers by human serum alone
K. pneumoniae was cultured in YT broth (20 ml) at 37 °C with shaking (200 rpm) for approximately 16 h. K. pneumoniae was pelleted by centrifugation (5,500 × g, 10 min, 29 °C) and resuspended to an optical density (OD600 nm) of 2.0 in whole human serum (Sigma H4522, Sigma-Aldrich Corporation, St. Louis, MO, USA). The suspension was serially diluted 1:10 with fresh human serum in a total volume of 3 ml. Serially diluted cultures were incubated at 37 °C with shaking (200 rpm) and cell numbers were enumerated periodically by the method of Miles and Misra27 on LB agar in triplicate with three biological repeats. This was done to assay for any cell number reduction by serum alone.
Assaying reduction of B. bacteriovorus HD100 numbers by human serum alone
B. bacteriovorus predatory cultures (500 ml) prepared as described previously were passed through a 0.45 µm Millex filter (Millipore, Billerica, MA, USA) to remove any remaining starter-prey, E. coli S17-1. The filtered predatory culture was centrifuged (5,500 × g, 20 min, 29 °C) to pellet cells. The pellet was resuspended in 500 µl Ca/HEPES to give a 1000 × concentrated suspension typically containing approximately 4 × 1010 pfu/ml of B. bacteriovorus. The suspension was serially diluted 1:10 with Ca/HEPES buffer in a total volume of 500 µl. Serially diluted B. bacteriovorus suspension (450 µl) was added to 4.45 ml of whole human serum and suspensions were incubated at 37 °C with shaking (200 rpm). Cell numbers were determined periodically by serial dilution and enumeration of B. bacteriovorus plaques in a soft YPSC overlay agar (0.25 g/l magnesium sulphate, 0.5 g/l sodium acetate, 1.0 g/l Bacto peptone, 1.0 g/l yeast extract, pH 7.6; adjust to 0.25 g/l anhydrous calcium chloride after autoclaving). For YPSC bottom agar, add 10 g/l agar. For YPSC overlay agar, add 6 g/l agar) containing a lawn of E. coli S17-1 as described previously25. Enumerations were performed in duplicate (two technical repeats) with two biological repeats.
Predation assays in buffer and serum
Predatory cultures of B. bacteriovorus (50 ml) prepared as described earlier in methods were passed through a 0.45 µm Millex filter to remove any remaining E. coli S17-1 and the filtered predatory culture was centrifuged at 5,500 × g, 20 min, 29 °C. The pellet was resuspended in 1 ml of Ca/HEPES to give a 50 × concentrated preparation typically containing approximately 5 × 109 cfu/ml of predatory B. bacteriovorus. K. pneumoniae prey was cultured in YT broth (20 ml) at 37 °C with shaking (200 rpm) for approximately 16 h. Prey cells were pelleted by centrifugation (5,500 × g, 10 min, 29 °C) and resuspended to an optical density (OD 600 nm) of 1.0 (equivalent to approximately 5 × 109 cfu/ml) in 5 ml whole human serum (Sigma H4522, Sigma-Aldrich Corporation, St. Louis, MO, USA) or 5 ml Ca/HEPES buffer. Predation assays were started by combining 5 ml of prey suspension with 500 µl of concentrated B. bacteriovorus suspension or 500 µl of Ca/HEPES as a control in a sterile Falcon tube (50 ml) and incubated at 37 °C in a rotary shaker (200 rpm). Starting predator and prey numbers were determined by enumeration of both prey and predator populations. Changes in prey population were enumerated by method of Miles and Misra plating in triplicate. Changes in predator population were determined by serial dilution and enumeration of B. bacteriovorus plaques in a soft YPSC overlay agar.
Fluorescence microscopy
For fluorescence microscopy, predation assays in serum or buffer were prepared as described in methods. To be able to visualise small predators in the visually challenging serum environment, we used B. bacteriovorus HD100 expressing, constitutively from the chromosome, Bd0064mCherry fluorescent protein (named HD100 mCherry) described in Willis et al.28. Aliquots (10 µl) of predation assay were withdrawn for microscopy and placed on a solid 1% agarose pad prepared in Ca/HEPES buffer. Fluorescent microscopic images were acquired using a Nikon Ti-E Eclipse inverted microscope (excitation 555 nm / emission 640 nm, 1 s exposure).
Analysis of B. bacteriovorus morphology in serum
MicrobeJ image analysis software29 was used to detect fluorescent B. bacteriovorus HD100 mCherry cells and measure the roundness (4 × area / (π × major axis 2) of each individual cell. A roundness cut off of 0.65 was selected to distinguish between cells with typical B. bacteriovorus vibroid morphology (roundness score < 0.65) and those with an abnormal round morphology (roundness score ≥ 0.65). Manual inspection of cells near the cutoff validated that the cells were correctly called.
Analysis of K. pneumoniae cell area in serum
MicrobeJ image analysis software was used to detect K. pneumoniae prey cells in serum and measure the area of each individual cell. The mean cell area of the K. pneumoniae KPC population in serum was determined at 0 hours (n = 271 individual measurements) and 96 hours (n = 170 individual measurements).
Pre-exposure of predator and prey to serum followed by predation in buffer
To test whether delay effects seen in predation in serum were due predominately to serum-induced changes in predator or prey, additional experiments were performed. B. bacteriovorus predatory cultures (50 ml, typically containing approximately 1 × 108 pfu/ml) prepared as described were passed through a 0.45 µm Millex filter to remove any remaining E. coli S17-1 and the filtered predatory culture was centrifuged at 5,500 × g, 20 min, 29 °C. The pellet was resuspended in 5 ml of whole human serum and incubated at 37 °C with shaking (200 rpm) for 2 h. K. pneumoniae prey was cultured in YT broth (20 ml) at 37 °C with shaking (200 rpm) for approximately 16 h. Prey cells were pelleted by centrifugation (5,500 × g, 10 min, 29 °C) and resuspended to an optical density (OD 600 nm) of 1.0 (equivalent to approximately 5 × 109 cfu/ml) in 5 ml whole human serum and incubated at 37 °C with shaking (200 rpm) for 2 h. Following exposure to serum, predator and prey suspensions were pelleted by centrifugation (5,500 × g, 10 min, 29 °C). K. pneumoniae pellets were resuspended in 5 ml of Ca/HEPES and B. bacteriovorus HD100 pellets were resuspended in 500 µl of Ca/HEPES. To begin predation assays, 500 µl of B. bacteriovorus suspension was added to 5 ml of K. pneumoniae KPC suspension in Ca/HEPES and incubated at 37 °C with shaking (200 rpm). Predation was determined by enumeration of both prey and predator populations as described. Experiments were performed with three biological repeats. Prey enumerations were performed with technical triplicates and predator enumerations performed with technical duplicates.
Measure of complement activity during predation
To establish how complement levels changed during predation in serum, haemolysis assays were carried out using standard protocols as described earlier with minor modifications30, 31. Predation assays with B. bacteriovorus (approximate starting concentration 1 × 108 cfu/ml) and K. pneumoniae (approximate starting concentration 5 × 109 cfu/ml) in whole human serum were prepared as described previously and incubated at 37 °C with shaking (200 rpm). Aliquots of serum (1 ml) were removed at each time point and passed through a 0.2 µM Millex syringe driven filter to remove both predator and prey. Aliquots of filtered serum were immediately frozen (−20 °C) prior to determining remaining complement activity. The classical complement pathway activity was tested with sheep erythrocytes opsonised with Rabbit anti-sheep red blood cell stroma IgG antibodies (Sigma-Aldrich). Sheep erythrocytes were washed twice in 0.9% NaCl and twice in GVB++ (3.12 mM barbitone, 0.97 mM sodium barbiturate, 145 mM NaCl, 0.83 mM MgCl2, 0.25 mM CaCl2 with 0.1% gelatin). The erythrocytes were resuspended in GVB++ to 109 cells/ml and incubated with the antibody for 30 minutes at 37 °C, followed by incubation on ice for 45 minutes. The opsonized sheep cells (EA) were washed twice in GVB++ and resuspended to 108 cells/ml. The serum samples were serially diluted 2-fold in 50 µl of GVB++ and incubated with 25 µl of EA at 37 °C for 30 minutes. The assay was stopped by adding 125 µl of cold GVB++ buffer. The assay controls included 100% lysis control (25 μl of EA lysed in 175 of μl deionized water) and blank (25 μl of EA incubated in 175 μl of GVB++). The samples were centrifuged briefly (5000 x g, 2 minutes) to pellet intact erythrocytes and the haemolysis was measured spectrophotometrically as the amount of released haemoglobin (OD at 415 nm) in a microplate reader.
Determination of B. bacteriovorus HD100 burst size
The average number of predator progeny released from K. pneumoniae prey cells following infection and replication was determined by video microscopy to aid parameterization. Briefly, K. pneumoniae cells were prepared and infected with B. bacteriovorus HD100 following the protocol described for predation assays with K. pneumoniae cells resuspended in Ca/HEPES buffer. Predation assays were incubated at 37 °C with shaking (200 rpm) for 2 h and 10 µl of the suspension was then removed from the assay and placed on a solid 1% agarose pad surface prepared in Ca/HEPES buffer. Microscopic images were acquired over several hours at room temperature every 150 s as described previously32. Microscopic images were encoded into time-lapse movies and the number of progeny released from individual prey cells counted to give the average burst size (Supplementary material, Figure S1).
Mathematical Modelling
We developed mathematical models of K. pneumoniae KPC predation by B. bacteriovorus HD100 in both buffer and human serum. The motility of the bacteria and incubation in a rotary shaker led to a relatively homogenous environment, hence we used population based ODEs. The models we developed consider how the populations of predator and prey bacteria change over time in different environments. The models were influenced by previous modelling of B. bacteriovorus predation14, 18–20 as well as a model of the interaction of bacteria with serum33. The serum model presented here has ten variables (Table 1) and 27 parameters (Table 2). We use a reduced version of the serum model to simulate predation in buffer, discounting the serum specific variables and related parameters. The full model is shown diagrammatically in Fig. 1, with full details of the model given in the supplementary materials. Experimental results were used to set initial conditions for the model, estimate rate parameters for the mechanisms of predation, and validate the model for serum and for buffer (see supplementary materials for a detailed description). All model simulation, parameter fitting and analysis were performed in MATLAB34. Combinations of the model variables were used as reporting variables to emulate experimental prey and predator enumeration. Individual model variables were also analysed to explore predation dynamics that cannot be observed experimentally.
Table 1.
Model variables and definitions for both the buffer and human serum predation models.
| Variable | Definition |
|---|---|
| BD F | Free predator |
| P S | Free susceptible pathogen |
| P R | Free resistant pathogen |
| PBD O | Pathogen with predator (outside) |
| PBD I | Pathogen with predator (inside) |
| BD I | Inactivated predator |
| W N | Nutritious predation-derived waste |
| W M | Non-degradable predation-derived waste |
| I | Serum antimicrobials* |
| R | Serum growth resources* |
The variables marked with *are present in the serum model only.
Table 2.
Parameters used in both buffer and serum models with definitions and units.
| Parameter | Definition | Units |
|---|---|---|
| k 1 early | Early predator-pathogen binding rate | ml cell−1 s−1 |
| k 1 late | Late predator-pathogen binding rate | ml cell−1 s−1 |
| k 2 | Predator internalization rate | s−1 |
| k 3 | Predator reproduction rate (time from entry to exit) | s−1 |
| k 4 | Transient predator-pathogen binding rate | s−1 |
| V max | Maximum pathogen growth rate on serum | s−1 |
| Maximum pathogen growth rate on predation-derived waste | s−1 | |
| E | Efficiency of serum resource metabolism | arb. units |
| E w | Efficiency of predation-derived waste resource metabolism | arb. units |
| s | Serum growth resource at half maximal growth | arb. units |
| s w | Waste growth resource at half maximal growth | arb. units |
| d P | Rate of accumulation of predation-derived waste | s−1 |
| d BD | Debris produced from predator cell death | s−1 |
| d I | Natural antimicrobial inactivation rate | s−1 |
| Maximal predator removal rate due to dead predator debris | s−1 | |
| Baseline predator removal rate due to dead predator debris | s−1 | |
| h | Rate of change of predator removal rate | arb. units |
| a BD | Predator removal rate by antimicrobials | s−1 |
| a P | Prey removal rate by antimicrobials | s−1 |
| c BD | Predator dependant antimicrobial inactivation rate | s−1 |
| c P | Pathogen dependant antimicrobial inactivation rate | s−1 |
| f | Rate of prey transition to resistance | s−1 |
| n | Fractional index of pathogen-antimicrobial interaction | dimensionless |
| ρ | Predator progeny number | dimensionless |
| λ | Natural predator death rate | s−1 |
| ν | Proportion of viable invaded prey | dimensionless |
| δ | Reduced attachment length at early timepoints | h−1 |
| I 0 | Initial serum antimicrobial concentration | arb. units |
| R 0 | Initial serum growth resource concentration | arb. units |
Figure 1.

Schematic diagram of relationship between model variables based on biological mechanisms. The Ca/HEPES buffer model has eight variables: free B. bacteriovorus predators (BD F), uninfected predation-susceptible K. pneumoniae prey (P S), uninfected predation-resistant K. pneumoniae prey (P R), prey with predator attached outside (PBD O), invaded prey with predator on the inside (PBD I), inactivated predator (BD I), nutritious waste (W N) and non-degradable waste (W M). The serum model has an additional two variables serum growth resources (R) and serum antimicrobials (I). Detailed description of the biological mechanisms can be found in the supplementary materials.
Results
Predation of K. pneumoniae KPC in defined buffer
Experimentally, predation of K. pneumoniae KPC in a buffer environment at 37 °C was found to be highly reproducible (Fig. 2a) and resembled predation results reported for other Gram-negative bacteria such as E. coli, Acinetobacter spp. and Shigella spp. in buffer environments7, 17. Figure 2a shows a rapid reduction in the viability of K. pneumoniae in the presence of B. bacteriovorus HD100 with an approximate four log reduction of prey numbers by four hours. Reduction of prey was concomitant with an increase in predator numbers which is indicative of predation. These early time point results reflect the typical predatory lifescycle reported for B. bacteriovorus in lab strains of E. coli: an initial period of attachment and invasion is followed by prey death and filamentous replication of predator within a bdelloplast and lysis of the prey after 3–4 hours6. The timing and magnitude of the reduction in prey viability suggest that these initial predation events occurred, in buffer, with a high level of synchronicity which was further confirmed by additional microscopy (Supplementary Figure S2).
Figure 2.

Predation Experimentally Measured and Modelled in Ca/HEPES buffer. (a) Graphs show individual biological repeats- separate experiments (Expt) of the live bacterial counts from predation in buffer experiments, results from each individual biological experiment are represented by the same colour line on each graph. Changes in the population of both K. pneumoniae prey (Kp + HD100) and B. bacteriovorus HD100 predator (HD100 + Kp) were enumerated periodically during the predation assay. As a control, prey suspended in Ca/HEPES was enumerated periodically in the absence of B. bacteriovorus HD100 predator (Kp alone). (b) Prey concentration (green line) initially shows a 6 log10 reduction by 6 hours (see inset) before regrowth overtakes predation effects. Predator concentration (orange line) grows in line with the prey death for the first six hours then reduces, in line with experimental results. Experimental results reproduced from A shown as a bar plot, the model simulation lies within the error bars of the experimental results. (c) Model simulation results showing prey with a predator attached (PBDO), prey with a predator inside (PBDI) and inactivated predators (BDI) over time. Attachment drops off after 5 hours due to prey becoming resistant.(d) Model simulation of waste accumulation over time. Non-degradable waste increases monotonically leading to prey resistance whilst nutritious waste initially increases but is then depleted as it is used by the growing predominently resistant prey. Parameter values and further model validation are detailed in supplementary material. The model gives a good fit to the experimental data.
The mathematical model of predation of K. pneumoniae KPC in buffer by B. bacteriovorus HD100 gave a good fit to the experimental data (Fig. 2b). The model includes the effect of debris from predation (termed “predation-derived waste” model) on predators and prey. We consider that this waste will have two main effects, firstly some can be used by the prey for metabolism (WN), leading to growth of the prey cell population and secondly, any unused debris will remain in the media (WM) and contribute to the resistant phenotype suggested by the experimental data. The model followed the same trends seen experimentally, with an initial drop in prey numbers followed by a rapid regrowth within 24 hours. Such rapid regrowth is not reported for laboratory E. coli strains in buffer and may indicate proteolysis of predator by prey, liberating amino-acids for prey nutrition. The parameter estimation predicts an internalisation time of 11.5 minutes (k 2 in mins) and a replicated predator exit time of 4 hours (k 3 in hours) in K. pneumoniae, which are broadly in line with published times for E. coli 35, although species and strain specific variation is to be expected. In addition to predator and prey counts which can be measured experimentally, the model allows the examination of other quantities, such as bdelloplast numbers and total serum antimicrobial activity, which are difficult to measure quantitatively in vitro, allowing the underlying dynamics to be investigated further. The model, with best-fit parameters, predicts that in buffer, B. bacteriovorus concentration increases at early time points (up to approximately 8 hours), then declines slowly over time at a rate that matches the experimental data and is in line with previous studies14. Figure 2c shows predators attached to prey (PBDO), prey with predators inside (PBDI) and transiently attached predators (BDI) over time and declining PBDI and PBDO concentrations show that new predation is halted after the first 5 hours, in buffer, indicating that all the K. pneumoniae prey bacteria that have not been invaded by a predator are now resistant in the buffer model. The results from both the model and experimental work follow a very similar trend to experimental results for in-buffer predation of a different bacterium Erwina caratovora ssp. caratovora presented in Shemesh and Jurkevitch17, which show the same pattern of a large drop in prey numbers due to predation followed by an apparently resistant regrowth. Our model here supports their conclusions that plastic resistance induced by means of environmental changes could explain the experimental results and suggests that the amount of resistance is dependent upon the build-up of debris from successful predation.
To test the robustness of the model we conducted a sensitivity analysis at several key time points representing different stages of the simulation for the buffer model. Figure 3 shows a one-at-a-time sensitivity analysis for a 10% change in parameter values at 2, 6 and 24 hours, calculated as described in the supplemental material. This showed that the model is largely robust to 10% variations in parameter values. In the short term, the model is sensitive to small changes in ρ, k3 and P0, but the longer term dynamics are not affected. Variation in the natural death rate of predators, λ, has a long term effect on the B. bacteriovorus concentration. It should also be noted that while the system is sensitive to these parameters, they are not all directly amenable to manipulation in an experimental setting.
Figure 3.

One-at-a-time sensitivity analysis of a ± 10% change in each parameter at three different time points. The variation in parameter sensitivity at different times reflects the different mechanisms dominating predator and prey interactions at those times. The sensitivity coefficient is scaled such that a value of ± 1 represents a ± 10% change in concentration as a result of a ± 10% change in parameter value. Parameter values detailed in Supplementary Table S2.
In the first 6 hours of the simulation, K. pneumoniae concentration in the buffer model is sensitive to predation related parameters (ρ, k 3 and starting concentrations) but in the longer term, the model is not sensitive to these parameters. This can be explained by longer-term prey regrowth, shown experimentally in Fig. 2a. The 10% variation in these parameters changes the temporal dynamics of predation but the total amount of predation remains constant. We also looked at both large and smaller parameter changes and found the results to be the same (results not shown). Longer term (24 hours), the K. pneumoniae concentration is sensitive only to changes in the efficiency of metabolism of predation-derived waste (E W). This clearly demonstrates that in the closed experimental system it is the regrowth of K. pneumoniae that dominates experimental outcome. This occurs as a result of changing conditions over the course of the experiment, often neglected in previous work. Whilst the amount of prey-derived waste present (but not the efficiency of its usage) could possibly be manipulated experimentally in vitro, it cannot be altered in vivo, the host-relevant state, so we did not consider that further.
The sensitivity analysis highlights an important concept. Whilst we, and others previously, report on predation and factors affecting the amount and rate of predation, the closed batch experimental model may give predation dynamics that differ to what might be seen in a medically relevant circulation- perfused, physiological situation. However, no such quantitative perfused studies exist in vivo yet, as they are very experimentally challenging. Considering the dynamics of serum-based predation in batch is a crucial stepping stone to such future work.
Reduction of predator cell numbers in whole human serum alone
To better understand the effect of human serum on the reduction of B. bacteriovorus HD100 and K. pneumoniae and to define parameters for our mathematical model, we separately determined the viability of B. bacteriovorus HD100 and K. pneumoniae in a serum environment over a range of starting concentrations. Reduction of B. bacteriovorus in human serum was strongly dependent on the initial bacterial concentration (Fig. 4a). An initial B. bacteriovorus concentration of 3.67 × 107 pfu/ml was found to be cleared from human serum within 24 h. In contrast, B. bacteriovorus was found to persist up to 72 h in serum when present at a 100-fold higher initial concentration of 3.17 × 109 pfu/ml. To further investigate these dynamics, we developed a model of B. bacteriovorus survival. In order to obtain the concentration-dependent behaviour seen in the experimental results we made the B. bacteriovorus degradation rate proportional to time, serum antimicrobials and increasing dead B. bacteriovorus cell debris in the serum (This is not needed in the buffer model where B. bacteriovorus is not lysed by serum antimicrobials). The serum model gives a very early (1 hour) fall in B. bacteriovorus concentration over time that follows the same trends as the experimental data (Fig. 4a) for all starting concentrations we considered (Fig. 4c). In the model, initial removal of B. bacteriovorus is due to serum antimicrobials, and the model predicts that these are exhausted within the first hour of the simulation in the closed batch. However, reduction in B. bacteriovorus continues experimentally, (Fig. 4a), past 1 hour, over and above the natural death rate (λ). Removal of B. bacteriovorus at these times, in the model, is due to the effect of deleterious products released into the serum from the damaged predators. We hypothesise these products may include hydrolytic predatory enzymes detached from their usual internal predator self-protection proteins36. We also considered a simpler model for B. bacteriovorus reduction, with no antimicrobial-independent degradation. Whilst this gave a good fit to experimental data at a low initial concentration (3.67 × 107 pfu/ml), it was not able to account for B. bacteriovorus reduction at higher initial concentrations ( > 3.67 × 107 pfu/ml).
Figure 4.

(a) Reduction of B. bacteriovorus HD100 by whole human serum. A serially diluted preparation of B. bacteriovorus HD100 was inoculated into whole human serum and viability measured over a 72 h period. (b) Reduction of K. pneumoniae concentration by whole human serum. A serially diluted preparation of K. pneumoniae was inoculated into whole human serum and viability measured over a 72 h period by Miles and Misra. (c) Model simulations of B. bacteriovorus reduction in human serum for three different initial conditions. Solid line show simulation result whilst the circles show the experimental results (replicated from panel a). Parameter values are given in Supplementary Table S2, in the supplementary materials.
Reduction of prey cell numbers in whole human serum alone
The survival of K. pneumoniae prey in human serum was similarly investigated over a range of starting concentrations (~5 × 105 cfu/ml to ~9 × 109 cfu/ml). Again, removal of bacteria from human serum was found to be strongly dependent on the initial concentration of bacterial cells present (Fig. 4b). Despite initial drops in viability, starting concentrations of K. pneumoniae cells above 7.76 × 106 cfu/ml were found to recover and persist in serum following a period of regrowth. In contrast, lower starting concentrations swiftly dropped below our limit of detection (<1 × 103 cfu/ml) and did not recover by 72 h. We used all the values for experimentally-determined reduction of K. pneumoniae in serum (Fig. 5a) to fit the K. pneumoniae growth parameters in our serum model. We assumed an initial condition for serum antimicrobials (I) from the best-fit parameters we obtained from parameter fitting the B. bacteriovorus reduction data. This modelling and experimentation guided us in selecting appropriate predator to prey starting concentrations for predation assays in human serum, reported below.
Figure 5.

Predation in two different batches of whole human serum. (a) Serum batch SLBN9196V. (b) Serum batch SLBN8826V. Graphs show individual biological repeats of the live bacterial counts from predation in serum experiments, results from each individual biological experiment (Expt) are represented by the same colour line on each graph. Changes in the population of both K. pneumoniae prey (Kp + HD100) and B. bacteriovorus HD100 predator (HD100 + Kp) were enumerated periodically during the predation assay. As a control, prey suspended in serum was enumerated periodically in the absence of B. bacteriovorus HD100 predator (Kp alone). Despite illustrating some differences, similar patterns were seen across all 6 experimental repeats.
Predation of K. pneumoniae (KPC) in human serum
Having measured and modelled the interactions between serum prey and predators alone, and having quantified the dynamic responses that alter their phenotypes in response to the serum surroundings; we then compared predation in serum versus that in buffer.
To investigate and model potentially important differences between predation in buffer and biological fluid, we repeated our buffer based assays in batches of human serum where factors other than predator-prey interaction could ameliorate bacterial numbers. Selection of initial predator to prey ratio for predation experiments in human serum was informed by the viability of K. pneumoniae KPC and B. bacteriovorus in serum reduction assays reported herein (Fig. 4a and b), and were chosen to be high enough that K. pneumoniae would not be cleared by serum alone, as in an established infection. As no accurate OD600 nm can be measured for small B. bacteriovorus cells, accurate predator concentrations cannot be determined at the time of inoculation. Therefore, an approximate but tight range of initial predator concentrations were achieved by establishing a highly reproducible protocol. Accurate initial predator numbers for such experiments are enumerated by post-experiment viable counting with results available after the experiment (these are plotted in Fig. 5 and subsequently used in the mathematical modelling).
An initial concentration of approximately 5 × 109 cfu/ml was selected for K. pneumoniae KPC prey on the basis that initial concentrations of this size were found to persist in serum for the duration of the assay (96 h) with only a minimal, early loss of viability (Fig. 4b). Therefore, significant reductions in prey viability, compared to controls, could be attributed to B. bacteriovorus, rather than solely any antimicrobial effect of the serum environment. Conversely, an initial concentration of approximately 5 × 108 pfu/ml was chosen for B. bacteriovorus as starting concentrations of this size were found to steadily decrease in viability, in serum alone, with complete removal of predators occurring by 58 hours. This natural serum-related decrease in predators in the absence of prey allows us to attribute any rise in B. bacteriovorus, coupled with a decrease in K. pneumoniae, as an indicator of true predation causing predator replication. In addition the level of B. bacteriovorus used is similar to a topical dose applied to optical membranes of rabbit eyes, and reported as safe by Romanowski et al.37.
In contrast to assays performed in buffer, the dynamics of predation in serum were found to be more variable and the synchronicity observed in buffer was lost. However, some common trends did emerge. Predation and reduction of K. pneumoniae was observed in all six independent experiments using two different batches of human serum, (Fig. 5a and b) as determined by a decrease in prey population coupled with an increase in predator numbers, but predation was invariably followed by regrowth of the prey population. However, the length of time before measurable predation occurred and the extent to which it occurred, was found to be variable between the independent experiments, despite the small range of initial predator and prey concentrations, which ranged between 4.15 × 108−1.2 × 109 pfu/ml and 1.5–3.33 × 109 cfu/ml respectively. The length of delay before a measurable reduction in the prey population was seen correlated with the B. bacteriovorus viable count measured at eight hours. For example, in experiments 3 and 4, in which the least 8hr predator death occurred, (<1 Log10 reduction in predator concentration at eight hours), we see the earliest predation-effected decrease in prey concentration (between 24 and 32 hours). Conversely, in experiments 5 and 6, in which the most 8hr predator death occurs (>4 Log10 reduction in predator concentration at eight hours) we see the latest decrease in prey concentration (between 72 and 84 hours). This delay in predation, seen in all the independent experiments, is very different to the predation kinetics in buffer and important to appreciate when considering possible therapeutic applications.
The mathematical model (Fig. 6a) suggests that the delay in predation we observe in serum cannot be explained solely by the initial death of B. bacteriovorus, as predation then proceeds at a much slower rate than that measured and modelled in buffer. Nor can this decreased rate of predation be due to continuing clearance of the predator population by the antimicrobial component of the serum, as the model predicts serum antimicrobials are exhausted within a few minutes of the start of the predation experiments (Fig. 6b). This has been confirmed experimentally, (Fig. 7a) and is discussed in detail later. Instead, the mathematical model suggests that a period of reduced attachment of predators to prey is also required to give the dynamics observed. Therefore, to better understand the dynamics of predation in serum and to gain insights into potential causes for this delay, we performed a microscopic study of predation in serum.
Figure 6.
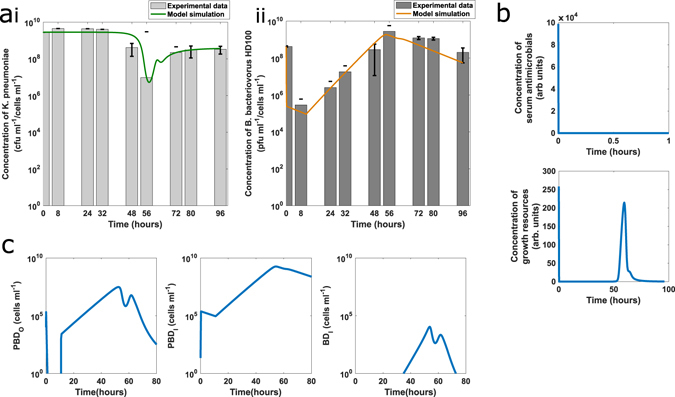
Model simulations of predation in human serum in comparison to experimental data, validating parameters estimated. (a) Model simulations with the best fit parameters (given in Supplementary Table S2) give a good fit to experimental data (mean of experimental repeats 1 and 2 from Fig. 5), with comparable dynamics. The drop in prey concentration due to predation and corresponding rise in predator numbers seen in the experimental data is replicated in the numerical simulation (b) The predators attached to prey (PBDO), predators inside prey i.e. bdelloplast (PBDI) and inactivated predators (BDI) are shown plotted against time. All three plots show a halting of predation for the first approximately 15 hours, then a continuing of the predatory cycle. By around 56 hours all the prey have become resistant. (c) The simulated serum antimicrobial concentrations showed that the antimicrobial activity of the serum is used up quickly (within the first 10 minutes of the simulation). Initial serum growth resources in the simulation are also quickly depleted, but an increase in growth resources is seen after 50 hours as lysis of bdelloplasts starts to release nutrients into the system.
Figure 7.
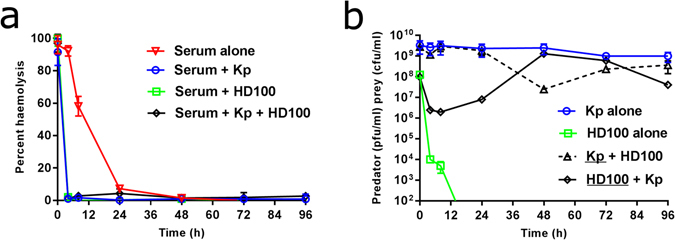
(a) Classical pathway complement activity of human serum incubated in presence of predator and/or prey as measured by percent haemolysis of sensitised erythrocytes. Human serum was incubated at 37 °C alone as a control (Serum alone) or in the presence of K. pneumoniae (Serum + Kp), in the presence of B. bacteriovorus HD100 (Serum + HD100) or in the presence of both K. pneumoniae and B. bacteriovorus HD100 (Serum + Kp + HD100). Serum was periodically sampled, filtered (0.2 µM) and diluted to 25% prior to measuring serum complement activity as determined by percent haemolysis of erythrocytes. CH50 serum values were calculated from these data and these are included in supplementary materials, Supplementary Table S1. (b) Enumeration of live bacterial populations incubated in human serum used for determining classical pathway complement activity. Predator and prey numbers in human serum were determined immediately prior to filtering (0.2 µM) and filtered aliquots were subsequently used for determining complement activity (panel a).
We used B. bacteriovorus HD100 expressing fluorescent mCherry protein (HD100 mCherry) in order to visualize small predator cells microscopically in the visually complex medium of serum. Predation of K. pneumoniae prey by HD100 mCherry was verified (as this is genetically a different predator to HD100) in Fig. 8a. These results again show a drop in the concentration of the predator population at 8 hours (approximately 5 Log10 reduction) followed by recovery of B. bacteriovorus concentration and an associated delayed drop in the prey population at 120 hours. Due to the altered genetic background of HD100 mCherry (chromosomal integration of the mCherry gene) and associated biological strain expressing the mCherry protein, the dynamics of predation may be different to the wildtype, but the overall behaviour is the same (Fig. 8a). Therefore these HD100 mCherry results were not used for modelling purposes.
Figure 8.

(a) Predation in whole human serum by HD100 mCherry fluorescent predator. Changes in the population of both K. pneumoniae prey (Kp + HD100 mCherry) and B. bacteriovorus HD100 mCherry predator (HD100 mCherry + Kp) were enumerated periodically during the predation assay. As a control, K. pneumoniae prey suspended in serum was enumerated periodically in the absence of B. bacteriovorus HD100 mCherry predator (Kp alone). (b) Microscopic analysis of B. bacteriovorus morphology in human serum with K. pneumoniae prey. MicrobeJ software was used to automatically detect mCherry tagged fluorescent B. bacteriovorus cells and measure the roundness (4 × area / (π × major axis 2) of each individual cell at each time point. A roundness cut off of 0.65 was selected to distinguish between cells with typical vibroid morphology (roundness score < 0.65) and those with an abnormal round morphology (roundness score ≥ 0.65). Number of cells analysed ranged between 1026 and 109 cells. Exact sample sizes for each time point are given in Supplementary material, Figure S4.
When predation events in serum were observed microscopically, we saw a rapid, early change in predator morphology. HD100 mCherry with typical vibroid morphology at 0 hours changed to a rounded morphology by two hours in serum with K. pneumoniae prey (Fig. 8b). Microscopic examples of fluorescent HD100 mCherry with altered morphologies are given in supplementary material Figure S3ab and the number of individual cells measured at each timepoint are provided in supplementary material Figure S4. The entire predator population began with a typical vibroid morphology (96.5% of all cells measured) but rapidly changed to a rounded morphology by two hours in serum (1.94% of all cells measured retained a vibroid morphology at 2 hours). The proportion of the predator population with a vibroid morphology remained low up to 8 hours (3.67%) before beginning to slowly recover from 24 hours onwards (Fig. 8b). This trend coincided with an increase in viable counts in the predator population (Fig. 8a), presumably due to successful predation. This change in predator morphology is likely to have a large impact on predation events, including attachment to prey.
The model fits the experimental data well using a reduced early attachment rate (k 1 early) and introducing a delay parameter, δ, to determine how long that reduced attachment continues. As can be seen from the experimental data (Fig. 5a,b), the length of the delay and the extent of the reduced attachment is variable between independent experiments in serum, and so studying the mean of all the experiments would be inappropriate as the smoothing effect would remove vital dynamics. For this reason, we began by fitting the parameters to the mean of experiments 1 and 2 only, which showed similar temporal dynamics in the experimental data, to obtain best-fit parameter values. We then set all the parameters to these best-fit values except k 1 early and δ which we fitted individually to each independent experiment. Figure 6a shows model simulations using best fit parameters for the model plotted using the mean of experiments 1 and 2. This serum model simulation (Figure 6a,i) shows, in line with experimental results, the drop in prey concentration at approximately 48 hours followed by regrowth to a stable concentration of approximately 2 × 108 cells/ml. B. bacteriovorus concentration in this simulation also compares well with the experimental data. Figure 6c shows the corresponding concentrations of predators attached to prey (PBDO), predators inside prey (PBDI) and inactivated predators (BDI) in the model simulation. These plots show that predation is halted until approximated 10 hours (as given by the delay parameter δ), then proceeds. Attachment rapidly declines after 56 hours as the prey become resistant to predation in the model (and apparently are predation resistant in our experimental data (Fig. 5a,b)). The model predicts a B. bacteriovorus replication time of 10 hours, which is longer than predation in buffer and is in line with estimates based on the rate of B. bacteriovorus increases in the experimental data from all biological repeats (Fig. 5).
The growth resources available to prey in the model have two sources, an initial serum component, which is rapidly depleted in the first hours of the simulation, and a predation-effected component which becomes significant in the system at approximately 50 hours as prey-lysis products start to accumulate, giving the high level of K. pneumoniae regrowth seen after 56 hours. Sensitivity analysis of the serum model (Supplementary material, Figure S5) at key points in the time course (early phase; lowest prey concentration stage and regrowth stage) highlights the multifactorial nature of the serum experimental system. Whilst the model is robust to changes in many parameters, it is most sensitive to changes in parameters relating to the death of B. bacteriovorus due to serum. In particular the prey concentration at 60 hours is by far the most sensitive to this, suggesting that changes in the ability of predator to resist the serum effects could have a large effect on the drop in prey numbers at this point.
To validate the best fit parameters we use in the model we also repeated the parameter estimation using two other groups of experiments (experiments 3 and 4; experiments 5 and 6). In both cases the only significant difference between the parameters chosen was the early attachment rate k 1 early (Supplementary material, Figure S6) as discussed above. Estimating the early attachment rate (k 1 early) and delay time (δ) individually showed us that the different behaviours observed experimentally can be substantially explained, and the model fitted to the all experimental data (Fig. 9) despite the complexity of the experimental data. The model suggests a small early attachment rate for all experiments with some variation in the extent of this and the time it takes to return to higher rates, hence free B. bacteriovorus get little protection from the serum at early timepoints and many are killed by serum antimicrobials and predator debris/predatory enzymes, with only a limited number of predators successfully entering prey at this stage.
Figure 9.

Model simulations of predation of K. pneumoniae by B. bacteriovorus in human serum, with varying levels delayed successful attachment, allows the model to fit each independent experiment (see Fig. 5 for experimental results).The delayed attachment parameter values for each experiment are k1 early = [1.433 × 10−14; 1.204 × 10−14; 3.999 × 10−12; 2.175 × 10−11 ; 6.597 × 10−16; 2.304 × 10−16] and δ = [11.03; 10.29; 10.43; 10.55; 18.64; 17.49]. The remaining parameters which are fixed between experiments are given in Supplementary Table S2, in the supplementary material.
Notably, a relatively small variation in the length of reduced attachment leads to much larger variation in the timing of the significant drop in prey numbers, this would support the view that the rounded predator cell majority are permanently prevented from entering prey, hence the long delay leads to reduced predator reproduction and it takes time to build up the progeny number to effectively then reduce K. pneumoniae numbers. A model with no early predator attachment to prey did not fit the data (not shown). The model does (Figure 6aii) predict an active but small predatory population at early timepoints (approximately 0.05% of the initial predator input at 2 hrs) and this correlated with a low percentage of vibroid B. bacteriovorus (1.94%) at the same timepoint in Fig. 8b).
In order to investigate the potential for serum derived components to bind to the surface of predator or prey and possibly halt predation, we separately exposed B. bacteriovorus and K. pneumoniae to whole human serum, prior to subsequent predation in Ca/HEPES buffer (Fig. 10). Pre-exposure of both predator and prey separately to whole human serum for two hours was found to have no subsequent delaying effect on predation once B. bacteriovorus and K. pneumoniae were removed from serum and combined in Ca/HEPES buffer. Indeed, predation of K. pneumoniae (mean concentration at 0 hours 3.17 × 108 cfu/ml) by B. bacteriovorus (mean concentration at 0 hours 3.65 × 108 cfu/ml) following exposure to human serum proceeded in an almost identical fashion to that reported earlier in Ca/HEPES buffer (Fig. 10 vs Fig. 2a) with an approximate five Log10 drop in prey concentration by four hours incubation time. These results lead us to conclude that a binding of serum components, or early predator debris, to the cell surface of either K. pneumoniae or B. bacteriovorus, (that could inhibit attachment of active predators), is either highly reversible on washing in buffer, or unlikely to be the cause of the delay in predation we observe in our serum predation assays.
Figure 10.
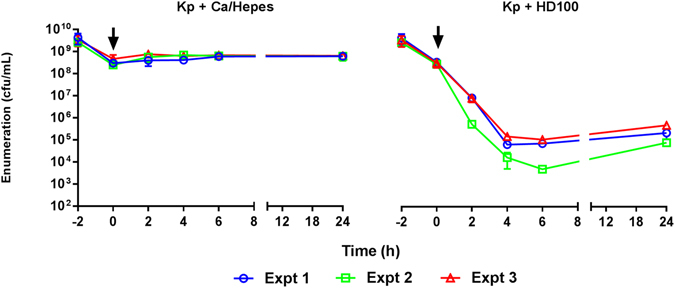
Predation in Ca/HEPES showing live bacterial numbers following pre-exposure of predator and prey to human serum. B. bacteriovorus HD100 and K. pneumoniae were exposed to human serum for two hours (t = −2 to 0 h). Predator and prey were then resuspended in Ca/HEPES and combined together in a predation assay and changes in the K. pneumoniae prey population enumerated periodically (Kp + HD100). As a control, K. pneumoniae prey previously exposed to human serum was resuspended in Ca/HEPES and enumerated periodically in the absence of B. bacteriovorus HD100 predator (Kp + Ca/HEPES). Arrows indicate the time at which bacteria were transferred from serum to buffer and subjected to respective treatment). B. bacteriovorus previously exposed to whole serum was added to K. pneumoniae to give a predator concentration at t = 0 h of approximately 5 × 108 pfu/ml.
Changes in serum complement activity
It is not possible to measure experimentally (from a predator-prey mixture) all the antimicrobial components of serum, but one of the major antibacterial factors in serum that we could readily measure was complement through the classical pathway, using a haemolysis assay with predators and prey alone or in combination (Fig. 7a)30, 31. Additionally, changes in viability of the bacterial populations incubated in serum were enumerated over the 96 hour time period (Fig. 7b). Serum incubated in the presence of B. bacteriovorus alone, K. pneumoniae alone or in the presence of both predator and prey was depleted of classical complement pathway activity by 4 hours, the earliest time point measured. This experimental result fits with our mathematical model, which predicts the depletion of serum antimicrobial activity in less than 1 hour. This rapid loss of complement activity highlights one of the limitations of using a closed experimental system and should be considered when extrapolating results to some in vivo scenarios such as the circulatory system38, but is of relevance to isolated wound sites. It is possible that in a more refreshed system, where serum is replenished by bodily circulation that once prey numbers are reduced below a threshold level by B. bacteriovorus predation the antimicrobial activity of the serum, in combination with leukocytes, may clear the pathogen and the pathogen regrowth we observe in our closed model systems would not occur. We have established that we have a model that describes the additional complexity of predation in serum and accurately fits the extensive experimental results. Moreover, the modelling has guided experimentation to clarify the mechanisms involved in predator-prey-serum interactions.
Discussion
Antimicrobial resistance has emerged as a global health issue and has stimulated us to test and model how predatory B. bacteriovorus prey upon K. pneumoniae in this study. Multidrug resistant Gram-negative bacterial infections, especially those as a result of so-called ESKAPE pathogens39, including K. pneumoniae, are a real problem in developing and developed countries and in the healthcare setting13. Many of these infections include multidrug-resistant strains, at different bodily sites, designated currently ‘untreatable’ using conventional antibiotics40, 41. As a response, alternatives to antibiotics are being sought42, 43. The use of predatory bacteria, such as B. bacteriovorus, as “living antibiotics” is an attractive prospect but it is crucial to investigate Bdellovibrio-pathogen interactions and kinetics of predation in host-relevant environments, and to be able to predict the effects of predator doses, if they are to be used as a therapeutic agents. In light of this, we chose to model pathogen-predation in buffer compared to that in human serum.
Research to date modelling and quantifying the activity of predatory bacteria has focused mostly on laboratory strains of prey bacteria in ideal (buffer or growth media) conditions. Separate research has tested the effects of pathogen treatments with predators in vivo without modelling. We need to improve our understanding of host relevant conditions under which B. bacteriovorus can prey on clinical, antibiotic-resistant strains of bacteria that cause untreatable infections. Although promising steps have been taken towards this with in vivo experimental studies of B. bacteriovorus as a therapeutic28, 44, including leukocyte interactions in Willis and co-workers, these experiments did not investigate underlying serum antibacterial mechanisms that may affect the dynamics of predation in a therapeutic setting. To address this, we set out to develop a model that captures the complexities of human serum and could therefore inform on predation dynamics in this setting and guide experimentation.
We have shown that B. bacteriovorus HD100 can effectively prey in both buffer and human serum on a clinical isolate of K. pneumoniae KPC, a major cause of carbapenem-resistant infections and problematic as a carrier status in patients45. We developed a mathematical model which fits the experimental data closely and we used this model to explore the underlying dynamics of predation. Through this interdisciplinary approach, we have shown that the interactions of this predator and pathogen in the human serum environment, and their responses to it are markedly different to that seen in buffer. These differences are indicative of the inherent problems of simplifying clinical processes to laboratory standards.
The mathematical model of the buffer-based predation experiments included terms relating to predation, prey growth and previously proposed “plastic resistance” phenotypes17, gave a good fit to the experimental data for both predator and prey concentrations over a 96 hour period and was validated with an independent data set. The parameter estimates for predator internalisation and time to predator-progeny release in buffer were within the range of observed times in E. coil 35, and the estimated natural predator death rate was also in line with previous studies in buffer14. These factors lead us to believe we have a functional mechanistic model of B. bacteriovorus predation that includes the dominant mechanisms.
This paper shows for the first time that predation of a specific clinical pathogen by a predator in serum is markedly different to predation in buffer, and through the development of mathematical models directing further experimental validation, where experimentally tractable, we have addressed why this is the case. We show that, in the case of the predation in serum model, reduced initial predator attachment for a period up to 19 hours, compared to the buffer model, makes it possible for the model to fit the serum-predation experimental results. Our model assumes that every attachment event results in successful predation, therefore the term “reduced attachment” as used here, represents an increase in unsuccessful predator-prey attachment events that do not result in predation or simply less frequent attachment events occurring. To determine the cause of reduced early predator attachment, as suggested by the model, serum pre-incubation of predators or prey separately, prior to predation tests in buffer, were performed. These experiments suggested that the delay in predation observed was not as a result of direct binding of serum components to either the predator or prey cell surface, blocking attachment, as predation following exposure to human serum proceeded in an almost identical fashion to that reported earlier in Ca/HEPES buffer. Nor were the predators or prey irreversibly changed by the serum pre-treatment.
Serum is a complex mixture which is not amenable to microscopic work with small, unlabelled, predatory bacteria. However, the need to visualise predators during this delayed predation period led us to make additional microscopic observations of predation in serum using fluorescent HD100 mCherry predators. This showed that B. bacteriovorus rapidly changed from a typical vibroid morphology to a rounded morphology on initial contact with serum. At population level this altered morphology was temporary and predators were seen to have returned back to vibroid over the time course of the study. Vibroid morphology was associated with an increase in concentration of the predator population, presumably due to successful predation. Predator rounding may be more likely in a predator that has bound to and sensed prey but proving this experimentally is beyond the scope of this study. These results do suggest that preconditioning B. bacteriovorus to human serum is relevant for any future therapeutic use.
The detection of the predation delay in serum and directed experimentation, described above, emphasises the utility of our mathematical model, it provides insights into the population dynamics of predation that are difficult or impossible to determine experimentally. Predation events, such as attachment, are short lived and difficult to measure accurately, en masse, by direct microscopic observation, a task which becomes even more arduous in a complex granular medium such as serum. We therefore hypothesize that the delay in predation we observe in serum is a result of reduced successful attachment events arising from a change in predator morphology, which we observe experimentally. This morphology returns to vibroid over time, correlating with increasing B. bacteriovorus cell numbers after rare predation events have allowed predator replication. The observed delay in predation should be further explored experimentally, and studies investigating the transcriptional response of both predator and prey adapting to a serum environment are underway and may provide answers to this complex issue, but are beyond the scope of this work.
After accumulation of predators via rounds of replication there is a significant drop in the K. pneumoniae prey numbers but later there is a period of regrowth (Fig. 5). We make the assumption that prey cells accumulating in a serum-based population are becoming resistant to predation following initial prey bursting and exit in their neighbourhood. We postulate that this resistance is a plastic phenotypic resistance, as this has been reported previously for the predation of another gammaproteobacterium, Erwinia carotovora in buffer17. It should be stressed that this plastic resistance is believed to transient in nature rather than due to genetic mutation of the prey. Shemesh and Jurkevitch hypothesised that this phenotypic resistance may be due to modification they describe as “hardening of the prey cell wall” resulting from the release of low levels of lytic enzymes by predators during ineffective prey cell encounters. In our model, the development of prey phenotypic resistance in serum is dependent upon the amount of prey cell lysis resulting from predation. Lysed prey will release “waste”:- including a mixture of both intracellular prey enzymes and predator lytic enzymes, produced to effect bdelloplast exit. These free lytic enzymes may act on neighbouring cells, possibly resulting in the “hardening effect” described, or there may be other “anti-predation” modifications which await characterisation. This explanation would support the work of Shemesh and Jurkevitch, that predatory enzymes may play an indirect role in producing a transiently resistant prey population17. Alternatively, this “predation-waste”, containing the remnants of prey cell membranes and walls left over from lysis46, may act as a physical barrier to attachment. We note, however, that the formulation of the model allows flexibility such that the model is not restricted to a particular underlying biological mechanism(s) for this prey regrowth after predation. In a replenished circulatory system, in vivo, rather than the batch system we tested, we would not expect such waste accumulation and so such a “resistant” prey response may not be seen in privileged or minimally perfused body sites. The zebrafish infection study of Willis et al.28 may be a relevant example.
Predator-prey-serum interaction kinetics are important if B. bacteriovorus are ever to be used as topical (e.g. on tracheotomies), ingested (to attempt gut clearance for patients who are carriers), applied to wounds (surgically or environmentally caused) or inhaled (for lung infection) therapies for human Klebsiella infections. Recently Shatzkes et al.44 have highlighted the potential of one of these approaches, showing that repeated intranasal administration of. B. bacteriovorus 109 J can reduce the bacterial burden of K. pneumoniae in rat lungs. Their study was able to measure prey, but not predator, viability over a 24 hour period reporting a reduction of K. pneumoniae prey in the majority of animals.
The extended live predator and prey count data in our study emphasise the importance of building upon such studies44 to understand the dynamics of predation beyond any initial reduction in the prey population. The prey regrowth we observe in buffer and serum, presumably due to plastic resistance, could impede convalescence after B. bacteriovorus treatment and is important to address, hence our inclusion of a resistance mechanism in the model. Whilst the potential to treat an initial K. pneumoniae infection with B. bacteriovorus has been established44, preventing pathogen regrowth is just as crucial and needs further research. Recently, Willis et al.28 demonstrated for the first time that injected B. bacteriovorus can be used to cure an otherwise lethal bacterial infection in living animals. They found that the synergistic action of the innate immune system with the predatory bacteria were responsible for reduced pathogen numbers and increased survival in zebrafish larvae. Our serum predation modelling is an important precursor to a further study including leukocytes and as a “3Rs” strategy to reduce the use of unnecessary animals in medical research.
The approach we take here, combining modelling with experimental verification of growth response by a clinical pathogen in human serum, shows that pathogen regrowth after predation can be modelled. This encourages future quantitative research in more complex bodily settings, such as wounds, with innate immune cells acting also. This approach may aid dosing and viable predator-persistence considerations for more systemic studies such as the bloodstream injections of predators recently published by Shatzkes et al.38.
Our work here and future models will assist the design of a treatment regime aimed at preventing such regrowth, such as exploring whether multiple doses of B. bacteriovorus could synergise with the immune system to clear an infection.
In summary, in this work we have shown for the first time that B. bacteriovorus can effectively prey on the antibiotic-resistant, human-pathogen K. pneumoniae KPC in human serum, as well as buffer. There can be later regrowth of the pathogen, post-predation, which needs to be researched further. We have shown that the dynamics of predation in human serum are complex and differ significantly from the comparatively simple dynamics of predation in buffer. The synergy of mathematical modelling and experimental science has provided new theories for the mechanisms behind these differences. These insights guide future experimentation to mitigate delayed predation in serum and prey regrowth, for efficient therapeutic administration of B. bacteriovorus in serum-rich settings, such as infected wounds. Also a study of how, or if, predators and prey secrete compounds that change serum composition will be valuable. Future strategies to produce the most active predator doses may come from pre-adaptation of predators in serum prior to administration, or the development of serum resistant B. bacteriovorus strains through forced evolution. This study has highlighted the importance of using realistic, bodily-derived media, extended time-course studies and mathematical modelling as we take steps towards validating predatory bacteria as living antibiotics.
Electronic supplementary material
Acknowledgements
We thank Rob Till and all the members of our laboratory and the A-Team Technicians in our department for provision of media and sterile supplies. We thank Dr Carey Lambert for helpful discussions and he and Professor Markus R Owen for critical comments on the manuscript. This work was funded by the US Army Research Office and the Defense Advanced Research Projects Agency under Cooperative Agreement Number W911NF-15-2-0028 to R.E.S. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the Army Research Office, DARPA, or the U.S. Government.
Author Contributions
D.N. carried out the experimental work with experimental contributions from P.R., C.M. and J. Tyson. D.R. carried out the complement activity work. M.B. and J. Twycross developed and analysed the mathematical models. R.E.S. supervised the experimental and, with J. Twycross, the modelling work. G.C. and M.D. provided and characterised the Klebsiella pneumoniae EARSSU271 isolate. M.B., D.N. and R.E.S. drafted the manuscript with comments from D.R., P.R., C.M., P.R., G.C., M.D., J. Tyson and J. Twycross. All authors gave final approval for publication.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Michelle Baker and David Negus contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08060-4
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hidron AI, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 2.Gaynes R, Edwards JR. & National Nosocomial Infections Surveillance, S. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41:848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 3.van Duijn PJ, Dautzenberg MJ, Oostdijk EA. Recent trends in antibiotic resistance in European ICUs. Curr Opin Crit Care. 2011;17:658–665. doi: 10.1097/MCC.0b013e32834c9d87. [DOI] [PubMed] [Google Scholar]
- 4.Mauldin PD, Salgado CD, Hansen IS, Durup DT, Bosso JA. Attributable hospital cost and length of stay associated with health care-associated infections caused by antibiotic-resistant gram-negative bacteria. Antimicrob Agents Chemother. 2010;54:109–115. doi: 10.1128/AAC.01041-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vincent JL, et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 6.Sockett RE, Lambert C. Bdellovibrio as therapeutic agents: a predatory renaissance? Nat Rev Micro. 2004;2:669–675. doi: 10.1038/nrmicro959. [DOI] [PubMed] [Google Scholar]
- 7.Dashiff A, Junka RA, Libera M, Kadouri DE. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J Appl Microbiol. 2011;110:431–444. doi: 10.1111/j.1365-2672.2010.04900.x. [DOI] [PubMed] [Google Scholar]
- 8.Starr MP, Baigent NL. Parasitic interaction of Bdellovibrio bacteriovorus with other bacteria. J Bacteriol. 1966;91:2006–2017. doi: 10.1128/jb.91.5.2006-2017.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stolp H, Starr MP. Bdellovibrio Bacteriovorus Gen. Et Sp. N., a Predatory, Ectoparasitic, and Bacteriolytic Microorganism. Antonie Van Leeuwenhoek. 1963;29:217–248. doi: 10.1007/BF02046064. [DOI] [PubMed] [Google Scholar]
- 10.Kessel M, Shilo M. Relationship of Bdellovibrio elongation and fission to host cell size. J Bacteriol. 1976;128:477–480. doi: 10.1128/jb.128.1.477-480.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fratamico PM, Whiting RC. Ability of Bdellovibrio bacteriovorus 109J to lyse gram-negative food-borne pathogenic and spoilage bacteria. Journal of Food Protection®. 1995;58:160–164. doi: 10.4315/0362-028X-58.2.160. [DOI] [PubMed] [Google Scholar]
- 12.Boucher HW, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 13.Mehrad B, Clark NM, Zhanel GG, Lynch JP. Antimicrobial Resistance in Hospital-Acquired Gram-Negative Bacterial Infections. Chest. 2015;147:1413–1421. doi: 10.1378/chest.14-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varon M, Zeigler BP. Bacterial Predator-Prey Interaction at Low Prey Density. Applied and Environmental Microbiology. 1978;36:11–17. doi: 10.1128/aem.36.1.11-17.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lotka AJ. UNDAMPED OSCILLATIONS DERIVED FROM THE LAW OF MASS ACTION. Journal of the American Chemical Society. 1920;42:1595–1599. doi: 10.1021/ja01453a010. [DOI] [Google Scholar]
- 16.Volterra V. Variations and fluctuations of the number of individuals in animal species living together. J. Cons. Int. Explor. Mer. 1928;3:3–51. doi: 10.1093/icesjms/3.1.3. [DOI] [Google Scholar]
- 17.Shemesh Y, Jurkevitch E. Plastic phenotypic resistance to predation by Bdellovibrio and like organisms in bacterial prey. Environmental Microbiology. 2004;6:12–18. doi: 10.1046/j.1462-2920.2003.00530.x. [DOI] [PubMed] [Google Scholar]
- 18.Marchand A, Gabignon O. [Theoretical model of the predator-prey interaction kinetics between “Bdellovibrio bacteriovorus” and “escherichia coli” (author’s transl)] Ann Microbiol (Paris) 1981;132 B:321–336. [PubMed] [Google Scholar]
- 19.Wilkinson MHF. Predation in the Presence of Decoys: An Inhibitory Factor on Pathogen Control by Bacteriophages or Bdellovibrios in Dense and Diverse Ecosystems. Journal of Theoretical Biology. 2001;208:27–36. doi: 10.1006/jtbi.2000.2197. [DOI] [PubMed] [Google Scholar]
- 20.Hobley L, King JR, Sockett RE. Bdellovibrio Predation in the Presence of Decoys: Three-Way Bacterial Interactions Revealed by Mathematical and Experimental Analyses. Applied and Environmental Microbiology. 2006;72:6757–6765. doi: 10.1128/AEM.00844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dattner, I. et al. Modelling and parameter inference of predator–prey dynamics in heterogeneous environments using the direct integral approach. Journal of The Royal Society Interface14, doi:10.1098/rsif.2016.0525 (2017). [DOI] [PMC free article] [PubMed]
- 22.Gilg O, Hanski I, Sittler B. Cyclic Dynamics in a Simple Vertebrate Predator-Prey Community. Science. 2003;302:866. doi: 10.1126/science.1087509. [DOI] [PubMed] [Google Scholar]
- 23.Murray, J. D. Mathematical Biology: I. An Introduction. (Springer New York, 2011).
- 24.Rendulic S, et al. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 25.Lambert, C. & Sockett, R. E. Laboratory maintenance of Bdellovibrio. Curr Protoc Microbiol Chapter 7, Unit 7B 2, doi:10.1002/9780471729259.mc07b02s9 (2008). [DOI] [PubMed]
- 26.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nature biotechnology. 1983;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 27.Miles AA, Misra SS, Irwin JO. The estimation of the bactericidal power of the blood. J Hyg (Lond) 1938;38:732–749. doi: 10.1017/S002217240001158X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willis AR, et al. Injections of Predatory Bacteria Work Alongside Host Immune Cells to Treat <em>Shigella</em> Infection in Zebrafish Larvae. Current Biology. 2016;26:3343–3351. doi: 10.1016/j.cub.2016.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nature Microbiology. 2016;1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klerx JP, Beukelman CJ, Van Dijk H, Willers JM. Microassay for colorimetric estimation of complement activity in guinea pig, human and mouse serum. Journal of immunological methods. 1983;63:215–220. doi: 10.1016/0022-1759(83)90425-8. [DOI] [PubMed] [Google Scholar]
- 31.Morgan, B. P. in T Complement Methods and Protocols Vol. 150 Methods in Molecular Biology 61–71 (2000). [DOI] [PubMed]
- 32.Fenton AK, Kanna M, Woods RD, Aizawa SI, Sockett RE. Shadowing the actions of a predator: backlit fluorescent microscopy reveals synchronous nonbinary septation of predatory Bdellovibrio inside prey and exit through discrete bdelloplast pores. J Bacteriol. 2010;192:6329–6335. doi: 10.1128/JB.00914-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ankomah P, Levin BR. Exploring the collaboration between antibiotics and the immune response in the treatment of acute, self-limiting infections. Proceedings of the National Academy of Sciences. 2014;111:8331–8338. doi: 10.1073/pnas.1400352111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MATLAB, Optimization and Parallel computing toolbox Release 2015a (Natick, Massachusetts, United States., 2016).
- 35.Lerner TR, et al. Specialized Peptidoglycan Hydrolases Sculpt the Intra-bacterial Niche of Predatory Bdellovibrio and Increase Population Fitness. PLOS Pathogens. 2012;8:e1002524. doi: 10.1371/journal.ppat.1002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert C, et al. Ankyrin-mediated self-protection during cell invasion by the bacterial predator Bdellovibrio bacteriovorus. Nat Commun. 2015;6:8884. doi: 10.1038/ncomms9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romanowski EG, et al. Predatory bacteria are nontoxic to the rabbit ocular surface. Sci Rep. 2016;6:30987. doi: 10.1038/srep30987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shatzkes K, et al. Examining the efficacy of intravenous administration of predatory bacteria in rats. Sci Rep. 2017;7:1864. doi: 10.1038/s41598-017-02041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert review of anti-infective therapy. 2013;11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 40.Snitkin ES, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Science translational medicine. 2012;4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elemam A, Rahimian J, Mandell W. Infection with panresistant Klebsiella pneumoniae: a report of 2 cases and a brief review of the literature. Clinical infectious diseases. 2009;49:271–274. doi: 10.1086/600042. [DOI] [PubMed] [Google Scholar]
- 42.Czaplewski L, et al. Alternatives to antibiotics—a pipeline portfolio review. The Lancet Infectious Diseases. 2016;16:239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- 43.Moloney MG. Natural Products as a Source for Novel Antibiotics. Trends in Pharmacological Sciences. 2016;37:689–701. doi: 10.1016/j.tips.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Shatzkes, K. et al. Predatory Bacteria Attenuate Klebsiella pneumoniae Burden in Rat Lungs. mBio7, doi:10.1128/mBio.01847-16 (2016). [DOI] [PMC free article] [PubMed]
- 45.Lübbert C, et al. Long-term carriage of Klebsiella pneumoniae carbapenemase–2-producing K pneumoniae after a large single-center outbreak in Germany. American Journal of Infection Control. 2014;42:376–380. doi: 10.1016/j.ajic.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Carey Lambert, et al. Interrupting peptidoglycan deacetylation during Bdellovibrio predator-prey interaction prevents ultimate destruction of prey wall, liberating bacterial-ghosts. Scientific Reports6(1) (2016). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


