Abstract
The establishment of the infant gut microbiota is a highly dynamic process dependent on extrinsic and intrinsic factors. We characterized the faecal microbiota of 4 breastfed infants and 4 formula-fed infants at 17 consecutive time points during the first 12 weeks of life. Microbiota composition was analysed by a combination of 16S rRNA gene sequencing and quantitative PCR (qPCR). In this dataset, individuality was a major driver of microbiota composition (P = 0.002) and was more pronounced in breastfed infants. A developmental signature could be distinguished, characterized by sequential colonisation of i) intrauterine/vaginal birth associated taxa, ii) skin derived taxa and other typical early colonisers such as Streptococcus and Enterobacteriaceae, iii) domination of Bifidobacteriaceae, and iv) the appearance of adultlike taxa, particularly species associated with Blautia, Eggerthella, and the potential pathobiont Clostridium difficile. Low abundance of potential pathogens was detected by 16S profiling and confirmed by qPCR. Incidence and dominance of skin and breast milk associated microbes were increased in the gut microbiome of breastfed infants compared to formula-fed infants. The approaches in this study indicate that microbiota development of breastfed and formula-fed infants proceeds according to similar developmental stages with microbiota signatures that include stage-specific species.
Introduction
The neonatal intestine is considered to become colonised with the first microbes immediately after birth1, 2, although recent studies have shown that in utero the environment may not be completely sterile3, 4. Directly after birth, the infant intestine is colonised by a succession of a variety of microorganisms. The development of the intestinal microbiota is highly dynamic due to initial low stability, limited bacterial richness and low diversity of the ecosystem that is establishing2, 5, 6. This process differs considerably between individual infants, and depends upon multiple extrinsic factors including mode of delivery, type of nutrition, use of antimicrobials, and gestational age7–10. With regard to development of organ systems of the infant, the colonising microorganisms do not only play a key role in driving post-natal maturation of the infant gut but also in development of the mucosal immune system11–16. As such, deviating microbial colonisation patterns or distortion of the microbial ecology early in life, which may be caused by nutritional changes, pathogen challenges, or antibiotic treatment, may elicit aberrant immune development processes that potentially cause long-lasting effects on the host organism, including susceptibility for a variety of developmental disorders and diseases17–19.
In recent years, amplicon based sequencing of the 16S rRNA gene has enabled the analysis of hundreds of samples from different origins at high phylogenetic resolution and much greater depth than previously possible. Additionally, shotgun metagenomics can provide an unprecedented functional view of the microbiota in the context of health and disease. A recent study by20, applying shotgun metagenomics, revealed that cessation of breastfeeding was one of the key factors driving maturation of the gut microbiome into an adultlike composition and functionally encoded capacity.
In this study, the development of the infant intestine microbiota was determined in the first 12 weeks of life, employing a high sampling frequency to enable in depth phylogenetic reconstruction of the de novo-colonisation of the newborn when fed a milk-based diet only. Although only a relatively small cohort of 4 breastfed and 4 formula-fed infants was included, the frequent sampling and the use of different molecular technologies (i.e. 16S profiling, qPCR targeting specific pathogens and supplementary FISH targeting specific phylogenetic groups) enabled the assessment of individuality as well as the impact of time and diet on the early-life microbiota colonisation pattern, including the evaluation of specific pathogens and their interactions with typical commensal taxa in the first 12 weeks of life.
Results
Subjects
Five breastfed (BF) and five formula-fed (FF) infants were initially enrolled. All infants were of Caucasian origin and were born vaginally during the winter season. After sample collection was completed, two infants (one breastfed and one formula-fed infant) were excluded a priori from further analysis because of too many missing samples and/or collection of insufficient faecal material. The relevant clinical characteristics of the eight infants included in the analysis are given in Supplementary Table 1. Baseline characteristics at time of recruitment were not significantly different between the two groups. All children in the formula-fed group were given the same brand (Nutrilon) of formula, containing a blend of the prebiotics GOS and FOS (scGOS:lcFOS (9:1); 0.6 g/100 ml). BF4 and BF3 received complementary formula (same brand) from 10 and 11 weeks onwards, respectively.
General bacterial population dynamics during the first weeks of life
In total 1,390,768 bacterial 16S rDNA sequences, with on average 11,307 (±6,355; SD) reads of 399 nt (±85; SD) per sample, were analysed. Irrespective of the type of feeding from birth onwards, infants generally developed a gut microbiota characterized by sequential colonisation of intrauterine/vaginal birth associated first colonisers, followed by skin derived taxa and subsequent rapid domination of Bifidobacteriaceae around 3 weeks of age. After 3 weeks, facultative anaerobes such as the genera Staphylococcus, Streptococcus and members of the phylum Proteobacteria generally declined, as visualised in Supplementary Figure S1. In order to characterize the development of microbiota composition during early life, multiple multivariate statistical analyses were employed to separately identify the impact of individual, time and diet on the overall community development. PCA based on the relative abundance of species revealed that the aforementioned variables accounted for 59.5% of the variation in microbiota composition. Microbiota signatures clustered predominantly by individual and were less dependent on feeding type or sampling time, although these latter variables did contribute to microbiota composition clustering (Fig. 1). Importantly, the centroids of the sampling time points during the first 21 days were clearly distinct from each other as well as from the later sampling points that also clustered more closely together (Fig. 1). The latter finding implies that the most dynamic developmental changes in gut microbiota composition take place during the first 3 weeks of the three months time frame that was analysed. Partial Redundancy Analysis (RDA), in which the effects of individual and type of feeding were removed, confirmed that the first 3 weeks were different from the subsequent 9 weeks (variation explained 7.1%, P = 0.002). The importance of individuality in microbiota composition in this dataset was confirmed by a partial RDA, revealing that individuality alone (i.e., after removal of the effects of feeding and time) explained 46.5% of the variation observed, and separation of individuals was shown to be significant by Monte Carlo permutation testing (P = 0.002) (Fig. 2). Notably, individuality was also the most important, and significant parameter, that explained 28.0% of the variation in microbial taxa as assessed by FISH (Supplementary Figure S2). Unsupervised clustering using Principal Coordinates Analysis (PCoA) of unweighted UniFrac distance matrices indicated that individuality was more pronounced in breastfed infants compared to formula-fed infants (Fig. 3A). This higher degree of individuality in the breastfed group appeared to be consistently present after day 3 (Fig. 3B).
Figure 1.
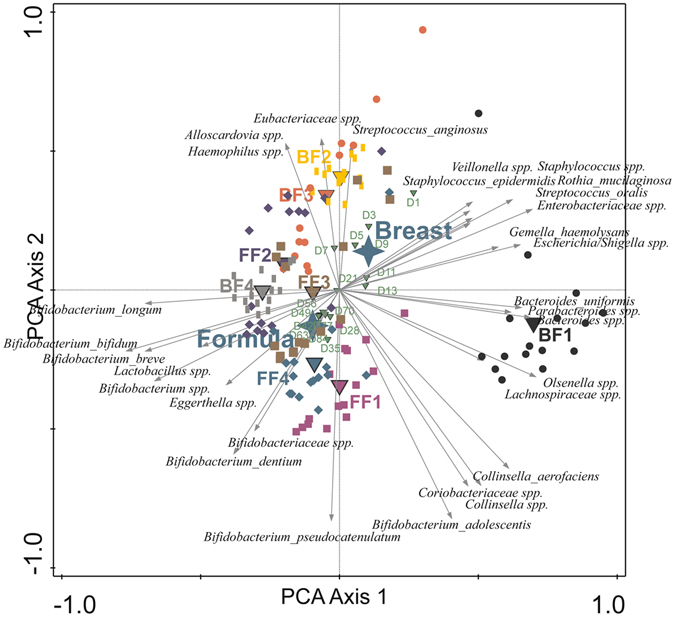
Principal component analysis (PCA) based on the species level gut microbiota composition of breast and formula-fed infants from day 1 to 12 weeks of age. Nominal environmental variables are indicated by large down triangles (infants: BF1-4 and FF1-4), small down triangles (time points: D1-D84) and stars (diet: formula and breast). Reads that could not be assigned on the species level were included on the genus (genus unspecified) or family level (family unspecified) depending on the phylogenetic resolution. Reads were always assigned to the most specific taxonomic level possible, taxa that could not be assigned on the species level were included as genera or families unspecified.
Figure 2.
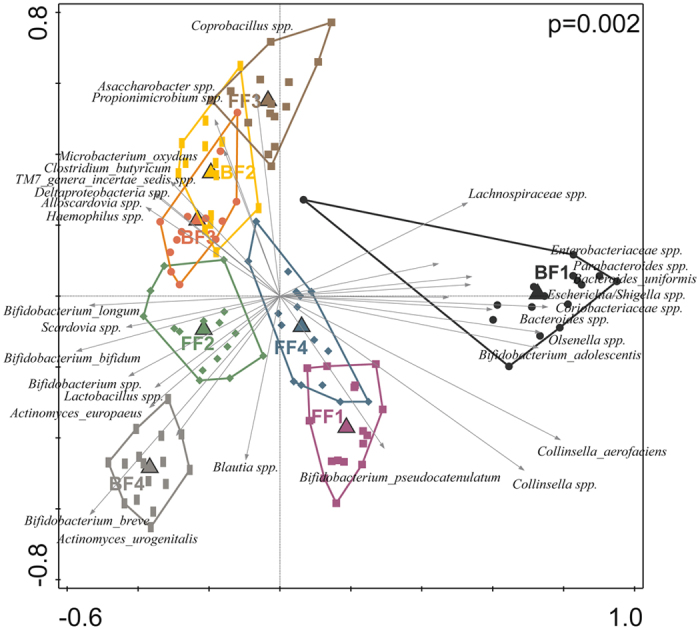
Triplot of partial RDA based on the relative abundance of detected species in individuals after removing the effects of time and type of feeding. Constrained explanatory variables are indicated by triangles: BF1-4 represents infants being breastfed and FF1-4 represents infants being formula-fed. The arrows indicate the 30 species which had the highest amount of variability in their values explained by the canonical axes. Upper right shows the P-value of Monte Carlo Permutation testing. Covariates time and feeding were first fitted by regression and then partialled out (removed) from the ordination. Taxa that could not be assigned on the species level were included as genera or families unspecified.
Figure 3.
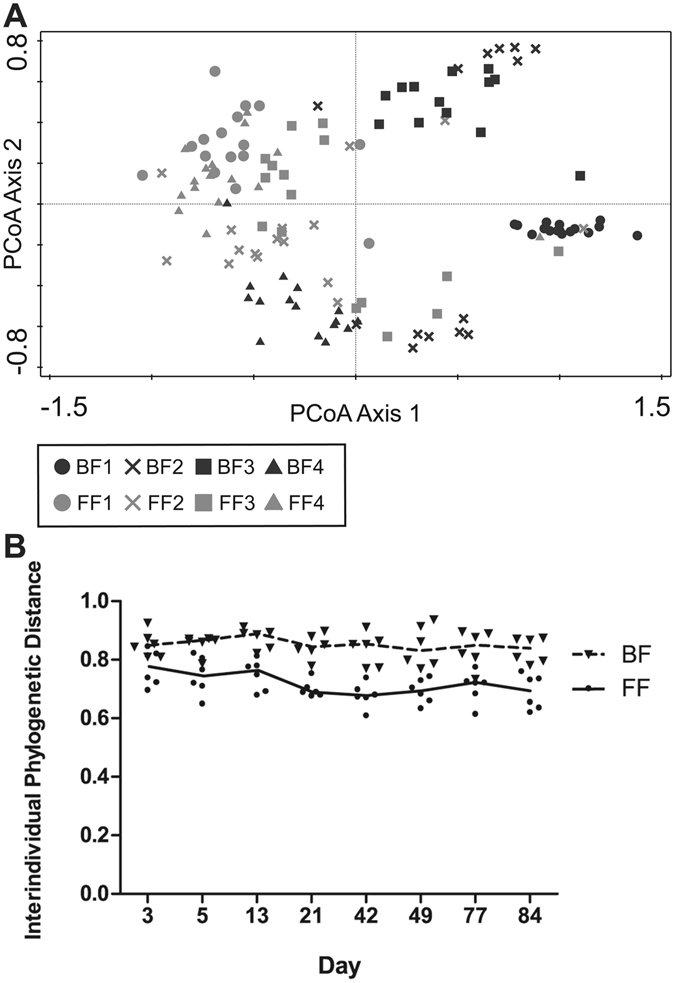
Individuality in breastfed infants versus formula-fed infants. (A) Principal Coordinates Analysis (PCoA): unweighted UniFrac interindividual distances of breast- and formula-fed infants are plotted based on similarities/dissimilarities among samples. (B) Interindividual distances from day 1 till 12 weeks of age. Unweighted UniFrac interindividual distances are plotted over time for breastfed (BF) infants and formula-fed (FF) infants. Curves represent the median.
To identify microbiota signatures that represent significant developmental progression of the microbiota composition over time, both the effects of individuality and feeding type were removed (partial RDA) and enabled the recognition of time-resolved, early life microbiota developmental trajectories with high significance (Fig. 4A; explained variation 10.4%, P = 0.002). Within these trajectories, microbiota compositional transitions can be recognized on basis of discriminatory taxa, being intrauterine/vaginal birth associated first colonisers, such as Lactobacillus crispatus, Corynebacterium pseudogenitalium, Sphingomonadales spp., Enterococcaceae spp., Pelomonas spp. and Ochrobactrum spp. (day 1 to 3)7, 21–24. These first colonizers were followed by the cumulative colonisation of typical early life taxa such as members of the Enterobacteriaceae family, Streptococcus spp. and skin derived taxa such as Staphylococcus spp. during the day 3 to day 21, which decreased at later stages and was followed by an increasing domination of Bifidobacterium spp. (day 21 to 35) for the remainder of the sampling period and the appearance of low-abundant adult-like taxa (e.g. the genera Eggerthella and Blautia) at around three months of age. Separate analyses of breast- and formula-fed infants (Fig. 4, panels B and C, respectively) revealed that the effect of time was visible in both groups. The time-resolved developmental progression pattern was more homogeneous in the formula-fed infants (explained variation is 18.4%, P = 0.002), which support our finding that microbiome development of the breastfed infants is more individually determined (explained variation is 7.0%, P = 0.02, see also Fig. 4B,C).
Figure 4.
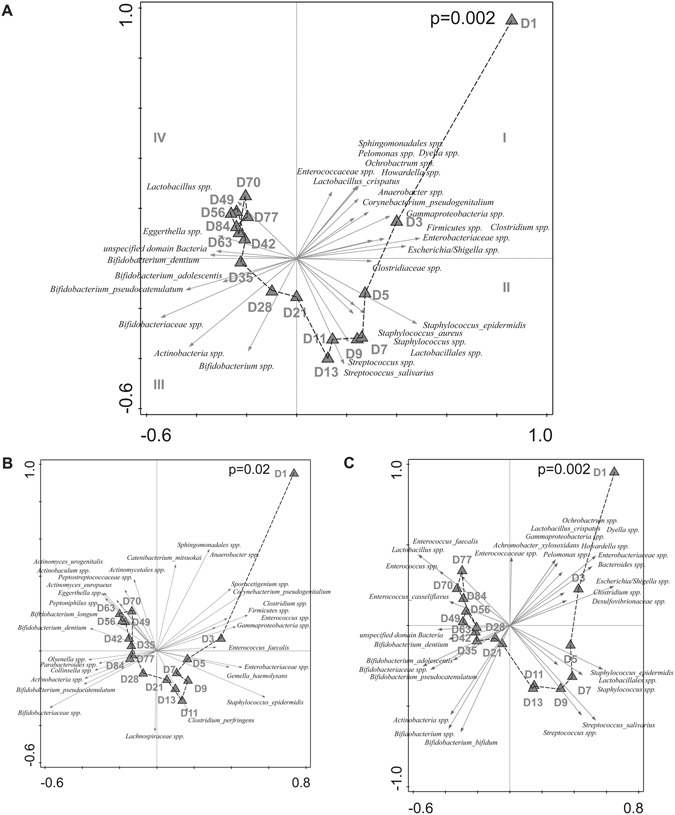
(A,B,C) Triplot of partial RDA based on the relative abundance of detected species in individuals over time (day 1 until 12 weeks of age). Constrained explanatory variables are indicated by triangles: day 1 (D1) until day 84 (D84). Impact of time on microbiota composition was assessed in (A) all breastfed and formula-fed infants, after removing the effects of individual and type of feeding, (B) Breastfed infants only, after removing the effects of individual, and (C) Formula-fed infants only, after removing the effects of individual. The arrows indicate the 30 species which had the highest amount of variability in their values explained by the canonical axes. Upper right shows the p-value of Monte Carlo Permutation testing. Taxa that could not be assigned on the species level were included as genera, families, orders, classes or phyla unspecified.
Classification of the 16S sequences down to the species level revealed the presence of the potential gastrointestinal tract pathogens C. perfringens and C. difficile, but also the upper oro-gastrointestinal and respiratory tract pathogens H. parainfluenzae, K. pneumonia and S. pneumoniae. Their identity and presence was confirmed by a targeted qPCR approach (Fig. 5), which as a consequence of the higher sensitivity of qPCR, identified additional positive samples as compared to the 16S-based profiling. All infants had at least one faecal sample with detectable levels of S. pneumoniae and H. parainfluenza, albeit at very low relative abundance, during the first 12 weeks of life. During these episodes of carriage there were no concurrent parental reports of upper respiratory illness. The parents of infant BF3 reported a cold on day 13, which coincided with the detection of K. pneumoniae from days 14–56, a microorganism that was not detected in any of the other infants. C. difficile was detected in all formula-fed infants and in BF3 that received complementary formula from week 11. This pathogenic taxon typically emerged in the second to third month of life. High levels of C. perfringens were detected from day 9 onwards in BF1, in which also higher levels of typical adultlike taxa belonging to the Bacteroidaceae, Lachnospiraceae and Enterobacteriaceae families were detected, which might potentially reflect a dysbiotic community state related to the C. perfringens colonisation observed during the first weeks of life. As a consequence, BF1 showed most distinct profile compared to the other subjects (Fig. 1). Another taxon important for the observed individuality is Bifidobacterium, the presence and absence of identified members (phylotypes) of this bacterial genus were strong drivers for the separation of the different individuals, as visible in Fig. 2.
Figure 5.
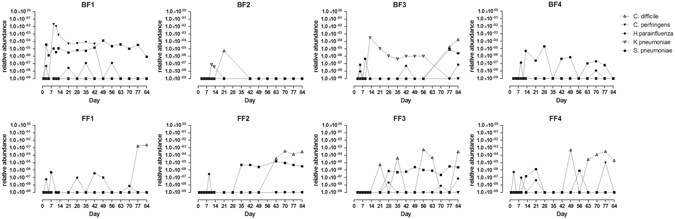
Relative abundance of potential pathogens of the oro-gastrointestinal and respiratory tract as detected by qPCR in the faeces of breastfed and formula-fed infants from day 1 till 12 weeks. Relative abundance was calculated as a fraction of the calculated copy numbers for total bacterial counts (16S universal primers) and targeted pathogens. The detection threshold for all qPCRs performed was considered to be 100 target copies of DNA.
Bacterial population development and the influence of feeding
Although the time-resolved developmental signature is comparable between breastfed and formula-fed infants, several diet-associated discriminating signature taxa are characteristic for the subsequent microbiota transitions observed. Streptococcus and Enterococcus species were discriminatory signature taxa associated with formula feeding in the first month and second-to-third month of life, respectively. Notably, the enrichment of Enterococcus phylotypes was significantly associated with the presence of the potential pathogen C. difficile (Supplementary Figure S3). In addition, Random Forest analyses revealed that among the top 20 microbial classifiers predictive for breastfeeding, 5 skin-associated genera (Staphylococcus, Actinomyces, Propionibacterium, Corynebacterium and Gemella, ranked by importance) were present (Supplementary Figure S4). The prevalence and dominance of typical skin colonisers that were selected on basis of previously identified skin-associated taxa25, was particularly more pronounced and more persistent during the different microbiota developmental stages in breastfed infants (Supplementary Figure S5). Next to these skin-associated microbial groups, Random Forest analysis also identified the relative abundance of several Bifidobacterium species as a classifier for the discrimination of the infants’ microbiota in relation to the feeding regime. In particular, a phylotype most similar to Bifidobacterium dentium was identified as being present in higher relative abundance in formula-fed infants compared to those that were breastfed (Supplementary Figure S6). Taken together, these data show that diet-associated gut microbial signatures were present, but that a similar effect of time on gut microbial development was present in both dietary groups.
Discussion
Birth coincides with the newborn’s entry into a world densely populated by bacteria, resulting in the dynamic development of a microbial community in the infant gut. Aberrations of this colonisation process very early in life, e.g. through antibiotics, have been shown to have long-lasting consequences for the developing microbiota26 that have been associated with increased risk of developing metabolic or immunological disease in later life27–31. To allow improved protection of this fragile colonisation process, e.g. through nutritional approaches, a more detailed insight into the developmental stages of early life microbial intestinal communities is warranted.
We investigated the dynamics of the faecal microbiota during the first 12 weeks of life of vaginally delivered, healthy breastfed and formula-fed infants, using an integrative approach comprising complementary molecular technologies, namely 16S profiling, qPCR and FISH. The large number of faecal samples collected per infant (total 136; 17 samples per infant, over the first 12 weeks of life) allowed detailed analyses of the progression of bacterial colonisation of the GI-tract in a time-resolved manner. In this small sample group, individuality and age-related maturation were the two most important factors explaining the variation in microbiota composition observed during the first 3 months of life. Although the initial process of colonisation is very dynamic and complex, already the first faecal samples following the meconium defecation, contained key taxa which were unique to the individual and persisted in the ecosystem during the consecutive 12 weeks. Overall, individuality was the strongest predictor of microbiota composition, which is in line with previous findings of a slightly larger study 8. Intriguingly this uniqueness in microbial composition appeared to be more pronounced in breastfed infants, which we hypothesize to be due to the mother-specific and fluctuating composition of breast milk. Particularly the high variety of human milk oligosaccharides (HMOs), which has been reported to be quite diverse among mothers and that also differs over time within a single breastfeeding mother32, is likely to play an important determining role in gut microbial development and composition33, 34. Also the mother’s milk microbiota may play an important role in the establishment of the gut microbiota of a breastfed infant35. Consequently, breast milk can drive immune-developmental effects in the infants via either direct or indirect mechanisms, involving direct changes in the infant’s intestinal mucosal responses to dietary ingredients, or involving selective modulation of the microbiota composition and its development, respectively32. Conversely, infant formula contains a relatively simple oligosaccharide composition that is stable over time and does not involve e.g. a milk microbiota, skin-associated taxa and maternal antibodies. It may thereby select more similar microbial communities in the formula-fed infants. Nowadays, different prebiotic oligosaccharides, such as galacto-oligosaccharides, fructo-oligosaccharides, polydextrose, and mixtures of these are present in virtually all commercially available infant formula preparations. These prebiotics bring infant formula one step closer to breast milk and may support the similarity of the microbiota development signature congruency between breast- and formula-fed infants that we observed. However, the slightly lower degree of individuality observed in the formula-fed infants may be a consequence of the fact that prebiotics lack the structural diversity of HMOs, and therefore cannot be considered their functional equivalent.
The present study confirmed earlier observations that microbial succession dynamics is a non-random process and that infants are rapidly colonised by microbes from different environments they are exposed to7, 9, 20. We show that mother-associated microbes were among the first colonisers, which was most apparent in the meconium samples. These samples are phylogenetically most distinct in our time-resolved analysis and contain taxa that are representative of the intrauterine/vaginal environment. Interestingly, some signature genera associated with these initial communities were uniquely enriched in the first faecal samples (days 1–3), which may suggest that they were non-viable contaminations in the infant intestine that are derived from the birth canal, or that they were initially viable but failed to colonise substantially in the infant gastrointestinal tract. In contrast, other members of these initial communities displayed prolonged colonisation and persistence in the gastrointestinal tract of newborns, which was in particular observed for the genus Lactobacillus. The bacterial taxa that succeeded these initial communities included facultative anaerobic bacteria such as Enterobacteriaceae members and Streptococcus spp., confirming previously reported dynamics of the infant microbiota8, 36. These bacteria have been proposed to pave the way for strict anaerobes through consumption of the available oxygen10, 37. In addition, our study highlighted the presence of typical skin associated microorganisms in the faecal microbiome during the first two weeks of life. Our analyses detected a third developmental phase characterized by progressive domination of the microbiota by Bifidobacteriaceae, a finding on which existing literature is conflicting. Several studies have reported that Bifidobacteriacea almost always dominate the intestinal community of breastfed neonates in early life38–40, but paucity of Bifidobacterial species by culture-independent investigations has also been described, likely due to technical biases40. In our study-population, the last phase of intestinal colonisation included the appearance of typical adult-like strict anaerobes like Blautia and Eggerthella after 8 weeks and onwards, which could represent the starting point for the development towards an adultlike composition, of which the stabilization has been proposed to occur around 3 years of age41.
Remarkably, in the present study population the sequential signatures of intestinal colonisation were detected in both breastfed and formula-fed infants, suggesting that diet was not the primary successional mechanism involved in gut microbiota maturation, but that other factors such as physiochemical and immunological development of the newborn were likely to be more critical determinants in this study. Although the developmental signature between both diet groups was quite congruent, there were differences between the signature taxa of the different developmental stages. For example, the taxa typically associated with human skin, like Propionibacterium, Staphylococcus, Gemella and Corynebacterium were found to be more dominant and persistent members of the microbiota in breastfed infants, which might be a reflection of the more frequent and intense contact of breastfed infants with the mother’s skin42.
In the present study, formula-fed infants harboured higher levels of a specific bifidobacterial taxon that is phylogenetically closely related to B. dentium compared to breastfed infants. Recently, shotgun metagenomics that allows annotation down to the species level, showed that B. adolescentis appeared to be enriched in formula-fed infants20. These findings can likely be explained by the differential carbon source availability in the two feeding regimes. Particularly, the relatively large fraction of the HMOs that is built on basis of Lacto-N-biose cannot be utilized by B. dentium and B. adolescentis, but is readily utilized by various other Bifidobacterium species43, including the HMO-specialist B. longum 44. Other late signature taxa for formula-fed infants were Enterococccus as detected by 16S profiling and C. difficile as identified by qPCR. Although C. difficile was also detected in one breastfed infant from 11 weeks onward, this occured after the introduction of complementary formula feeding. Higher prevalence and relative abundance of C. difficile in formula-fed infants have been reported before20, 45. Possibly breast milk contains a factor (e.g. maternal antibodies or antimicrobial HMOs46) that prevents C. difficile colonisation, or supports a microbiota development pattern that exerts higher endogenous colonisation resistance against C. difficile. Interestingly, a recent study in a single infant of 6 months old showed opposite results when the infant switched from mother milk to cow milk (not formula)47. This study differs from previous studies and our study in that the infant was a few months older and on solid foods already. Intruigingly, we found that the appearance of detectable levels of C. difficile was significantly associated with enrichment of four Enterococcus phylotypes. Enterococcus spp. have previously also been positively associated with C. difficile in the gut microbiota of patients treated for malignancy, a finding that is supported by infection studies in mice48. C. difficile colonisation that we and others observed appears to depend on diet-microbiota interactions. This may eventually provide novel leads towards prevention or suppression of this opportunistic pathogen in the human intestinal tract, if warranted. We however note that the significance and effects of asymptomatic C. difficile colonisation on the gut microbiome, infections and allergic disease in neonates and later in life remain to be determined49, 50.
Although the number of infants in this study was small and limits the statistical power of comparisons, the large number of faecal samples collected per infant (17 samples per infant, during the first 12 weeks of life) allowed detailed analysis of the progression of bacterial colonisation of the intestinal tract per individual. Importantly, time-resolved microbial signatures were congruently detected in the pool of 8 infants and appeared in both feeding regimes. Nevertheless, differences depending on the feeding regime were detected. To provide leads for further optimization of infant formula, the importance of these differences and their impact on the interplay of the microbiota with the developing mucosal immune and metabolic functions remains to be determined and require larger study cohorts. Further knowledge of the relationship of the ‘normal’ colonisation patterns in healthy infants with the development of appropriate mucosal immune function, compared to aberrant colonisation in e.g. antibiotic treated infants, is paramount and to fuel intervention strategies aimed at reducing the risk of immune system associated diseases in later stages of life.
Material and Methods
Subjects and study design
For this 12 weeks longitudinal study, two times five infants were recruited that were exclusively breastfed or formula-fed from birth onward, with the exception that 2 breastfed infants were complementary fed with formula from week 10 or 11 onward. All infants were born after more than 39 weeks of gestation, were apparently healthy, and were all vaginally delivered at the hospital (Nieuwegein, The Netherlands). Each infant stayed in the hospital for a maximum of 24 hours after delivery. Signed informed consent was obtained from each infant’s parents. The St. Antonius Hospital local Ethics Committee approved the study and methods were carried out in accordance with the relevant guidelines and regulations.
Formula-fed infants were allowed to receive the parents’ choice of formula, and the brand was recorded. All parents decided on the same brand (Nutrilon) of formula, containing a blend of the prebiotics GOS and FOS (scGOS:lcFOS (9:1); 0.6 g/100 ml). All children were vaccinated according to the National Vaccination Schedule and the specific dates were documented during the study. Parents were instructed to keep a journal recording key events in the categories of illness, medication, and dietary change.
Faecal sample collection
Infant faecal samples were collected by the parents at specific time points during the first 12 weeks of life; every other day during the first two weeks of life, and after that, every week until the infant was three months old (17 samples in total).
Parents were provided with collection devices and diapers (Pampers, Procter & Gamble) for the complete study period. Latter approach was used to ensure minimal variation in faeces absorption by the diaper and unambiguous collection of faecal samples. After immediate collection, parents stored the faecal samples in their home freezers (at −20 °C), and the samples were subsequently transferred and stored in the hospital freezers until the end of the collection period of the study. Finally, all samples were transported on dry ice to NIZO Food Research BV (Ede, the Netherlands) and stored at −20 °C until further processing. Each transportation step of the faecal samples was carried out in frozen state, strictly preventing intermediate thawing of the samples. Batches of 48 samples were thawed overnight on ice at 4 °C after which aliquots were prepared and frozen at −80 °C until DNA extraction. Aliquots for FISH were processed immediately, as described in the supplementary material and methods.
FISH
The FISH method is described in the supplementary material and methods.
16S rRNA gene sequencing analysis
DNA extraction
DNA isolation from faeces was performed as previously51, 52. Briefly, after bead-beating, DNA was purified by maximal 3 consecutive phenol-chloroform extractions (until a lucid solution was obtained), followed by isopropanol precipitation of the DNA. The resulting pellets were washed with 70% (v/v) ethanol, and dissolved in 100 ml TE buffer by overnight incubation at 4 °C. DNA samples were stored at −20 °C until further processing.
Library preparation for 16S rRNA pyrosequencing
For the preparation of the amplicon pool for pyrosequencing, the following universal primers were applied for amplification of the V3-V6 region of the 16S rRNA gene: (i) forward primer, 5′-CCATCTCATCCCTGCGTGTCTCCGACTAGNNNNNNACTCCTACGGGAGGCAGCAG-3′ (the italicised sequence is the 454 Life Sciences primer A, and the bold sequence is the broadly conserved bacterial primer 338 F; NNNNNN designates the sample-specific six-base barcode used to tag each PCR product in combination with (ii) reverse primer 5′-CCTATCCCCTGTGTGCCTTGG CAGTCTCAGCRRCACGAGCTGACGAC-3′ (the italicised sequence is the 454 Life Sciences primer B, and the bold sequence is the broadly conserved bacterial primer 1061 R). PCR amplification mixture contained: 1 μL faecal DNA, 1 μL barcoded forward primer (10 μM), 15 μL master mix [1 μL KOD Hot Start DNA Polymerase (1 U/μL; Novagen, Madison, WI, USA), 5 μL KOD-buffer (10 ×), 3 μL MgSO4 (25 mM), 5 μL dNTP mix (2 mM each), 1 μL (10 μM) of reverse primer] and 33 μL sterile water (total volume 50 μL). PCR conditions were: 95 °C for 2 minutes followed by 35 cycles of 95 °C for 20 s, 55 °C for 10 s, and 70 °C for 15 s. The approximately 750 bp PCR amplicon was subsequently purified using the MSB Spin PCRapace kit (Invitek) and the concentration was checked with a Nanodrop 1000 spectrophotometer (Thermo Scientific). A composite sample for pyrosequencing was prepared by pooling 200 ng of the purified PCR products obtained for each sample. The approximately 750 bp mixed amplicon in the pooled sample was purified from gel (1% agarose) with the MinElute Gel Extraction kit (Qiagen, Venlo, The Netherlands). The concentration of the gel-extracted pooled-amplicon was determined and 50 μl (14.5 ng/μl purified PCR product) was submitted for pyrosequencing of the V3-V4 region of the 16S rRNA gene on the 454 Life Sciences GS-FLX platform using Titanium sequencing chemistry (GATC Biotech, Konstanz, Germany).
16S rRNA gene sequence bioinformatic analysis
Pyrosequencing data were analyzed with a workflow based on the Quantitative Insights Into Microbial Ecology (QIIME) v1.253, using settings as recommended in the QIIME 1.2 tutorial, with the following exceptions: reads were filtered for chimeric sequences using Chimera Slayer54, and OTU clustering was performed with settings as recommended in the QIIME newsletter of December 17th 2010 (http://qiime.wordpress.com/2010/12/17/new-default-parameters-for-uclust-otu-pickers/) using an identity threshold of 97%. Diversity metrics were calculated as implemented in QIIME 1.2. To determine the amount of diversity shared between two communities (beta diversity) the UniFrac metric was employed55. UniFrac distances are based on the fraction of branch length shared between two communities within a phylogenetic tree constructed from the 16S rRNA gene sequences. With unweighted UniFrac, only the presence or absence of lineages is considered (community membership), whereas weighted UniFrac, also relative abundances of lineages within communities are considered (community structure). Additional data handling was performed using in-house developed Python and Perl scripts.
qPCR
Real-time detection was performed on DNA samples targeting the following set of pathogens that were detected by 16S profiling: Clostridium difficile, Clostridium perfringens, Haemophilus parainfluenzae, Klebsiella pneumonia and Streptococcus pneumonia. The target organisms, their respective genetic loci targeted by qPCR and the primer (and probe) sequences used are listed in Supplementary Table 2. Specific PCR conditions employed were adjusted per target locus and was based on the PCR protocols provided in the respective literature references from which the primer (and probe) sequences were obtained (Supplementary Table 2). Amplification and detection were performed using the ABIPRISM 7300-PCR sequence detection system (Applied Biosystems. Foster City, CA), detecting the fluorescent products in the last step of each amplification cycle. After amplification, melting curve analysis was employed to establish PCR-product specificity in case of SYBR Green based detection. Positive and negative controls (other bacterial species) were included in each qPCR run. Gene copy numbers per gram of faeces were extrapolated for each sample, using positive-control template DNA and standard curves generated in triplicate and linear C t-value regression in serial 10-fold template dilutions.
Statistical analysis
Clinical characteristics of the infants and potential differences between groups were assessed by Mann-Whitney-U test. All analyses were performed using SPSS version 20.0 (SPSS Inc., Chicago, IL, USA). Principal Coordinate Analyses (PCoA; unsupervised), Principal Component Analysis (PCA; unsupervised) and Redundancy analysis (RDA; supervised) was performed using CANOCO for Windows 5.0 (Microcomputer Power, USA) according to the manufacturer’s instructions56 where the UNIFRAC distances (PCoA) and the relative abundance of the 168 identified species through 16S sequencing and the 7 phylogenetic groups targeted by FISH were used as responsive variables (PCA and RDA). RDA is the canonical form of PCA analyses and is a multivariate linear regression model where several response parameters are related to the same set of environmental (explanatory) variables. Partial RDA was employed to analyse the effect of time and individual after covariance attributable to diet (always) and individual and time, respectively, was first fitted by regression and then partialled out (removed) from the ordination as described in the Canoco 5 manual56. Variation explained by the explanatory variables corresponds to the classical coefficient of determination (R2), but was adjusted for degrees of freedom (for explanatory variables) and the number of cases56. Statistical significance was assessed by the Monte Carlo permutation procedure (MCPP) with 499 random permutations under the full model56. Machine learning algorithms (Random Forest) were used to predict to what extent each taxon can predict type of feeding received.
Electronic supplementary material
Acknowledgements
The authors would like to thank the parents and infants enrolled for this study. We also want to thank Ingrid van Alen, Erwin Raangs, Saskia van Schalkwijk and Iris I. van Swam for excellent technical assistance.
Author Contributions
H.M.T., N.B.M.M.R., A.M.V., J.B., D.M.S., M.K.2. and G.T.R. designed the study. N.B.M.M.R., D.M.S., E.F., H.J.M.H. and A.M.V. executed the study and performed measurements. H.M.T., J.B., D.M.S. and G.A.M.K. performed data analysis. Interpretation of the data was done by H.M.T., N.B.M.M.R., J.B., D.M.S., G.A.M.K. H.J.M.H., A.M.V., M.K.2. and G.T.R. H.M.T., N.B.M.M.R., J.B., G.A.M.K., A.M.V., M.K.2. and G.T.R. wrote and/or revised the manuscript. All authors, including M.K.1. and N.C., reviewed and approved the final manuscript.
Competing Interests
Authors declare conflicts of interest. This work was funded, in part, by Wyeth Nutrition. Nikhat Contractor and Martin Kullen were employed by Wyeth Nutrition during conduction of the study.
Footnotes
Harro M. Timmerman, Nicole B. M. M. Rutten, Jos Boekhorst, Michiel Kleerebezem and Ger T. Rijkers contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08268-4
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vaishampayan PA, et al. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castanys-Munoz E, Martin MJ, Vazquez E. Building a Beneficial Microbiome from Birth. Adv Nutr. 2016;7:323–330. doi: 10.3945/an.115.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nuriel-Ohayon M, Neuman H, Koren O. Microbial Changes during Pregnancy, Birth, and Infancy. Front Microbiol. 2016;7:1031. doi: 10.3389/fmicb.2016.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aagaard K, et al. The placenta harbors a unique microbiome. Science translational medicine. 2014;6:237ra265. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adlerberth, I. Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutr Workshop Ser Pediatr Program62, 13–29; discussion 29–33, doi:10.1159/000146245 (2008). [DOI] [PubMed]
- 6.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penders J, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 10.Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere MF. Development of intestinal microbiota in infants and its impact on health. Trends in microbiology. 2013;21:167–173. doi: 10.1016/j.tim.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 12.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 13.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 14.Sjogren YM, et al. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin Exp Allergy. 2009;39:1842–1851. doi: 10.1111/j.1365-2222.2009.03326.x. [DOI] [PubMed] [Google Scholar]
- 15.Gronlund MM, Arvilommi H, Kero P, Lehtonen OP, Isolauri E. Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0-6 months. Arch Dis Child Fetal Neonatal Ed. 2000;83:F186–192. doi: 10.1136/fn.83.3.F186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brugman S, Perdijk O, van Neerven RJ, Savelkoul HF. Mucosal Immune Development in Early Life: Setting the Stage. Archivum immunologiae et therapiae experimentalis. 2015;63:251–268. doi: 10.1007/s00005-015-0329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vael C, Desager K. The importance of the development of the intestinal microbiota in infancy. Curr Opin Pediatr. 2009;21:794–800. doi: 10.1097/MOP.0b013e328332351b. [DOI] [PubMed] [Google Scholar]
- 18.Alm B, et al. Neonatal antibiotic treatment is a risk factor for early wheezing. Pediatrics. 2008;121:697–702. doi: 10.1542/peds.2007-1232. [DOI] [PubMed] [Google Scholar]
- 19.Torow N, Marsland BJ, Hornef MW, Gollwitzer ES. Neonatal mucosal immunology. Mucosal immunology. 2017;10:5–17. doi: 10.1038/mi.2016.81. [DOI] [PubMed] [Google Scholar]
- 20.Backhed F, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Human Microbiome Project, C Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verstraelen H, et al. Characterisation of the human uterine microbiome in non-pregnant women through deep sequencing of the V1-2 region of the 16S rRNA gene. PeerJ. 2016;4:e1602. doi: 10.7717/peerj.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno, I. et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol, doi:10.1016/j.ajog.2016.09.075 (2016). [DOI] [PubMed]
- 25.Zeeuwen P, et al. Microbiome Dynamics of Human Epidermis Following Skin Barrier Disruption. Journal of Investigative Dermatology. 2012;132:S111–S111. doi: 10.1038/jid.2012.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azad MB, et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG. 2016;123:983–993. doi: 10.1111/1471-0528.13601. [DOI] [PubMed] [Google Scholar]
- 27.Dogra, S. et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. MBio6, doi:10.1128/mBio.02419-14 (2015). [DOI] [PMC free article] [PubMed]
- 28.Arrieta MC, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Science translational medicine. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 29.Azad MB, Bridgman SL, Becker AB, Kozyrskyj AL. Infant antibiotic exposure and the development of childhood overweight and central adiposity. Int J Obes (Lond) 2014;38:1290–1298. doi: 10.1038/ijo.2014.119. [DOI] [PubMed] [Google Scholar]
- 30.Risnes KR, Belanger K, Murk W, Bracken MB. Antibiotic exposure by 6 months and asthma and allergy at 6 years: Findings in a cohort of 1,401 US children. Am J Epidemiol. 2011;173:310–318. doi: 10.1093/aje/kwq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trasande L, et al. Infant antibiotic exposures and early-life body mass. Int J Obes (Lond) 2013;37:16–23. doi: 10.1038/ijo.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22:1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M, et al. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J Pediatr Gastroenterol Nutr. 2015;60:825–833. doi: 10.1097/MPG.0000000000000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis ZT, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:13. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pannaraj, P. S. et al. Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA pediatrics, doi:10.1001/jamapediatrics.2017.0378 (2017). [DOI] [PMC free article] [PubMed]
- 36.Favier CF, de Vos WM, Akkermans AD. Development of bacterial and bifidobacterial communities in feces of newborn babies. Anaerobe. 2003;9:219–229. doi: 10.1016/j.anaerobe.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Jost T, Lacroix C, Braegger CP, Chassard C. New insights in gut microbiota establishment in healthy breast fed neonates. PloS one. 2012;7:e44595. doi: 10.1371/journal.pone.0044595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scholtens PA, Oozeer R, Martin R, Amor KB, Knol J. The early settlers: intestinal microbiology in early life. Annu Rev Food Sci Technol. 2012;3:425–447. doi: 10.1146/annurev-food-022811-101120. [DOI] [PubMed] [Google Scholar]
- 39.Favier CF, Vaughan EE, De Vos WM, Akkermans AD. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002;68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turroni F, et al. Diversity of bifidobacteria within the infant gut microbiota. PloS one. 2012;7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jimenez E, et al. Staphylococcus epidermidis: a differential trait of the fecal microbiota of breast-fed infants. BMC microbiology. 2008;8:143. doi: 10.1186/1471-2180-8-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao JZ, et al. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl Environ Microbiol. 2010;76:54–59. doi: 10.1128/AEM.01683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schell MA, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Penders J, et al. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol Lett. 2005;243:141–147. doi: 10.1016/j.femsle.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 46.Bode L. The functional biology of human milk oligosaccharides. Early human development. 2015;91:619–622. doi: 10.1016/j.earlhumdev.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Davis MY, Zhang H, Brannan LE, Carman RJ, Boone JH. Rapid change of fecal microbiome and disappearance of Clostridium difficile in a colonized infant after transition from breast milk to cow milk. Microbiome. 2016;4:53. doi: 10.1186/s40168-016-0198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bridgman SL, et al. High fecal IgA is associated with reduced Clostridium difficile colonization in infants. Microbes Infect. 2016;18:543–549. doi: 10.1016/j.micinf.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 50.Lees EA, Miyajima F, Pirmohamed M, Carrol ED. The role of Clostridium difficile in the paediatric and neonatal gut - a narrative review. Eur J Clin Microbiol Infect Dis. 2016;35:1047–1057. doi: 10.1007/s10096-016-2639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zoetendal EG, et al. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat Protoc. 2006;1:870–873. doi: 10.1038/nprot.2006.142. [DOI] [PubMed] [Google Scholar]
- 52.Bron PA, Meijer M, Bongers RS, de Vos WM, Kleerebezem M. Dynamics of competitive population abundance of Lactobacillus plantarum ivi gene mutants in faecal samples after passage through the gastrointestinal tract of mice. J Appl Microbiol. 2007;103:1424–1434. doi: 10.1111/j.1365-2672.2007.03376.x. [DOI] [PubMed] [Google Scholar]
- 53.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haas BJ, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.ter Braak, C. & Šmilauer, P. Canoco reference manual and user’s guide: software for ordination (version 5.0). (Microcomputer Power, 2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


