Abstract
Chiari malformation type I (CMI) is a congenital abnormality of the cranio-cerebral junction with an estimated incidence of 1 in 1280. CMI is characterized by underdevelopment of the occipital bone and posterior fossa (PF) and consequent cerebellar tonsil herniation. The presence for a genetic basis to CMI is supported by many lines of evidence. The cellular and molecular mechanisms leading to CM1 are poorly understood. The occipital bone formation is dependent on complex interactions between genes and molecules with pathologies resulting from disruption of this delicate process. Whole-exome sequencing of affected and not affected individuals from two Italian families with non-isolated CMI was undertaken. Single-nucleotide and short insertion–deletion variants were prioritized using KGGSeq knowledge-based platform. We identified three heterozygous missense variants: DKK1 c.121G>A (p.(A41T)) in the first family, and the LRP4 c.2552C>G (p.(T851R)) and BMP1 c.941G>A (p.(R314H)) in the second family. The variants were located at highly conserved residues, segregated with the disease, but they were not observed in 100 unaffected in-house controls. DKK1 encodes for a potent soluble WNT inhibitor that binds to LRP5 and LRP6, and is itself regulated by bone morphogenetic proteins (BMPs). DKK1 is required for embryonic head development and patterning. LRP4 is a novel osteoblast expressed receptor for DKK1 and a WNT and BMP 4 pathways integrator. Screening of DKK1 in a cohort of 65 CMI sporadic patients identified another missense variant, the c.359G>T (p.(R120L)), in two unrelated patients. These findings implicated the WNT signaling in the correct development of the cranial mesenchyme originating the PF.
Introduction
Chiari malformation type 1 (CMI) (OMIM 118420) is currently defined as a downward herniation of the cerebellar tonsils across the foramen magnum of at least 5 mm.1 The prevalence of CMI is estimated to be in the 1/1280.2 CMI causes neurological dysfunction by direct compression of the neural tissue at the cranio-vertebral junction. One of the more severe conditions concurrent with CMI is syringomyelia (SM), an abnormal collection of cerebrospinal fluid (CSF) in a fluid-filled sac within the spinal cord or brain stem, reported with an incidence rate as high as 70–80%.3 Although acquired CMI has been reported to occur after trauma, existing cranio-cervical tumors, many cases are suspected to be congenital. The pathogenesis of CMI is incompletely understood. Although multiple mechanisms have been proposed, ‘classical’ CMI is thought to be caused by a mesodermal defect involving sclerotomes implicated in the development of the occipital bone, resulting in a posterior fossa (PF), which is too small and shallow to accommodate the normal sized cerebellum.4, 5 A genetic contribution in at least a subset of CMI cases comes from at least three sources: familial aggregation, twin studies and co-segregation with known genetic conditions.6, 7 Most likely, the pattern of inheritance of CMI is oligogenic with variable penetrance.8, 9 The association between CMI and craniosynostosis, the premature fusion of calvarial sutures, has been recognized for several decades.10, 11 CMI is a frequent finding in syndromic and multisuture craniosynostosis, characterized by early fusion of lambdoid sutures and cranial base synchondroses, such as Crouzon’s syndrome.12 Cases of non-syndromic single-suture craniosynostosis such as scaphocephaly (premature closure of sagittal suture) associated with CMI seems to be less frequent.10, 13 As proposed by Pouratian et al,14 the mechanism for the association of single-suture sagittal synostosis and CMI may be a craniocephalic disproportion resulting from an increased head circumference or an unrecognized underlying common genetic alteration.
The vertebrate cranial base is a complex structure composed of bone, cartilage and other connective tissues. During development, the cranial base first appears as a sheet of undifferentiated mesoderm-derived mesenchymal cells that condense and chondrify to form the chondrocranium.15 The orchestration of the developing romboencephalon and cranio-cervical junction is critical for the proper neuro–osseus relationships. Alteration of these processes can result in hindbrain herniation.
Here we present the results of the exome sequencing of two Italian families in which non-isolated CMI segregated. Exome analysis identified rare missense variants in two components, LRP4 and DKK1, of the canonical WNT signaling pathway that is essential for cranial bone patterning and development. Sequencing analysis of DKK1 in a cohort of 65 sporadic CMI patients identified another very rare missense variant demonstrating that DKK1 may have a role also in a minority of isolated CMI cases.
Methods
Subjects
We ascertained CMI families at the Neurosurgery Clinic of the Giannina Gaslini Children’s Hospital, Genova, Italy. Diagnosis of CMI was based on MRI demonstration of downward herniation >5 mm of the cerebellar tonsils on a mid-sagittal T1-weighted image in the presence of signs or symptoms indicating neural compression at the cranio-vertebral junction, syringo-hydromyelia, cerebellar dysfunction or intracranial hypertension. Detailed morphometric measurements of the MRI records, as well as blood samples were obtained with written informed consent from all participating patients. This study was approved by the local ethical committee. A total of 2 familial cases and 65 sporadic forms of isolated CMI were enrolled for the study. For controls, we recruited 100 age-matched individuals who were subjected to MRI at the Giannina Gaslini Hospital for medical conditions other than malformations of PF. MRI records will be examined for confirmation of affected and unaffected status by a neuroradiologist. Clinical characteristics of sporadic cases are presented in the Supplementary Table 1.
Analysis of posterior cranial fossa volume (PCFV)
PCFV was determined in the two twins of pedigree #VED and in the son of #DRN pedigree and compared with those of 29 pediatric control subjects matched for age and sex. Controls were randomly selected from the Department of Radiology database at Giannina Gaslini Hospital and were selected if they had available head MRI and had no history of intracranial abnormalities. Details on the lengths measured on the mid-sagittal T1-weighted images taken in consideration for PCFV determination are presented in the Supplementary Table 2. PCFV was calculated using the software ITK-SNAP3.4.0 (http://www.itksnap.org/pmwiki/pmwiki.php), a tool to segment structures in 3D medical images. The statistical analysis was performed using one-way ANOVA and commercial statistical software (SPSS, SPSS Inc., Chicago, IL, USA). A probability value of <0.05 was considered statistically significant.
Whole-exome sequencing
Peripheral blood genomic DNA samples of each of member (affected and not affected) of the families were prepared for whole-exome sequencing (WES). Exome capture and sequence library construction were performed using the SureSelect Human All Exon V4/V5 (Agilent Technologies, Santa Clara, CA, USA) and the Paired-End Multiplexed Sequencing library kit (Illumina Inc., San Diego, CA, USA). The captured DNA was sequenced with an Illumina HiSeq 2000 (2 × 100 bp, three samples per lane) at the McGill University and Genome Quebec Innovation Center (Montréal, Canada). Genome analysis Toolkit (GATK) (Broad Institute, Cambridge, MA, USA) (v.3.2-2, Broad Institute, Cambridge, MA, USA) haplotype caller was used to recalibrate the alignments and to call single-nucleotide variants (SNVs) and short insertion–deletion (Indel) variants.16, 17 We annotated and prioritized the SNVs and Indel using a software known as a knowledge-based mining platform for Genomic and Genetic studies using Sequence data (KGGSeq) (http://grass.cgs.hku.hk/limx/kggseq/). This procedure combined a series of functions and valuable resources at three different levels (genetic level, variant-gene level and knowledge level) to filter and highlight variants from exome sequencing data.18 Biological knowledge (gene expression, functional annotations, protein–protein interactions, clinical/knockout phenotypes relevant to the diseases) was used to fish out remaining variants. We then further prioritized variants on the basis of PPI with genes involved in human cranio-facial development (MSX1/MSX2, FGFR1/FGFR2/FGFR3, WNT3, TWIST1, SIX3, DLX3).19 Sanger sequencing was used to validate the candidate SNVs selected, their segregation in the families and also to sequence the entire coding region of DKK1 gene. All primers were designed using Primer3 online tool (v. 0.4.0; Whitehead Institute for Biomedical Research, Cambridge, MA, USA). PCR products were purified with ExoSAP-it (Termo Fisher Scientific, Waltham, MA, USA), sequenced on both strands using Big Dye Terminator Cycle Sequencing Kit v3.1 Kit (Applied Biosystems, Foster City, CA, USA) and resolved on an automated sequencer (ABI 3130xl Genetic Analyzer, Applied Biosystems). Primer sets used for the amplification are presented in Supplementary Table S3. Candidate SNVs were evaluated using a custom TaqMan genotyping assay (Life Technologies, Carlsbad, CA, USA) in a cohort of 100 in-house controls. We have submitted data for the variants to the gene variant database at http://databases.lovd.nl (patient IDs: 00100621; 00100647; 00100655; 00100656).
Results
Clinical findings
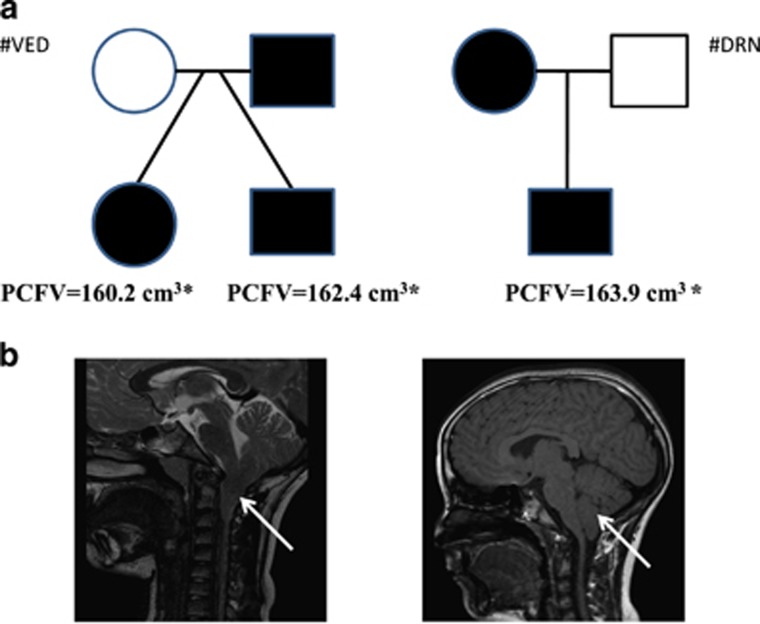
Index cases of #VED family (Figure 1a) were two 15 years old dizygotic twins, born from non-consanguineous parents. Family history was unremarkable with the exception of the paternal uncle suffering from chronic headache. The female presented at 8 years of age with headache, emesis, nystagmus and strabismus. MRI scan showed a CMI with cervico-medullary kinking (bulbar variant of CMI) with no SM, basilar invagination and a mild scaphocephaly (Figure 1b). Subsequently, she underwent to PF decompression. Two years after her surgery, she presented with headache and congestion of retinal vessels. The twin brother presented with a severe scaphocephaly and although he had no neurological symptoms, a brain MRI scan was performed because of the findings in his sister. It revealed a CMI abnormality with caudal displacement of the cerebellar tonsils and SM. He underwent surgical decompression treatment at another institution. MRI scans of the twins’ parents, showed a reduced PF volume in the father, although there was no signs of caudal displacement of the cerebellar tonsils below the foramen magnum.
Figure 1.
Pedigrees of the two Italian families. (a) Squares denote males and circles denote females. Filled symbols denote individuals with confirmed affected status. Exomes of all the individuals were sequenced. Volume calculated using the software ITK-SNAP3.4.0 (http://www.itksnap.org/pmwiki/pmwiki.php). The mean value in pediatric controls was 173.5 (166.8–187.9) cm3. *The value was found statistically lower respect to controls. (b) T2/TSE sagittal MR image of the affected daughter of #VED pedigree (left) and T1/TSSE sagittal MR image of affected son of #DRN pedigree (right) showing pathologic tonsillar ectopia (arrow) through the foramen magnum before surgical decompression.
In the #DRN family (Figure 1a), both the mother and the son were affected by CMI. The son was a 12 years old who came to the attention of the clinics for neck backache after a tonsillectomy. He presented with facial dysmorphisms as prominent head, hypertelorism, epicanthus, clinodactyly and psychomotor delay. The mother was also affected by CMI. MRI study of the son revealed the presence of CMI malformation, a reduced PF volume and abnormal CSF flow (Figure 1b).
Morphometric analysis revealed that both twins of #VED pedigree and the son of #DRN pedigree had a PCFV smaller than age- and sex-matched pediatric controls (P<0.05) suggesting underdevelopment of the PF in these patients (Figure 1a).
Whole-exome sequencing
For the #VED pedigree, we first evaluated the possibility that the disorder could be caused by known craniosynostosis genes, but we failed to identify any mutations. Given the affected status of both twins and the small PF of the father, we considered the autosomal dominant as the most likely inheritance model, even if the recessive homozygous and the recessive compound heterozygous were also taken in consideration. However, these two latter models did not identify any candidate variants. For #DRN given, the affected status of the mother, we considered only the autosomal dominant model. Using this model, the filtered data were narrowed to eight SNVs of biologically plausible genes in the two families. All the candidate variants were missense variants (Table 1).
Table 1. Finally identified variants in CMI families.
| Gene | Gene functiona | Variant | SIFTb | Polyphen-2c | Mutation Tasterd | Expressione | Animal models |
|---|---|---|---|---|---|---|---|
| Family #VED | |||||||
| FBN3 | Extracellular matrix constituent | c.143G>A:p.(R48Q) | D | PD | P | Connective tissue | — |
| ITGA10 | Glycoprotein involved in cell adhesion | c.1681C>G:p.(Q561E) | T | B | DC | Lung, heart, chondrocytes | Mild chondrodysplasiaf |
| EP300 | Transcriptional co-activator | c.7168G>A:p.(A2390T) | T | PD | DC | Bone marrow | — |
| DKK1 | Inhibitor of the WNT pathway | c.121G>A:p.(A41T) | D | PD | DC | Chondrocytes, osteocytes | Lethal, absence of the head, digits anomaliesg |
| MATN3 | Cartilage matrix component | c.526G>T:p.(V176L) | T | B | P | Primitive streak, chondrocytes | Abnormalities of the knee and joint cartilagesh |
| Family #DRN | |||||||
| LRP4 | Inhibitor of the WNT pathway | c.2552C>G:p.(T851R) | D | B | D | Osteoblasts, osteocytes | Bone and cranium anomaliesi |
| EXT2 | Glycosyltransferases involved in the synthesis of heparan sulfate | c.1178G>A: p.(R393K) | T | B | D | Ubiquitous | Ectopic bone growthj |
| BMP1 | Peptidase capable to induce cartilage formation | c.941G>A:p.(R314H) | D | D | D | Ubiquitous | Lethal, failure of the closure of body wallk |
Abbreviations: B, benign; D, damaging; DC, disease causing; PD, probably damaging; T, tolerated.
The DNA mutation and amino-acid numbering systems are based on the following cDNA sequences accession numbers: FBN3 NM_032447; ITGA10 NM_003637; EP300 NM_001429; DKK1 NM_012242; MATN3 NM_002381; LRP4 NM_002334, EXT2 NM_000401; BMP1 NM_001199 and protein sequences accession numbers: DKK1 NP_036374: FBN3 NP_115823; ITGA10 NP_003628; EP300 NP_001420; MATN3 NP_002372; LRP4 NP_002325 EXT2 NP997005; BMP1 NP_001190.
According to GeneCards Database (http://www.genercards.org).
SIFT, the Sorting Intolerant from Tolerant algorithm (http://www.jcvi.org/).
Polyphen-2, polymorphism phenotyping 2 (http://genetics.bwh.harvard.edu/).
According to GeneCards Database.
Bengtsson et al.45
Mukhopadhyay et al.24
Yang et al.46
Johnson et al.29
Stickens et al.47
Suzuki et al.33
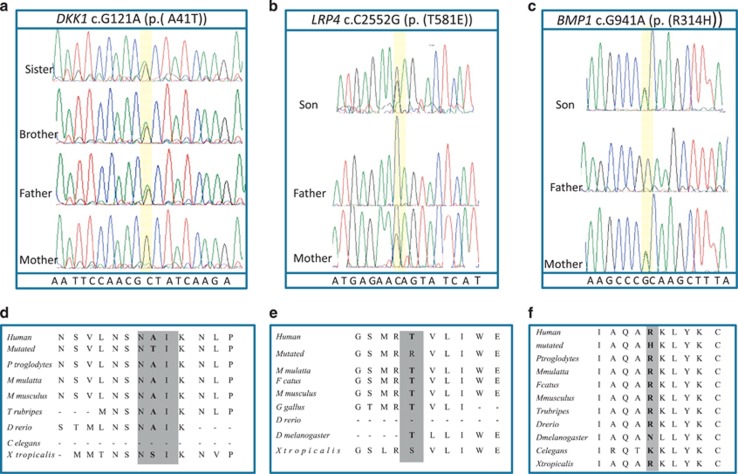
We conducted validation for these eight potential candidate variants in all family members using standard DNA dye terminator cycle sequencing and in 100 unrelated healthy controls using TaqMan genotyping assay (Life Technologies). The FBN3 c.143G>A (p.(R48Q)) was present in 2 of the 100 controls and was thus considered a polymorphism. The EP300 c.7168G>A (p.(A2390T)) SNV was not validated by direct sequencing and was thus categorized as calling error. The DKK1 c. 121G>A (p.(A41T)), LRP4 c.2552C>G (p.(T851R)) and BMP1 c.941G>A (p.(R314H)) that segregated in all affected individuals and were absent in the controls remained the most likely candidate genes (Figure 2). The ITGA10, MATN3, EXT2 SNVs, although perfectly segregating with the disease phenotype, were predicted to affect the protein function only by one out three in silico tools (SIFT, Polyphen-2 and Mutation Taster) (Table 1) and, thus, they were considered less relevant.
Figure 2.
Family segregation of the DKK1, BMP1 and LRP4 SNVs. Sanger sequencing chromatograms showing the segregation of the heterozygous (a) DKK1 c.121G>A variant of the pedigree #VED, (b) LRP4 c.2552CC>G and (c) BMP1 c.941G>A in the pedigree #DRN. Multiple sequence alignment showing conservation of protein sequence across species of human DKK1 (d) and LRP4 (e) BMP1 (f). The NAI motif of DKK1 protein is gray highlighted. The DNA variants and amino-acid numbering systems are based on the following cDNA sequences accession numbers: DKK1 NM_012242; LRP4 NM_002334, BMP1 NM_001199 and protein sequences accession numbers: DKK1 NP_036374: LRP4 NP_002325; BMP1 NP_001190.
DKK1 is a 29 kDa protein belonging to the Dickkopf (DKK) family.20 DKK1 is involved through its inhibition of the WNT signaling pathway in the vertebrate embryogenesis including head induction, skeletal development and limb patterning.20 In the adult, DKK1 is implicated in bone formation, arthritis and cancer.21 DKK1 encodes for a secreted protein that is a high-affinity ligand for LRP6.22 DKK1 blocks Lrp6-mediated Wnt/beta-catenin signaling by interacting with domains that are distinct from those required for Wnt/frizzled interaction.23 (Figure 3b). Dkk1 was identified in a screen for head inducers in Xenopus.20 Overexpression of Dkk1 in early Xenopus blastomeres led to embryos with enlarged anterior head structures, whereas injection of antibody to Dkk1 lead to microcephaly.23 DKK1 mutant mice die at the birth, exhibiting absence of cephalic neural crest-derived head structures and forelimb malformations ranging from fusion of the distal-most limb elements to the appearance of ectopic preaxial and postaxial digits.24 DKK1 c.121G>A (Figure 2a) variant was absent in the 1000 Genomes Project and was reported at very low frequency (MAF: 0.001-0.003) in other public databases. Although the DKK1 p.A41T substitution falls in a functionally uncharacterized portion of the protein, it lies in a conserved amino-acid motif, NAIKN (amino acids 40–44) that is present in multiple Dkk proteins and is conserved among species (Figure 3b).22 A similar motif appears in two other proteins regulating Wnt signaling, namely SOST25 and WISE.26 The core motif would appear to be ‘NXI,’ where ‘X’ is a small residue (Ala, Ser) or Trp. The ‘NXI’ motif is important for binding of DKK1 to LRP5/6/4 receptors. All in silico tools consistently predicted a deleterious effect of the DKK1 c. 121G>A (Table 1).
Figure 3.
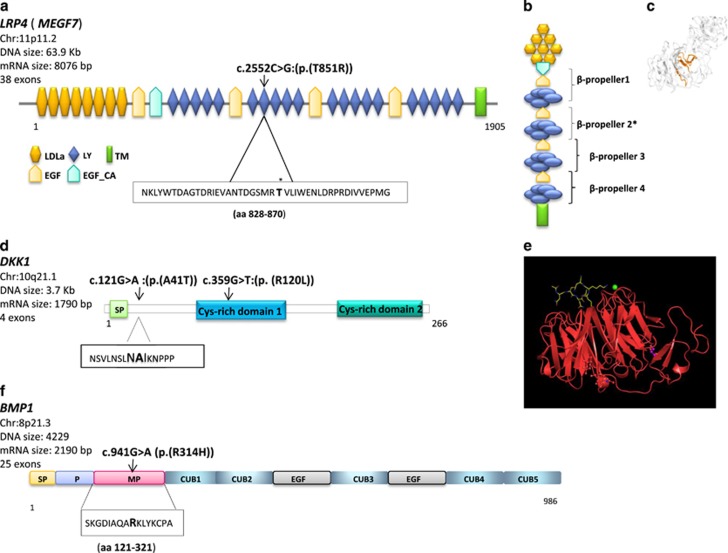
Localization of the LRP4, DKK1 and BMP1 SNVs. (a) Schematic representation of the domains of the full-length LRP4 protein. LRP4 harbors 8 low-density lipoprotein receptor (LDLR) domain class A (LDLa), 4 epidermal growth factor (EGF)-like domains, a calcium-binding EGF-like domain (EGF-Ca), 20 LDLR class B repeat (LY), a transmembrane domain (TM) and an intracellular domain. The LRP4 c.2552C>G affected the seventh LY repeat. (b) The LDLR class B repeat contains five tandem repeats of an YWTD motif to build a β-propeller structure protein. The LRP4 c.2552C>G affected the second β-propeller domain*. (c) Crystal structure of aa 743–1348 of LRP4. The seventh LY domain is shown in orange. The 3D structure is from (http://www.proteinmodelportal.org/). (d) Representation of the domains of DKK1 protein. The DKK1 c.121G>A (p.(A41T)) affects the NAIKN motif (amino acids 40–44) that is present in multiple DKK proteins and is conserved among species. The ‘NXI’ core is important for binding of DKK1 to LRP5/6/4 receptors. SP: signal peptide. The DKK1 c.359G>T affected the Cys-rich 1 domain. (e) Crystal structure showing the interaction of E1 (first β-propeller and following EGF-like domain; Uniprot 075581, aa 20–326, red) of LRP6 with a DKK1 peptide containing the NAI motif (Uniprot 094907, aa 38–44, yellow). Structure deposited into Protein Data Bank under accession codes 3SOQ (LRP6 E1/DKK1 peptide complex) by Bourhis et al.22 (f) Representations of BMP1 domains. BMP1 harbors a signal peptide (SP), a pro-domain (P) that needs to be removed, an astacin-like metallo proteinase domain, five CUB (Complement Uegf BMP1) domains, two EGF-like domains. The BMP1 c.941G>A affected the metallo proteinase domain. The DNA variants and amino-acid numbering systems are based on the following cDNA sequences accession numbers: DKK1 NM_012242; LRP4 NM_002334, BMP1 NM_001199 and protein sequences accession numbers: DKK1 NP_036374: LRP4 NP_002325; BMP1 NP_001190.
LRP4 (LDL receptor-related protein 4; MEGF7) belongs to the low-density lipoprotein receptor gene family, whose members are involved as signal transducers or modulators of several fundamental pathways, including bone morphogenetic protein (BMP), transforming growth factor β (TGF-β), PDGF and canonical Wnt signaling.27 LRP4 reduces Wnt signaling during bone development.28 Lrp4 binds the two secreted Wnt modulators Dkk1 and sclerostin, and is expressed by murine bone, specifically by osteoblasts.27 Mice carrying a functionally hypomorphic Lrp4 mutation have impaired bone growth and increased bone turnover, fusion of digits at the hind and fore limbs (polysyndactyly).29 LRP4 extracellular domain contains a signal peptide, eight LDLa domains (class A repeats), four β-propeller domains (class B repeats, or YWTD domains), six EGF-like domains and a domain for O-linked oligosaccharide modification. After a transmembrane domain, there is an intracellular domain containing an NPxY motif and a PDZ-interacting motif at the C-terminus (Figure 3a). The variant c.2552C>G affected the LDL receptor class B repeat 7 that is part of third β-propeller domain (Figures 2b and 3a). The LRP4 c.2552C>G variant was never reported and it is a patient-specific mutation. It is predicted to be damaging by two out of three in silico tools (Table 1).
BMP1 encodes a protein that is capable of inducing formation of cartilage in vivo.30 Although other BMPs are members of the TGF-β superfamily, this gene encodes a protein that is not closely related to other known growth factors. It cleaves the C-terminal propeptides of procollagen I, II and III.31 It may participate in dorso-ventral patterning during early development by cleaving chordin (CHRD). BMP1 is a metallo proteinase with a proteinase domain that is homologous to the crayfish enzyme astacin and a C-terminal domain comprising three CUB domains and one EGF-like domain (Figure 3c).32 BMP1 c.941G>A variant affects the metallo protease domain. This protein has been suggested to have essential roles in osteogenesis. Bmp1 homozygous knockout mice are perinatal lethal but do not show gross skeletal abnormalities except for reduced ossification of some skull bones.33 In human, mutations of BMP1 cause autosomal recessive form of osteogenesis imperfecta, a connective tissue disorder characterized by low bone mass, bone fragility and susceptibility to fractures after minimal trauma.34
Additional DKK1 variants in a cohort of CM1 sporadic patients
Given that DKK1 seemed to be the most strong candidate gene, we performed mutational analysis by Sanger sequencing of a cohort of 65 sporadic CMI cases and 100 controls.
Table 2 shows the results of the mutational analysis of DKK1. Notably, we could identify a missense variant in exon two of DKK1, the c.359G>T (p.(R120L)), (Supplementary Figure 1) in two sporadic CMI patients. Besides to CMI, one of the two patients also presented with SM (Supplementary Table 4). The c.359G>T was a de novo variant because it was not present in the parents and it is a patient-specific mutation because it was not found in controls subjects. The variant was identified at a very low frequency in public databases and it was reported in a patient with childhood-onset primary osteoporosis and in his affected sisters but not in the father who was also affected.35 Moreover, in silico tools predicted that the variants could have functional impact on the protein function because it affected in a very conserved sequence (the Cys-rich domain1) (Supplementary Figure 1). DKK1 c.359G>T could explain only a minority (3% 2/65) of the sporadic CMI cases.
Table 2. Genetic variants identified in exonic and flanking intronic regions of DKK1 in sporadic CMI patients.
| Mutation | AA change | Exon | Domain | Polyphena | SIFTb | Mutation Tasterc | PT/CT (65/100) |
|---|---|---|---|---|---|---|---|
| Variants absent in controls | |||||||
| c.359G>T | p.(R120L) | 2 | Cys-1 domain | PD | D | D | 2/0 |
| Variants present in controls | |||||||
| c.318A>G | p.(A106=) | 2 | Cys-1 domain | — | — | — | 21/32 |
| c.548-43A>G | — | Intron 3 | — | — | — | — | — |
Abbreviations: AA, amino acid; Polyphen, polymorphism phenotyping software; SIFT, sorting intolerant from tolerant software.
The DNA mutation and amino-acid numbering systems are based on DKK1 genomic sequence AH009834, DKK1 cDNA sequence NM_012242.2 and DKK1 protein sequence NP_036374.1.
Polyphen predictions for amino-acid changes: ‘PD’: probably damaging.
SIFT predictions for amino-acid changes: ‘D’: deleterious.
Mutation taster prediction: ‘D’: disease causing.
Discussion
In this study, we investigated the genetic causes of non-isolated CMI in two pedigrees by WES experiments. Volumetric analysis from preoperative MRI images of pediatric patients of the two families confirmed a smaller PF volume with respect to the age-related control group.
We identified two rare missense variations in two components of the canonical WNT signaling pathway, LRP4 and DKK1. The DKK1 c.121G>A (p.(A41T)) and LRP4 c.2552C>G (p.(T851R)) variants co-segregated completely with the disease, affected highly conserved amino acids and were predicted to be damaging or deleterious. A second missense change of DKK1, the c.359G>T (p.(R120L)), was found in another two independent sporadic cases of CMI.
A mesodermal defect involving sclerotomes implicated in the development of the occipital bone has been documented as a possible cause of CMI, resulting in an underdeveloped and small PF.4, 5 According to this hypothesis, the primary disease mechanism involves a compromised PF and tonsillar herniation occurs secondarily. Consistent with a genetic hypothesis for CMI, much of the PF morphology was found to be heritable.36 The occipital bone formation is dependent on complex interacts between genes and molecules with pathologies resulting from disruption of this delicate process. Speer et al9 postulated that an underlying gene responsible for CMI may have pleiotropic effects that influence PF volume, skull bone abnormalities, the extent of cerebellar tonsil herniation and the formation of SM. Mammalian skull is formed from cranial skeletogenic mesenchyme, which is derived from mesoderm. BMPs, TGF-β and WNTs are required in early stages of cranial bone development in the mouse and chick embryo.37
A large amount of data obtained in Xenopus, zebrafish, chick and mouse has provided evidence that the Wnt signaling is critical for proper development of the head and face, having important roles in various aspects of cranio-facial development ranging from axis formation to survival of cranial neural crest cells to patterning of the brain.37, 38 In this study, we presented the first association between DKK1 genetic variants and a human condition. The DKK1 c.121G>A (p.(A41T)) affects a pentapeptide motif (NAIKN; amino acids 40–44) conserved in all Wnt inhibitors and among species that is crucial for binding with LRPs. The DKK1 c.359G>T (p.(R120L)) variant is located on N-terminal cysteine-rich domain (Dkk-N) that, as reported by Lintern et al,39 does not affect Wnt signaling.
LRP4 encodes for a multifunctional member of the low-density lipoprotein receptor family. LRP4 has been proposed to function as an integrator of Bmp and Wnt signaling. Presence (or overexpression) of Lrp4 inhibits canonical Wnt signal. Variants in the LRP4 gene have been found to cause several allelic conditions both in humans and animals. LPR4 variants are implicated in neurological disorders including congenital myasthenic syndrome, myasthenia gravis, and diseases of kidney or bone (Supplementary Figure 2).40 Among these, two are characterized by a strong skeletal involvement, namely sclerosteosis and Cenani–Lenz syndrome (CLS). Mutations of LRP4 in CLS patients are located in the extracellular domain of the protein, in particular, in LDLa domain, as well as in the first β-propeller and third β-propeller domains.41 Apart the LRP4 p.L935P and the p.K801T* identified in two CLS cases, the LRP4 second β-propeller domain has few variants. The LRP4 c.2552C>G (p.(T851R)) identified in this study in a familial CMI patient affected the repeat 7 of the second β-propeller domain and lies in a low-rate region of LRP4 protein (Supplementary Figure 2). Our findings expand the clinical spectrum of LRP4-associated phenotypes because there is no signs of abnormal bone density and renal anomalies or neurological signs in patients harboring LRP4 variants. The only minimal bone anomaly of the proband of #DRN family, who was carrier of the LRP4 c.2552C>G, was the clinodactyly of the fifth digit. The boy also presented with prominent forehead and hypertelorism that are dysmorphic signs found in some patients with CLS. However, the boy has no the typical features of CLS such as syndactyly or polydactyly. Conversely, CLS is an autosomal recessive condition.
The twins of #VED family presented both uncorrected isolated craniosynostosis, but we did not find genetic variants in any of the three FGF receptor genes (FGFR1, FGFR2 and FGFR3) or in the transcription factor TWIST that account for many cases of craniosynostosis.19 Isolated craniosynostosis and CMI is a common association. Sgouros et al42 showed that cranial base growth is also altered in both multi- and single-suture craniosynostosis. We proposed that in this family, the brain malformation is associated with craniosynostosis as the result of a common genetic abnormality. It has been demonstrated by others that active canonical Wnt signaling is at least in part responsible for the patency of cranial sutures. Low levels of this signaling are ‘permissive’ for craniosynostosis.43 In the #VED kindred, the father has small PF but no CMI. Tonsillar herniation that is the gold standard by which individuals are diagnosed with CMI, was not found to be heritable lending further support to the hypothesis that may not be the best criterion for diagnosis as it likely occurs secondarily, does not correlate well with symptoms, and may not be necessary to cause symptoms.
Modifier genes and/or environmental factors have a major role in disease expression. The proband of #DRN pedigree was also carrier of the BMP1 c.941G>A variant. An extracellular protein, Wise, which binds BMP ligands and LRP4, acts to link BMP and WNT signaling.26 Thus, BMP1 signaling impairment could modulate the phenotype induced by LRP4 variants. We speculate that CMI, having both phenotypic and genetic heterogeneity, has oligogenic inheritance and is caused by more than one mutations in genes involved in cranio-facial development that is a complex process.
Even if we could not exclude a pleiotropic effect of the two major genes, LRP4 and DKK1, we speculated that other variants segregating with the disease (FBN3 c.143G>A, ITGA10 c.1681C>G, MATN3 c.526G>T in the pedigree #VED and ETX2 c.1178G>A in the pedigree #DRN) with a minor impact on the protein function may cooperate with mutations with major impact to modulate the clinical spectrum of the disease.
In conclusion, exome sequencing of families with non-isolated CMI has implicated inhibitors of WNT signaling is of crucial importance for the normal structural integrity of the developing cranial PF. We enlarged the clinical spectrum of conditions associated to LRP4 human mutations and provided evidence of the role of DKK1 in head morphogenesis in humans as in animal models.
Improved understanding of the pathogenesis of head and facial birth defects could offer new and exciting possibilities for their prevention and therapy. Give the critical roles of Wnt signaling in both embryonic and postnatal bone formation and homeostasis, modulating this pathway is widely recognized as an important area of therapeutic development for many bone diseases. In this light, it is conceivable that therapeutic agents specifically targeting the WNT pathway may be used to stimulate osteogenic differentiation in patients with cranio-facial birth defects. For example, Dkk1-induced inhibition of WNT signaling is required for the reentry into the cell cycle of human adult mesenchymal stem cells (MSC) from bone marrow.44 Thus, treatment of patient-derived adult MSC with small-molecule WNT inhibitors could represent a useful means of stimulating expansion of hMSCs in vivo and thereby enhancing their tendency to differentiate in osteoblasts under appropriate stimuli.
Acknowledgments
We thank the families who agreed to participate to the research study. PDM is funded by Fondazione Gerolamo Gaslini and Trust Aletti. EM was funded by Trust Aletti. AIMA CHILD (Associazione Italiana Malformazione di Chiari Child) funded the project.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Barkovich AJ, Wippold FJ, Sherman JL, Citrin CM: Significance of cerebellar tonsillar position on MR. Am J Neuroradiol 1986; 7: 795–799. [PMC free article] [PubMed] [Google Scholar]
- Meadows J, Kraut M, Guarnieri M, Haroun RI, Carson BS: Asymptomatic Chiari type I malformations identified on magnetic resonance imaging. J Neurosurg 2000; 92: 920–926. [DOI] [PubMed] [Google Scholar]
- Milhorat TH, Chou MW, Trinidad EM et al: Chiari I malformation redefined: clinical and radiographic findings for 364 symptomatic patients. Neurosurgery 1999; 44: 1005–1017. [DOI] [PubMed] [Google Scholar]
- Nishikawa M, Sakamoto H, Hakuba A, Nakanishi N, Inoue Y: Pathogenesis of Chiari malformation: a morphometric study of the posterior cranial fossa. J Neurosurg 1997; 86: 40–47. [DOI] [PubMed] [Google Scholar]
- Marin-Padilla M, Marin-Padilla TM: Morphogenesis of experimentally induced Arnold–Chiari malformation. J Neurol Sci 1981; 50: 29–55. [DOI] [PubMed] [Google Scholar]
- Stovner LJ: Headache and Chiari type I malformation: occurrence in female monozygotic twins and first-degree relatives. Cephalalgia 1992; 12: 304–307. [DOI] [PubMed] [Google Scholar]
- Herman MD, Cheek WR, Storrs BB: Two siblings with the Chiari 1 malformation. Pediatr Neurosurg 1990; 16: 183–184. [DOI] [PubMed] [Google Scholar]
- Speer MC, Enterline DS, Mehltretter L et al: Chiari type I malformation with or without syringomyelia: prevalence and genetics. J Genet Couns 2003; 12: 297–311. [DOI] [PubMed] [Google Scholar]
- Speer MC, George TM, Enterline DS, Franklin A, Wolpert CM, Milhorat TH: A genetic hypothesis for chiari i malformation with or without syringomyelia. Neurosurg Focus 2000; 8: E12. [DOI] [PubMed] [Google Scholar]
- Saldino RM, Steinbach HL, Epstein CJ: Familial acrocephalosyndactyly (Pfeiffer syndrome). Am J Roentgenol Radium Ther Nucl Med 1972; 116: 609–622. [DOI] [PubMed] [Google Scholar]
- Strahle J, Muraszko KM, Buchman SR, Kapurch J, Garton HJL, Maher CO: Chiari malformation associated with craniosynostosis. Neurosurg Focus 2011; 31: E2. [DOI] [PubMed] [Google Scholar]
- Cinalli G, Renier D, Sebag G, Sainte-Rose C, Arnaud E, Pierre-Kahn A: Chronic tonsillar herniation in Crouzon’s and Apert’s syndromes: the role of premature synostosis of the lambdoid suture. J Neurosurg 1995; 83: 575–582. [DOI] [PubMed] [Google Scholar]
- Leikola J, Koljonen V, Valanne L, Hukki J: The incidence of Chiari malformation in nonsyndromic, single suture craniosynostosis. Childs Nerv Syst 2010; 26: 771–774. [DOI] [PubMed] [Google Scholar]
- Pouratian N, Sansur CA, Newman SA, Jane Jr JA, Jane JA: Sr: Chiari malformations in patients with uncorrected sagittal synostosis. Surg Neurol 2007; 67: 422–427. [DOI] [PubMed] [Google Scholar]
- McBratney-Owen B, Iseki S, Bamforth SD, Olsen BR, Morriss-Kay GM: Development and tissue origins of the mammalian cranial base. Dev Biol 2008; 322: 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E et al: The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MX, Gui HS, Kwan JS, Bao SY, Sham PC: A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res 2012; 40: e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie AOM, Morriss-Kay GM: Genetics of craniofacial development and malformation. Nat Rev Genet 2001; 2: 458–468. [DOI] [PubMed] [Google Scholar]
- Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C: Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 1998; 391: 357–362. [DOI] [PubMed] [Google Scholar]
- Pinzone JJ, Hall BM, Thudi NK et al: The role of Dickkopf-1 in bone development, homeostasis, and disease. Blood 2009; 113: 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhis E, Wang W, Tam C et al: Wnt antagonists bind through a short peptide to the first b-propeller domain of LRP5/6. Structure 2011; 19: 1433–1442. [DOI] [PubMed] [Google Scholar]
- Li Y, Lu W, King TD et al: Dkk1 stabilizes Wnt co-receptor LRP6: implication for Wnt ligand-induced LRP6 downregulation. PLoS One 2010; 5: e11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay M, Shtrom S, Rodriguez-Esteban C et al: Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev Cell 2001; 1: 423–434. [DOI] [PubMed] [Google Scholar]
- Semënov M, Tamai K, He X: SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Bio Chem 2005; 280: 26770–26775. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Jones CM, Mercurio S et al: Wise, a context-dependent activator and inhibitor of Wnt signaling. Development 2003; 130: 4295–4305. [DOI] [PubMed] [Google Scholar]
- Choi HY, Dieckmann M, Herz J, Niemeier A: Lrp4, a novel receptor for Dickkopf 1 and sclerostin, is expressed by osteoblasts and regulates bone growth and turnover in vivo. PLoS One 2009; 4: e793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Zheng JJ, Wu D: The structural basis of DKK-mediated inhibition of Wnt/LRP signaling. Sci Signal 2012; 5: pe22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EB, Hammer RE, Herz J: Abnormal development of the apical ectodermal ridge and polysyndactyly in Megf7-deficient mice. Hum Mol Genet 2005; 14: 3523–3538. [DOI] [PubMed] [Google Scholar]
- Wang RN, Green J, Wang Z et al: Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis 2014; 1: 87–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigue-Antar L, Barker C, Kadler KE: Identification of amino acid residues in bone morphogenetic protein-1 important for procollagen C-proteinase activity. J Biol Chem 2001; 276: 26237–26242. [DOI] [PubMed] [Google Scholar]
- Kronenberg D, Vadon-Le Goff S, Bourhis JM et al: Strong cooperativity and loose geometry between CUB domains are the basis for procollagen c-proteinase enhancer activity. J Biol Chem 2009; 284: 33437–33446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Labosky PA, Furuta Y et al: Failure of ventral body wall closure in mouse embryos lacking a procollagen C-proteinase encoded by Bmp1, a mammalian gene related to Drosophila tolloid. Development 1996; 122: 3587–3595. [DOI] [PubMed] [Google Scholar]
- Cho SY, Asharani PV, Kim OH et al: Identification and in vivo functional characterization of novel compound eterozygous BMP1variants in osteogenesis imperfecta. Hum Mutat 2015; 36: 191–195. [DOI] [PubMed] [Google Scholar]
- Korvala J, Löija M, Mäkitie O et al: Rare variations in WNT3A and DKK1 may predispose carriers to primary osteoporosis. Eur J Med Genet 2012; 55: 515–519. [DOI] [PubMed] [Google Scholar]
- Markunas CA, Enterline DS, Dunlap K et al: Genetic evaluation and application of posterior cranial fossa traits as endophenotypes for Chiari type I malformation. Ann Hum Genet 2014; 78: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y, Snider TN: Neural crest cell signaling pathways critical to cranial bone development and pathology. Exp Cell Res 2014; 325: 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani P, Jarrell A, Myers J, Atit R: Visualizing canonical Wnt signaling during mouse craniofacial development. Dev Dyn 2010; 239: 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintern KB, Guidato S, Rowe A, Saldanha JW, Itasaki N: Characterization of wise protein and its molecular mechanism to interact with both Wnt and BMP signals. J Biol Chem 2009; 284: 23159–23168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Xiong W-C, Me L: LRP4 in neuromuscular junction and bone development and diseases. Bone 2015; 80: 101–108. [DOI] [PubMed] [Google Scholar]
- Ohkawara B, Cabrera-Serrano M, Nakata T et al: LRP4 third b-propeller domain mutations cause novel congenital myasthenia by compromising agrin-mediated MuSK signaling in a position-specific manner. Hum Mol Genet 2014; 2: 1856–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgouros S, Hockley AD, Goldin JH, Wake MJ, Natarajan K: Intracranial volume change in craniosynostosis. J Neurosurg 1999; 91: 617–625. [DOI] [PubMed] [Google Scholar]
- Behr B, Longaker MT, Quarto N: Differential activation of canonical Wnt signaling determines cranial sutures fate: a novel mechanism for sagittal suture craniosynostosis. Dev Biol 2010; 344: 922–940. [DOI] [PubMed] [Google Scholar]
- Gregory CA, Singh H, Perry AS, Prockop DJ: The Wnt signaling inhibitor Dkk-1 is required for re-entry into the cell cycle of human adult stem cells from bone marrow (hMSCs). J Biol Chem 2003; 278: 28067–28078. [DOI] [PubMed] [Google Scholar]
- Bengtsson T, Aszodi A, Nicolae C et al: Loss of alpha10beta1 integrin expression leads to moderate dysfunction of growth plate chondrocytes. J Cell Sci 2005; 118: 929–936. [DOI] [PubMed] [Google Scholar]
- Yang X, Trehan SK, Guan Y et al: Matrilin-3 inhibits chondrocyte hypertrophy as a bone morphogenetic protein-2 antagonist. J Biol Chem 2014; 289: 34768–34779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickens D, Zak BM, Rougier N, Esko JD: Mice deficient in Ext2 lack heparan sulfate and develop exostoses. Development, 2005; 132: 5055–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.