Abstract
The SDC25 C-domain is a very active guanine nucleotide dissociation stimulator (GDS) isolated from Saccharomyces cerevisiae which acts equally well on Ha-ras p21 and yeast RAS2. These properties make the SDC25 C-domain a suitable tool to study the basic mechanism of a GDS. The action of the SDC25 C-domain was analysed by mutation of structurally important regions of p21. Substitutions that influence the coordination of Mg2+.GDP or the interaction of the guanine ring were found to stimulate the intrinsic dissociation of GDP and suppress the action of the SDC25 C-domain. No relevant effects were observed with mutations in the phosphate binding loop L1 or by deleting the last 23 C-terminal residues of p21. Substitutions in the switch region 1 (loop L2) and 2 (loop L4) of p21 strongly impaired the action of this GDS; however, we show that this effect is not related to a decreased affinity of the SDC25 C-domain for the mutated p21. No functional competition could be found between this GDS and the catalytic domain of the human GTPase activating protein (GAP). This indicates that GDS and GAP bind to different sites of the p21.nucleotide complex, even though the same mutations in loops L2 and L4 regions affect the activity of both effectors. Since these two regions appear not to be involved directly in the interaction with GDS, we conclude that the negative effect induced by their mutation is related to their function as switches of selective conformations during the GDP to GTP exchange reaction catalysed by GDS.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adari H., Lowy D. R., Willumsen B. M., Der C. J., McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988 Apr 22;240(4851):518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- Amann E., Ochs B., Abel K. J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988 Sep 30;69(2):301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Beitel G. J., Clark S. G., Horvitz H. R. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature. 1990 Dec 6;348(6301):503–509. doi: 10.1038/348503a0. [DOI] [PubMed] [Google Scholar]
- Bischoff F. R., Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991 Nov 7;354(6348):80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990 Nov 8;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Boy-Marcotte E., Damak F., Camonis J., Garreau H., Jacquet M. The C-terminal part of a gene partially homologous to CDC 25 gene suppresses the cdc25-5 mutation in Saccharomyces cerevisiae. Gene. 1989 Apr 15;77(1):21–30. doi: 10.1016/0378-1119(89)90355-7. [DOI] [PubMed] [Google Scholar]
- Calés C., Hancock J. F., Marshall C. J., Hall A. The cytoplasmic protein GAP is implicated as the target for regulation by the ras gene product. Nature. 1988 Apr 7;332(6164):548–551. doi: 10.1038/332548a0. [DOI] [PubMed] [Google Scholar]
- Crechet J. B., Parmeggiani A. Characterization of the elongation factors from calf brain. 2. Functional properties of EF-1 alpha, the action of physiological ligands and kirromycin. Eur J Biochem. 1986 Dec 15;161(3):647–653. doi: 10.1111/j.1432-1033.1986.tb10489.x. [DOI] [PubMed] [Google Scholar]
- Créchet J. B., Poullet P., Camonis J., Jacquet M., Parmeggiani A. Different kinetic properties of the two mutants, RAS2Ile152 and RAS2Val19, that suppress the CDC25 requirement in RAS/adenylate cyclase pathway in Saccharomyces cerevisiae. J Biol Chem. 1990 Jan 25;265(3):1563–1568. [PubMed] [Google Scholar]
- Créchet J. B., Poullet P., Mistou M. Y., Parmeggiani A., Camonis J., Boy-Marcotte E., Damak F., Jacquet M. Enhancement of the GDP-GTP exchange of RAS proteins by the carboxyl-terminal domain of SCD25. Science. 1990 May 18;248(4957):866–868. doi: 10.1126/science.2188363. [DOI] [PubMed] [Google Scholar]
- Downward J., Riehl R., Wu L., Weinberg R. A. Identification of a nucleotide exchange-promoting activity for p21ras. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5998–6002. doi: 10.1073/pnas.87.15.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano O., Aldrich T., Tamanoi F., Taparowsky E., Furth M., Wigler M. Analysis of the transforming potential of the human H-ras gene by random mutagenesis. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4008–4012. doi: 10.1073/pnas.81.13.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano O., Crechet J. B., De Vendittis E., Zahn R., Feger G., Vitelli A., Parmeggiani A. Yeast mutants temperature-sensitive for growth after random mutagenesis of the chromosomal RAS2 gene and deletion of the RAS1 gene. EMBO J. 1988 Nov;7(11):3375–3383. doi: 10.1002/j.1460-2075.1988.tb03210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frech M., John J., Pizon V., Chardin P., Tavitian A., Clark R., McCormick F., Wittinghofer A. Inhibition of GTPase activating protein stimulation of Ras-p21 GTPase by the Krev-1 gene product. Science. 1990 Jul 13;249(4965):169–171. doi: 10.1126/science.2164710. [DOI] [PubMed] [Google Scholar]
- Garreau H., Camonis J. H., Guitton C., Jacquet M. The Saccharomyces cerevisiae CDC25 gene product is a 180 kDa polypeptide and is associated with a membrane fraction. FEBS Lett. 1990 Aug 20;269(1):53–59. doi: 10.1016/0014-5793(90)81117-7. [DOI] [PubMed] [Google Scholar]
- Gibbs J. B., Schaber M. D., Allard W. J., Sigal I. S., Scolnick E. M. Purification of ras GTPase activating protein from bovine brain. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5026–5030. doi: 10.1073/pnas.85.14.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroyoshi M., Kaibuchi K., Kawamura S., Hata Y., Takai Y. Role of the C-terminal region of smg p21, a ras p21-like small GTP-binding protein, in membrane and smg p21 GDP/GTP exchange protein interactions. J Biol Chem. 1991 Feb 15;266(5):2962–2969. [PubMed] [Google Scholar]
- Huang Y. K., Kung H. F., Kamata T. Purification of a factor capable of stimulating the guanine nucleotide exchange reaction of ras proteins and its effect on ras-related small molecular mass G proteins. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8008–8012. doi: 10.1073/pnas.87.20.8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D. A., Fukui Y., Yamamoto M. Homologous activators of ras in fission and budding yeast. Nature. 1990 Mar 22;344(6264):355–357. doi: 10.1038/344355a0. [DOI] [PubMed] [Google Scholar]
- Hwang Y. W., Miller D. L. A study of the kinetic mechanism of elongation factor Ts. J Biol Chem. 1985 Sep 25;260(21):11498–11502. [PubMed] [Google Scholar]
- John J., Frech M., Wittinghofer A. Biochemical properties of Ha-ras encoded p21 mutants and mechanism of the autophosphorylation reaction. J Biol Chem. 1988 Aug 25;263(24):11792–11799. [PubMed] [Google Scholar]
- John J., Schlichting I., Schiltz E., Rösch P., Wittinghofer A. C-terminal truncation of p21H preserves crucial kinetic and structural properties. J Biol Chem. 1989 Aug 5;264(22):13086–13092. [PubMed] [Google Scholar]
- John J., Sohmen R., Feuerstein J., Linke R., Wittinghofer A., Goody R. S. Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry. 1990 Jun 26;29(25):6058–6065. doi: 10.1021/bi00477a025. [DOI] [PubMed] [Google Scholar]
- Jones S., Vignais M. L., Broach J. R. The CDC25 protein of Saccharomyces cerevisiae promotes exchange of guanine nucleotides bound to ras. Mol Cell Biol. 1991 May;11(5):2641–2646. doi: 10.1128/mcb.11.5.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavounis C., Verrotti A. C., De Vendittis E., Bozopoulos A., Di Blasi F., Zahn R., Crechet J. B., Parmeggiani A., Tsernoglou D., Fasano O. Role of glycine-82 as a pivot point during the transition from the inactive to the active form of the yeast Ras2 protein. FEBS Lett. 1991 Apr 9;281(1-2):235–239. doi: 10.1016/0014-5793(91)80401-n. [DOI] [PubMed] [Google Scholar]
- Krengel U., Schlichting I., Scherer A., Schumann R., Frech M., John J., Kabsch W., Pai E. F., Wittinghofer A. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990 Aug 10;62(3):539–548. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- Maruta H., Holden J., Sizeland A., D'Abaco G. The residues of Ras and Rap proteins that determine their GAP specificities. J Biol Chem. 1991 Jun 25;266(18):11661–11668. [PubMed] [Google Scholar]
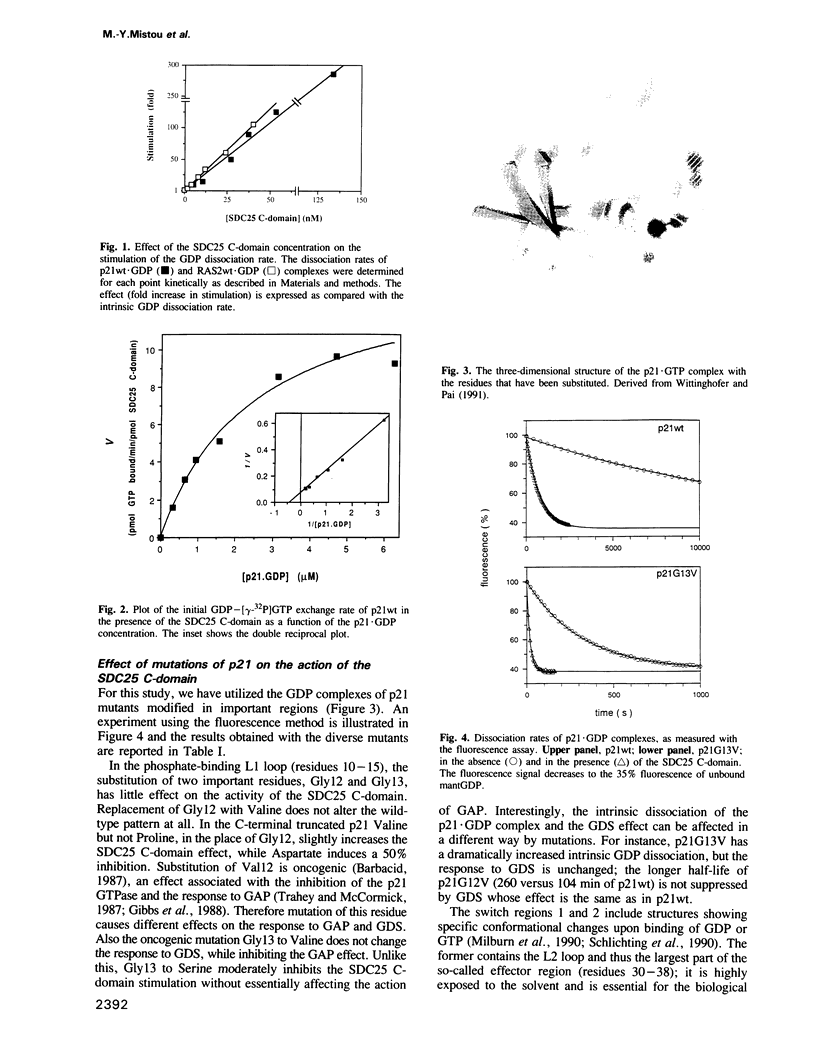
- Milburn M. V., Tong L., deVos A. M., Brünger A., Yamaizumi Z., Nishimura S., Kim S. H. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990 Feb 23;247(4945):939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- Pai E. F., Kabsch W., Krengel U., Holmes K. C., John J., Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989 Sep 21;341(6239):209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- Pai E. F., Krengel U., Petsko G. A., Goody R. S., Kabsch W., Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990 Aug;9(8):2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinstein J., Schlichting I., Frech M., Goody R. S., Wittinghofer A. p21 with a phenylalanine 28----leucine mutation reacts normally with the GTPase activating protein GAP but nevertheless has transforming properties. J Biol Chem. 1991 Sep 15;266(26):17700–17706. [PubMed] [Google Scholar]
- Rey I., Schweighoffer F., Barlat I., Camonis J., Boy-Marcotte E., Guilbaud R., Jacquet M., Tocque B. The COOH-domain of the product of the Saccharomyces cerevisiae SCD25 gene elicits activation of p21-ras proteins in mammalian cells. Oncogene. 1991 Feb;6(2):347–349. [PubMed] [Google Scholar]
- Schaber M. D., Garsky V. M., Boylan D., Hill W. S., Scolnick E. M., Marshall M. S., Sigal I. S., Gibbs J. B. Ras interaction with the GTPase-activating protein (GAP). Proteins. 1989;6(3):306–315. doi: 10.1002/prot.340060313. [DOI] [PubMed] [Google Scholar]
- Scherer A., John J., Linke R., Goody R. S., Wittinghofer A., Pai E. F., Homes K. C. Crystallization and preliminary X-ray analysis of the human c-H-ras-oncogene product p21 complexed with GTP analogues. J Mol Biol. 1989 Mar 5;206(1):257–259. doi: 10.1016/0022-2836(89)90540-8. [DOI] [PubMed] [Google Scholar]
- Schlichting I., Almo S. C., Rapp G., Wilson K., Petratos K., Lentfer A., Wittinghofer A., Kabsch W., Pai E. F., Petsko G. A. Time-resolved X-ray crystallographic study of the conformational change in Ha-Ras p21 protein on GTP hydrolysis. Nature. 1990 May 24;345(6273):309–315. doi: 10.1038/345309a0. [DOI] [PubMed] [Google Scholar]
- Shih T. Y., Stokes P. E., Smythers G. W., Dhar R., Oroszlan S. Characterization of the phosphorylation sites and the surrounding amino acid sequences of the p21 transforming proteins coded for by the Harvey and Kirsten strains of murine sarcoma viruses. J Biol Chem. 1982 Oct 10;257(19):11767–11773. [PubMed] [Google Scholar]
- Simon M. A., Bowtell D. D., Dodson G. S., Laverty T. R., Rubin G. M. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991 Nov 15;67(4):701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- Tong L. A., de Vos A. M., Milburn M. V., Kim S. H. Crystal structures at 2.2 A resolution of the catalytic domains of normal ras protein and an oncogenic mutant complexed with GDP. J Mol Biol. 1991 Feb 5;217(3):503–516. doi: 10.1016/0022-2836(91)90753-s. [DOI] [PubMed] [Google Scholar]
- Tong L., Milburn M. V., de Vos A. M., Kim S. H. Structure of ras proteins. Science. 1989 Jul 21;245(4915):244–244. doi: 10.1126/science.2665078. [DOI] [PubMed] [Google Scholar]
- Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987 Oct 23;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Tucker J., Sczakiel G., Feuerstein J., John J., Goody R. S., Wittinghofer A. Expression of p21 proteins in Escherichia coli and stereochemistry of the nucleotide-binding site. EMBO J. 1986 Jun;5(6):1351–1358. doi: 10.1002/j.1460-2075.1986.tb04366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia A., Chardin P., Wittinghofer A., Sander C. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry. 1991 May 14;30(19):4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- Valencia A., Kjeldgaard M., Pai E. F., Sander C. GTPase domains of ras p21 oncogene protein and elongation factor Tu: analysis of three-dimensional structures, sequence families, and functional sites. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5443–5447. doi: 10.1073/pnas.88.12.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willumsen B. M., Vass W. C., Velu T. J., Papageorge A. G., Schiller J. T., Lowy D. R. The bovine papillomavirus E5 oncogene can cooperate with ras: identification of p21 amino acids critical for transformation by c-rasH but not v-rasH. Mol Cell Biol. 1991 Dec;11(12):6026–6033. doi: 10.1128/mcb.11.12.6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittinghofer A., Pai E. F. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem Sci. 1991 Oct;16(10):382–387. doi: 10.1016/0968-0004(91)90156-p. [DOI] [PubMed] [Google Scholar]
- Wolfman A., Macara I. G. A cytosolic protein catalyzes the release of GDP from p21ras. Science. 1990 Apr 6;248(4951):67–69. doi: 10.1126/science.2181667. [DOI] [PubMed] [Google Scholar]