Abstract
Moyamoya angiopathy (MMA) is a cerebral angiopathy affecting the terminal part of internal carotid arteries. Its prevalence is 10 times higher in Japan and Korea than in Europe. In East Asian countries, moyamoya is strongly associated to the R4810K variant in the RNF213 gene that encodes for a protein containing a RING-finger and two AAA+ domains. This variant has never been detected in Caucasian MMA patients, but several rare RNF213 variants have been reported in Caucasian cases. Using a collapsing test based on exome data from 68 European MMA probands and 573 ethnically matched controls, we showed a significant association between rare missense RNF213 variants and MMA in European patients (odds ratio (OR)=2.24, 95% confidence interval (CI)=(1.19–4.11), P=0.01). Variants specific to cases had higher pathogenicity predictive scores (median of 24.2 in cases versus 9.4 in controls, P=0.029) and preferentially clustered in a C-terminal hotspot encompassing the RING-finger domain of RNF213 (P<10−3). This association was even stronger when restricting the analysis to childhood-onset and familial cases (OR=4.54, 95% CI=(1.80–11.34), P=1.1 × 10−3). All clinically affected relatives who were genotyped were carriers. However, the need for additional factors to develop MMA is strongly suggested by the fact that only 25% of mutation carrier relatives were clinically affected.
Introduction
The cerebral moyamoya angiopathy (MMA) is characterized by a progressive stenosis of the intracranial internal carotid arteries (ICA) and their bifurcation proximal branches, associated with the development of thin collateral vessels at the base of the brain.1 This rare condition (prevalence close to 6/100 000 in Japan, around ten times less in Europe) leads to ischemic and hemorrhagic stroke and affects both adults and children.2, 3 Its pathophysiology is so far unknown. The term moyamoya disease (MMD) refers to the isolated and idiopathic form of the condition. By contrast, the term moyamoya syndrome (MMS, also called quasi-moyamoya) is used when this angiopathy is associated with an acquired condition (eg, head irradiation) or a broad spectrum of diseases with a genetic component, either polygenic, chromosomal or monogenic. Among those are several Mendelian diseases with a highly variable penetrance of MMA, ranging from <2% in RASopathies to >75% in BRCC3-mutated patients.4
Although sporadic cases are largely preponderant in MMD, the implication of genetic factors has long been suspected, based on the strong ethnicity-related effect and a prevalence of familial cases close to 10–15%.1 The pattern of inheritance in familial MMD cases is unclear and most likely heterogeneous. The number of affected members in multiplex MMD families is usually limited, suggesting an oligogenic or multifactorial inheritance. However, a few large MMD families consistent with an autosomal dominant pattern of inheritance have been reported in Japan.5
RNF213, which encodes an enzyme with a RING finger and AAA+ ATPase domains, is the sole MMD susceptibility gene identified so far.6, 7 A very strong association between a sequence variant in this gene (c.14429G>A, p.(R4810K), rs112735431) and MMD exists in East Asian countries (Japan, Korea and, to a lesser degree, China), where nearly 80% of MMD patients carry at least one mutated allele, as compared to <1.5% in healthy controls with the same ethnic background.6, 7, 8, 9 Recently, this p.(R4810K) variant has also been shown in East Asia to increase the risk to develop MMA in various MMS, including neurofibromatosis type 1, suggesting that it could act as a modifier.10, 11
This p.(R4810K) variant has not been detected in European patients, strongly suggesting a founder-effect in Japan and East-Asia. No major susceptibility variant has been identified to date in Caucasian MMD patients.12 Several rare and non-disruptive RNF213 variants distinct from p.(R4810K) have been reported in MMD patients of European ancestry.6, 13, 14, 15, 16, 17 Some of these rare variants have been tested in controls and showed to be absent or present at a very low frequency. However, in most studies, controls have not been sequenced on the whole gene, precluding the comparison of frequencies of rare variants as a whole in cases and controls. Indeed, rare variant association analysis requires to use specific gene-based statistical approaches, called burden tests or collapsing tests, and not variant based tests.18, 19
In the present study, we aimed to determine whether rare RNF213 variants are associated with an increased risk of MMA in patients of Caucasian ancestry. We used a RNF213 gene burden based strategy comparing a cohort of 68 unrelated MMA probands of European ancestry with a large cohort of ethnically matched controls.
Materials and methods
Cases and controls
Seventy-three unrelated Caucasian MMA probands of self-reported European ancestry were included. They were referred to i) the department of genetics at Lariboisière Hospital (Paris, France), ii) the French national center for rare vascular diseases of the brain and the eye (CERVCO), iii) the French national center for pediatric stroke (Paris, France) or iv) the neurology department at Alfried Krupp Hospital (Essen, Germany), for diagnostic and clinical care of MMD or MMS of unknown etiology. Diagnosis of moyamoya was based on Magnetic Resonance Imaging or cerebral angiography showing a stenosis or an occlusion of the terminal part of the terminal ICA and/or the proximal portions of anterior and/or middle cerebral arteries associated to an abnormal deep vascular network.1 Patients diagnosed with both unilateral and bilateral MMA were included in the study. Clinical data, medical history and radiological findings were extracted from patient medical records or specific databases. Patients were classified as MMS of unknown etiology when, in addition to MMA, they showed dysmorphic or rare malformative features, or when they were affected by an acquired condition of particular severity or of an unusual age at onset. Given the abundant literature data about extracerebral vascular involvement in MMD patients, the sole presence of extracerebral artery steno-occlusion(s) in addition to MMA, was not considered as sufficient for classifying probands as MMS. In addition to these 73 probands, 39 affected or clinically healthy relatives were included. All patients and their participating relatives provided a written informed consent for genetic analysis according to the French and German laws. Genomic DNA was isolated from peripheral blood leukocytes according to standard protocols.
A total of 573 healthy controls, sampled in 6 different regions of France as part of the FREX project, were included. (FREX. The French Exome Project. https://www.france-genomique.org/spip/spip.php?article158.2016).
Whole exome sequencing analysis
Probands’ exomes were captured using Agilent SureSelect Human All Exon V4 (N=1), V5 (N=6) and Clinical Research Exome (N=66) kits (Agilent Technologies, Santa Clara, CA, USA). FREX control exomes were captured using Agilent SureSelect Human All Exon V5+UTR. Patient and FREX exome samples were processed through the same bioinformatics pipeline following GATK 3.3-0 Best Practices recommendations.20 Variants’ annotation was performed using Variant Effect Predictor (http://www.ensembl.org/Tools/VEP). For each detected RNF213 variant, prediction of pathogenicity was assessed using Polyphen2, SIFT and PROVEAN in-silico tools. Combined Annotation Dependent Depletion (CADD) Phred scores were obtained using the VEP plugins/CADD.pm. Cases variants have been deposited in the public ClinVar database under the submission name SGMMA001 (www.ncbi.nlm.nih.gov/clinvar/).
High-quality variants located in exons and in the ±50 bp around each coding exon of RNF213 (MIM 613768), based on NM_001256071.2, which encodes for the longest validated transcript, were extracted. Variants were considered of high-quality if they had a Variant Quality Score VQSLOD above −2 and if genotypes were missing in <5% of the patients or the FREX controls. Only individual genotypes with a genotype quality (GQ) score >20 were retained.
Principal component analysis
Principal component analysis (PCA) was performed on the genotypes of the patients and the controls for 16 670 common SNVs extracted from WES data, in order to ensure that cases and controls were ethnically matched. The top Principal Components (PC) coordinates between cases and controls (Supplementary Figure S1) were used in the association test as adjustment variables.
Statistics
Association between RNF213 variants found in cases and controls was tested using the fixed threshold collapsing method.21 Only missense, stoploss, stopgain, canonical splice site variants and small insertions-deletions (Indel) of RNF213 with a minor allele frequency (MAF) below 0.01 in ExAC non-Finnish European subpopulation were considered in the test. Association was then tested by comparing the number of cases and controls carrying at least one such variant in the RNF213 gene to avoid type-one error inflation due to linkage disequilibrium between variants. To correct for possible population stratification, we used the logistic regression framework and adjusted on the top 5 PCs from the PCA carried on the combined sample of cases and controls (Supplementary Figure S1). The R package glm was used for all computations (http://www.R-project.org/).
The likelihood ratio (LR) statistics proposed by Ionita-Laza et al.22 was used to test the presence of a region within RNF213 that would be significantly enriched in rare coding variants. We used the EBScan program developed by the authors and searched over different region sizes ranging from 10 to 25 kb.23
CADD Phred score of rare coding variants specific to cases and controls were compared using Wilcoxon Mann-Whitney tests.
Sanger sequencing
Standard PCR amplification and Sanger sequencing were used to confirm each RNF213 candidate variant detected in cases by WES analysis in probands and to test segregation of candidate variants in participating relatives.
Molecular modeling of the RNF213 RING finger domain
Templates that could serve for modelling the 3D structure of the RNF213 RING domain was searched for using Phyre2.24 The model was constructed using Modeller (v9.15).25 3D structures were manipulated using Chimera.26 RNF213 variants involving the RING-finger domain, and that have been identified either in our study or in previous moyamoya studies, have been represented on the model of the 3D structure of the RING domain.
Results
Four outlier patients detected in PCA were discarded from the analysis. Another patient was excluded because of an excess of missing data. In total, 68 Caucasian MMA probands and 573 ethnically-matched controls were included in the association study.
Demographic and clinical features of MMA patients
General characteristics of the MMA cohort are shown in Table 1. Fifty-nine patients had been referred for MMD and 9 for MMS of unknown etiology. Screening of these patients for known acquired or inherited causes of MMS was negative. Sex ratio was 2.8 females for 1 male and mean age at onset was 27.8 years (range 2-57 years). Seventeen probands presented their first MMA symptoms before 15 years of age. Eight probands had a familial moyamoya history. All the probands but two had bilateral steno-occlusive lesions on cerebral imaging. Clinical characteristics of the 9 included MMS probands are detailed in Supplementary table S1.
Table 1. Demographic, clinical and radiological characteristics of the 68 MMA patients included in the analysis.
| Characteristics | N patients (percentage) |
|---|---|
| Sex | |
| Male | 18 (26.5) |
| Female | 50 (73.5) |
| Adulthood (onset ≥15 years old) | 51 (75) |
| Childhood (onset <15 years old) | 17 (25) |
| Sporadic | 60 (88) |
| Familial | 8 (12) |
| MMD | 59 (87) |
| MMS | 9 (13) |
| Radiological findings | |
| Bilateral | 66 (97) |
| Unilateral | 2 (3) |
Abbreviations: MMD, moyamoya disease; MMS, moyamoya syndrome; N patients, number of patients.
RNF213 variants in patients and controls
After quality control, a total of 274 RNF213 variants were found in the combined sample of cases and controls; 224 of them were absent from ExAC Non-Finish Europeans or had a minor allele frequency below 0.01. Among these 224 low-frequency variants, 64 variants were missense, stoploss, stopgain, canonical splice site variants or Indel, and thus fulfilled the criteria to be included in the association tests. Twenty-three variants were found in cases, 49 in controls and 8 were shared by cases and controls (see Table 2 for the variants detected in MMA patients and Supplementary Table S2 for those detected in controls). Of note, the p.(R4810K) variant was absent from both cases and controls.
Table 2. Rare coding RNF213 variants identified among the 68 Caucasian MMA probands.
| gDNA change | cDNA change | Predicted protein change | Exon | Protein Domain | rs ID (dbSNP 144) | ExAC_NFE AF | FREX AF | MMA cases AF | MMA proband(s) ID | Polyphen2 | PROVEAN | SIFT | CADD-Phred |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr17:g.78261704T>C | c.352T>C | p.(Cys118Arg) | 4 | . | rs201620985 | 0.0022 | 0 | 0.007353 | M011_4 | Benign | Neutral | Tolerated | 0.02 |
| chr17:g.78261749C>A | c.397C>A | p.(Leu133Met) | 4 | . | rs149177904 | 0.0037 | 0.001745 | 0.007353 | M012_3 | Benign | Neutral | Tolerated | 2.7 |
| chr17:g.78261978T>A | c.626T>A | p.(Ile209Asn) | 4 | . | rs144769597 | 0.0023 | 0.001745 | 0.007353 | M012_3 | Benign | Neutral | Tolerated | 0.8 |
| chr17:g.78264440C>T | c.1184C>T | p.(Pro395Leu) | 7 | . | . | Absent | 0 | 0.007353 | M009_3 | Prob. dam. | Deleterious | Damaging | 24.2 |
| chr17:g.78302164C>T | c.3404C>T | p.(Ala1135Val) | 20 | . | rs201493500 | 0.0044 | 0.000873 | 0.007353 | M001_6 | Benign | Neutral | Tolerated | 3.3 |
| chr17:g.78313281C>A | c.5114C>A | p.(Thr1705Lys) | 26 | . | rs147868237 | 0.0054 | 0.009599 | 0.007353 | M004_3 | Prob. dam. | Deleterious | Damaging | 32 |
| chr17:g.78313329C>T | c.5162C>T | p.(Pro1721Leu) | 26 | . | rs189765261 | 0.0011 | 0.001745 | 0.007353 | M005_5 | Prob. dam. | Deleterious | Tolerated | 23 |
| chr17:g.78313697G>A | c.5530G>A | p.(Ala1844Thr) | 26 | . | rs545060242 | 0.0026 | 0.002618 | 0.007353 | M013_3 | Benign | Neutral | Tolerated | 18.7 |
| chr17:g.78337083G>A | c.11537G>A | p.(Arg3846His) | 40 | . | . | Absent | 0 | 0.007353 | M014_3 | Benign | Neutral | Tolerated | 11.7 |
| chr17:g.78338261G>A | c.11779G>A | p.(Ala3927Thr) | 42 | . | rs777402744 | 1.50E−05 | 0 | 0.007353 | M019_6 | Benign | Neutral | Damaging | 14.5 |
| chr17:g.78338279G>A | c.11797G>A | p.(Val3933Met) | 42 | . | . | Absent | 0 | 0.007353 | M006_3 | Prob. dam. | Neutral | Tolerated | 25.4 |
| chr17:g.78341825G>A | c.12037G>A | p.(Asp4013Asn) | 44 | RING | rs397514563 | 1.50E−05 | 0 | 0.007353 | M007_4 | Prob. dam. | Neutral | Tolerated | 27.6 |
| chr17:g.78341828C>A | c.12040C>A | p.(His4014Asn) | 44 | RING | . | Absent | 0 | 0.007353 | M004_3 | Prob. dam. | Deleterious | Damaging | 26.4 |
| chr17:g.78341843C>T | c.12055C>T | p.(Arg4019Cys) | 44 | RING | rs139265462 | 0.0008 | 0.002618 | 0.007353 | M008_4 | Pos dam. | Deleterious | Damaging | 24 |
| chr17:g.78341882T>C | c.12094T>C | p.(Cys4032Arg) | 44 | RING | . | Absent | 0 | 0.007353 | M001_6 | Prob. dam. | Deleterious | Damaging | 26.7 |
| chr17:g.78341886C>T | c.12098C>T | p.(Pro4033Leu) | 44 | RING | rs778482758 | Absent | 0 | 0.007353 | M010_10 | Prob. dam. | Deleterious | Damaging | 32 |
| chr17:g.78341912G>A | c.12124G>A | p.(Glu4042Lys) | 44 | . | . | Absent | 0 | 0.007353 | M008_4 | Benign | Neutral | Tolerated | 6.1 |
| chr17:g.78341940A>C | c.12152A>C | p.(His4051Pro) | 44 | . | . | Absent | 0 | 0.007353 | M011_4 | Pos dam. | Deleterious | Tolerated | 18.33 |
| chr17:g.78343331G>A | c.12185G>A | p.(Arg4062Gln) | 45 | . | . | Absent | 0 | 0.007353 | M002_3 | Pos dam. | Deleterious | Tolerated | 31 |
| chr17:g.78343607A>T | c.12365A>T | p.(Asp4122Val) | 46 | . | . | Absent | 0 | 0.007353 | M003_4 | Prob. dam. | Deleterious | Deleterious | 27.6 |
| chr17:g.78350110G>A | c.13195G>A | p.(Ala4399Thr) | 52 | . | rs148731719 | 0.0079 | 0.006108 | 0.014706 | M018_3 | Benign | Neutral | Tolerated | 12.88 |
| M015_7 | |||||||||||||
| chr17:g.78355371C>T | c.13822C>T | p.(Pro4608Ser) | 57 | . | . | Absent | 0 | 0.007353 | M016_4 | Benign | Deleterious | Tolerated | 4.5 |
| chr17:g.78355467G>A | c.13918G>A | p.(Gly4640Ser) | 57 | . | rs138223459 | 1.51E−05 | 0 | 0.007353 | M017_3 | Benign | Neutral | Tolerated | 5.4 |
Abbreviations: AF, allele frequency; CADD, Combined Annotation Dependent Depletion; ExAC, Exome Aggregation Consortium; ID, proband identifier; MMA, moyamoya angiopathy; NFE, Non-Finnish European; pos.dam, possibly damaging; prob.dam, probably damaging; RING, RING domain, based on NCBI data.
Exons and amino-acids positions are provided according to the NM_001256071.2 transcript and the NP_001243000.2 isoform. Genomic positions are given in Human Genome 19. Polyphen2, Provean, SIFT and CADD are in-silico predictive pathogenicity tools. Variants that are only found in MMA cases and not in FREX controls are highlighted in grey, and those found both in cases and FREX controls are left blank.
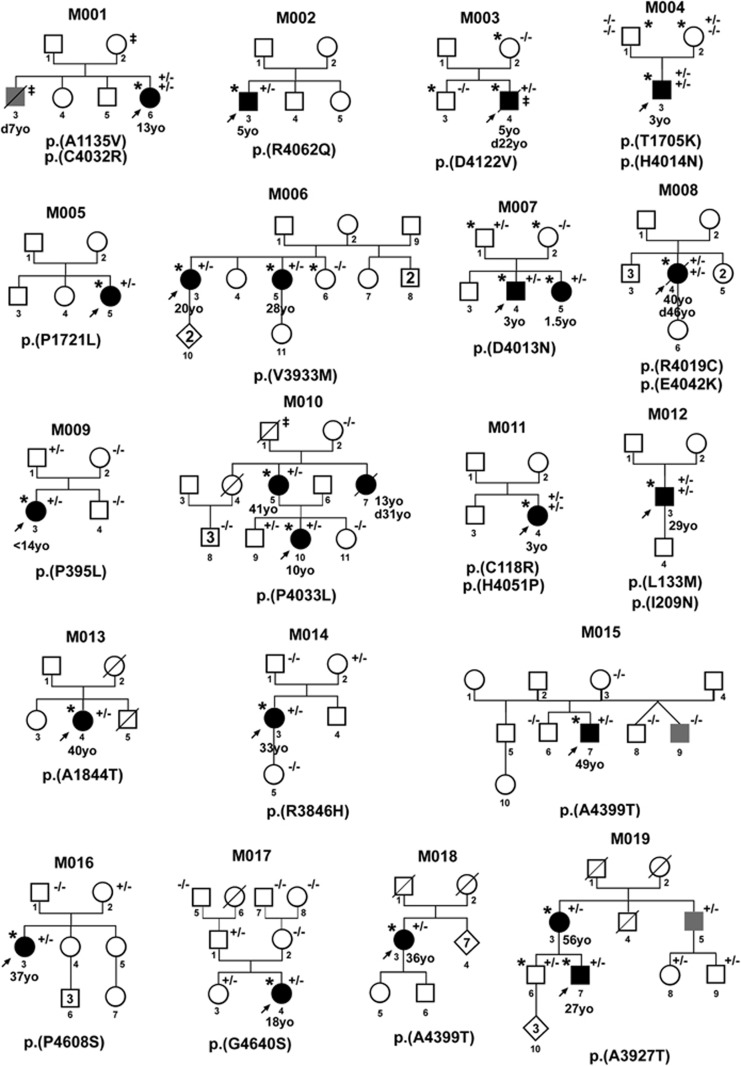
Nineteen of the 68 probands carried at least one rare coding variant in RNF213 versus 86 out of the 573 controls (27.9% versus 15%, respectively, with an odds ratio (OR)=2.19, 95% confidence interval (CI)=(1.21–3.85), P=0.008). The results remained significant when adjusted on the top 5 PCs (OR=2.24; 95% CI=(1.19–4.11), P=0.01). Family trees of the 19 mutated probands are shown in Figure 1 and clinical data are reported in Supplementary Table S3.
Figure 1.
Family trees of the 19 RNF213 mutated probands. Families of the mutated probands (arrow) are numbered from M001 to M019. Squares: males, circles: females, rhombs: individuals for whom we do not have sex information. Black filled symbols: moyamoya affected individuals; age at first symptoms is indicated below the symbol (yo, years old). Empty symbols: relatives without clinical symptoms suggestive of moyamoya based on past medical history. Grey-filled symbols: relatives presenting with either (i) an early-onset (<50 yo) but poorly characterized cerebrovascular condition, or (ii) a well characterized steno-occlusion of ICA terminal parts and/or circle of Willis but without neoangiogenic deep vessels, whatever the age. ‡, early-onset (<50 yo) extra-cerebral arterial disease. Black diagonal lines: deceased individuals, age of death in years is indicated below the symbol and preceded by ‘d’. *, whole exome sequencing performed. The RNF213 variant(s) identified in each family is (are) indicated under the corresponding family tree, and the status of the proband and his/her relatives, when sequenced, are noted +/− (heterozygous) or −/− (not mutated). Not all non-informative individuals have been shown in the family trees.
The 23 rare heterozygous variants identified in the 19 mutated probands were all missense substitutions. One of them was shared by two probands (Table 2 and Figure 2). None of the probands was homozygous for any mutated allele, but five patients carried two heterozygous variants in RNF213. The analysis of Binary Alignment/Map (BAM) files of proband M008-4 revealed that both variants were located on the same allele. Because of the large distance between RNF213 variants in the 4 other probands, BAM files analysis did not allow to determine if these patients were compound heterozygotes or not. A de novo mutation, p.(H4014N), was identified in one proband (M004-3).
Figure 2.
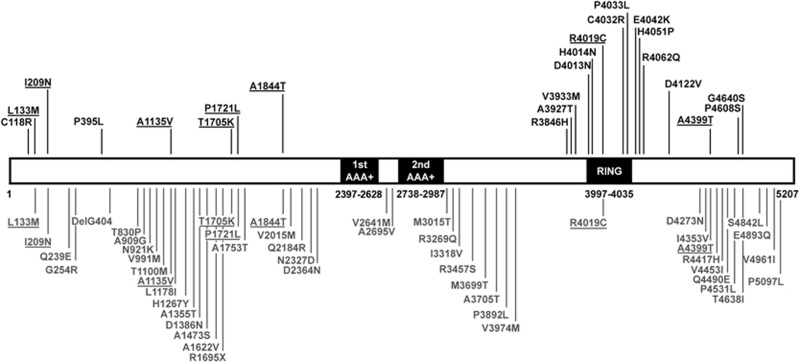
Schematic representation of the RNF213 protein and the RNF213 variants identified in the present study. This representation is based on the NP_001243000.2 isoform. The domain structure is based on NCBI data and Morito et al.42 Amino-acid boundaries of these protein domains are indicated below each domain. The RNF213 variants found among the Caucasian MMA probands are shown above the protein (in black). Those found in the French FREX cohort are represented below the protein (in gray). Variants found both in cases and in controls are underlined. AAA+: AAA-ATPase domains; RING: RING-finger domain.
Among the 49 heterozygous variants identified in 86 controls, 41 were found only in controls, including 39 missense variants, 1 in-frame deletion and 1 nonsense variant.
Rare coding variants specific to cases showed higher CADD Phred scores (mean and median values of 18.7 and 24.2 for case restricted variants versus 11.6 and 9.4 for control restricted variants, P=0.029).
When focusing the analysis on rare coding variants that are predicted to be deleterious by at least 2 softwares among Polyphen 2 (HumVar), SIFT and PROVEAN, we found a trend for accumulation of such variants among cases. Indeed, 9/68 cases carried at least 1 variant that meets these criteria versus 37/573 controls (13.2% versus 6.4%, P=0.045 without adjustment and P=0.077 after adjustment on top PCs).
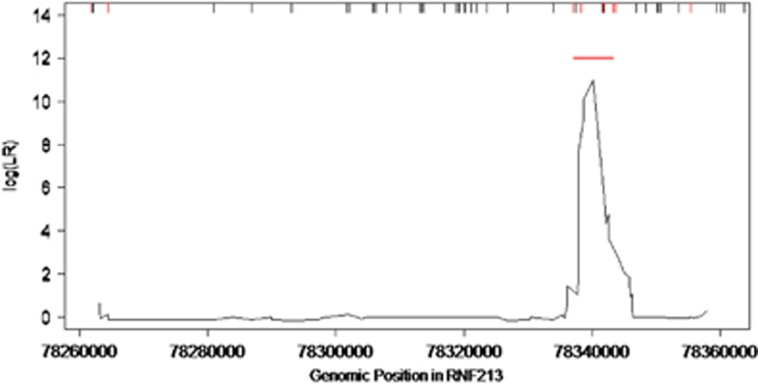
In contrast with variants identified in controls, those found in MMA probands preferentially clustered in the C-terminal part of the protein, mainly within or in close vicinity of the region encoding the RING finger domain (Figures 2 and 3). We identified a 6.2 Kb C-terminal region (chr17(hg19):78337083-78343331) where rare coding variants clustered significantly in cases (P<10−3 after correction for the different window sizes investigated, based on 1000 replicates). In this ‘hotspot’, 10 probands (14.7%) carried a total of 11 variants fulfilling inclusion criteria and 5 controls (0.9%) carried a total of 3 variants, leading to an OR of 19.19, 95% CI=(6.19–66.93); P=7.10−7. These data are shown in Figure 4. Case variants located in the hotspot showed higher pathogenicity prediction scores that those located outside, with a mean CADD Phred score of 22.2 versus 12.9 (P=0.039, Fisher Exact test). Of note, no variant has been identified in the AAA-ATPase domains of the protein, neither in cases nor in controls (Figure 2).
Figure 3.
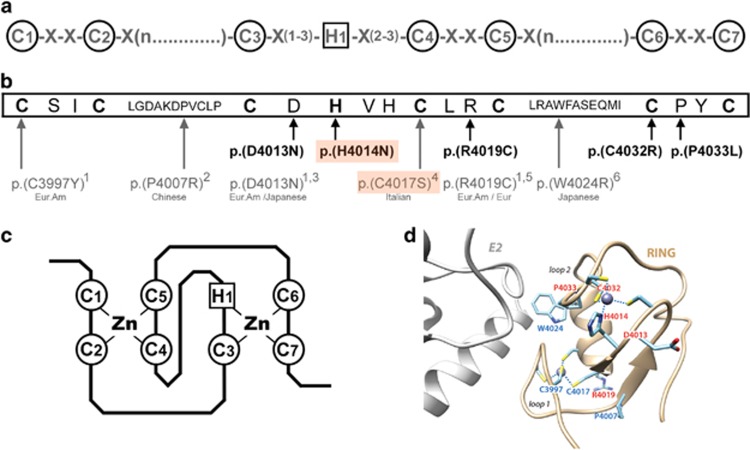
Rare RNF213 variants of MMA patients located in the RING-finger domain. (a) C3HC4 RING-finger consensus motif.27 Highly conserved cysteine and histidine residues involved in zinc-coordination are circled. (b) RNF213 RING-finger sequence (NP_001243000.2). Highly conserved cysteine and histidine residues are in bold. The rare RNF213 variants identified in MMA patients and involving the RING domain are shown below the RING-finger sequence. Variants found in our cohort of 68 Caucasian MMA are represented in bold. Those found in previously published MMA cohorts are represented in gray, and ethnicities of the corresponding cases are given below the variants. (1: Cecchi et al, Stroke, 2014; 2: Wu et al. PLoS One, 2012; 3: Liu et al. PLoS One, 2011; 4: Raso et al. J Neurosurg Sci, 2016; 5: Kobayashi et al. PLoS One, 2016; 6: Miyatake et al. Neurology, 2012). The p.(H4014N) and p.(C4017S) de novo variants are boxed in orange. Eur, European; Eur. Am, European American (c) Representation of the ‘cross-brace’ structure of the C3HC4 RING-finger domain, mediated through cysteine and histidine Zinc-binding. Panel D: Model of the 3D structure of the RNF213 RING finger (ribbon representation), based on the 3D structure of the RING domain of RNF146 (pdb 4qpl), solved in complex with its E2.43 Amino acids which are mutated in moyamoya patients are labeled in red (our study) and blue (previous studies). p.(R4019C) and p.(D4013N), found mutated both in our study and previous studies, are in red. The position of the putative E2 partner is shown in gray, in reference to the structure of the E2-E3 complex reported in DaRosa et al.,43 with which the model of the RN213 RING 3D structure was superimposed.
Figure 4.
Clustering of rare coding RNF213 variants detected in MMA cases. Scanning-window length is 10 kb. The rug plot on the top of the figure shows the distribution of rare variants in the region fulfilling the inclusion criteria for analysis. The rare variants only present in cases are shown in red, and those present only in controls are shown in black. The red line indicates the 6.2 kb region where rare coding variants clustered significantly in cases (P<10−3).
We then restricted our analysis to the 21 probands presenting with a childhood-onset MMA (N=17) and/or a familial MMA (N=8). Ten of these probands (47.6%) carried a total of 13 rare coding variants in RNF213. Comparison to the 86/573 (15%) mutated controls resulted in an OR of 5.15 (95% CI=(2.08; 12.58)), P=2.9 × 10−4). The results remained significant when adjusted on the top 5 PCs (OR=4.54; 95%CI=(1.80–11.34), P=1.1 × 10−3). Interestingly, eight out of these 10 mutated probands had one variant located in the mutational ‘hotspot’. Rare coding RNF213 variants specific to this subset of patients had higher CADD Phred scores compared to those specific to controls (mean and median values of 23.1 and 26.4 for case restricted variants versus 11.6 and 9.4 for control restricted variants, P=0.005).
Variant segregation analysis
A total of 39 relatives were genotyped. Among them, 16 carried the RNF213 variant found in the corresponding proband. Four out of these 16 mutated relatives were clinically affected with a MMA (M006_5, M007_5, M010_5, M019_3), indicating an incomplete penetrance of these variants. We did not identify any MMA-affected relative who did not carry the RNF213 mutation.
Clinical features of mutated cases
Eighteen out of the 19 mutated MMA probands had bilateral MMA. The proportion of mutants in the MMS group (2/9) and the MMD group (17/59) were similar, although the analysis of larger cohorts would be needed to firmly establish this point. Mutated probands M005-5 and M008-4 were diagnosed as MMS (Supplementary Table S2). The M005-5 proband has a left unilateral MMA diagnosed incidentally at 22 years of age. This MMA was associated with a congenital pituitary insufficiency and a left morning-glory syndrome. The M008-4 proband presented a bilateral MMA revealed at 40 years of age by a stroke; in addition she had a unilateral right thumb malformation and a mild facial dysmorphism. She died suddenly when 46 years old.
Molecular modeling of RNF213 RING finger domain and prediction of the impact of variants on its 3D structure
The C3HC4-type RING finger domain of RNF146 was proposed by Phyre2 as the best template for RNF213 RING finger domain modeling (Supplementary Figure S2). Considering our data and those from previously published studies, 9 rare coding variants located in the RING-finger domain have been reported in MMA patients to date, mostly in Caucasian patients (Figure 3). Four of these variants (the p.(H4014N) and p.(C4032R) variants identified in the present study, and the previously reported p.(C3997Y) and p.(C4017S) variants13, 17) affect key cysteine or histidine residues involved in zinc coordination. The amino acids C3997 and C4017 are within the first zinc coordination site, contributing to the conformation of the loop 1, whereas H4014 and C4032 are within the second site, contributing to the conformation of loop 2 (Figure 3d). Interestingly, the amino acid W4024, which is found to be mutated in Miyatake study (p.(W4024R)),8 is exposed at the surface of the RING domain, and located within the central helix that connects the two zinc coordination sites (Figure 3d). This helix participates with loops 1 and 2 in the interface that RING domains with E3 ligase activity form with E2s. In these cases, this position is often occupied by a tryptophane or bulky hydrophobic residues, and mutation of this residue impairs RING-E2 interaction essential for E3 ubiquitin ligase activity.27 The mutated amino acids D4013, R4019 and P4033, which are mutated herein and in previous studies,6, 13, 16 are located in the vicinity of the zinc-coordinating residues. Thus, the p.(R4019C) variant is predicted to have an impact on the conformation of loop 1, and the p.(D4013 N) and p.(P4033L) variants, on the conformation of loop 2. The p.(P4007R) variant, reported in Wu study,28 is also predicted to have an effect on the overall 3D structure of the RING domain, as this highly conserved residue is located at the beginning of one of the two beta-strands forming the core of the RING domain.
Discussion
We provided significant evidence for a positive association of rare RNF213 missense variants and MMA in patients of European ancestry. This association was even stronger in early onset and/or familial cases. These ‘Caucasian’ variants show higher pathogenicity predictive scores that those found in ancestry matched controls and significantly cluster in a C-terminal ‘hotspot’ encompassing the RING finger domain, contrasting with variants observed in controls that are evenly distributed along the gene.
Herein, we performed a RNF213 gene burden test, which collapsed the information for multiple rare variants into a single genetic score instead of testing each variant individually.18, 19 Several challenges have to be met when conducting such a test. In order to prevent stratification bias, a well-known genetic confounder of rare variant association studies,18, 19 we used a set of 573 controls of Caucasian French ancestry and restricted our analysis to ancestry matched cases and controls. In addition, we processed case and control exome data with a unique bioinformatic pipeline and excluded variants of poor quality to avoid technical confounders.29 This strategy allowed us to rigorously establish the association of rare RNF213 variants with MMA in Caucasian patients. To our knowledge, this study is the first gene-based burden analysis of rare (MAF <1%) RNF213 variants in Caucasians. An exome-wide collapsing test has been performed once in a Caucasian MMA cohort, but failed to detect a significant association with RNF213 variants.15 However, the use of a 5% MAF threshold might have introduced polymorphic neutral variants in both controls and patients, and thus might have diminished the power of the test. In Japanese MMD patients, where rare RNF213 variants distinct from p.(R4810K) have also been identified, a variable threshold collapsing test showed significant enrichment in rare RNF213 variants having a CADD Phred score >10.9 These data and our data strongly suggest that rare RNF213 variants distinct from p.(R4810K) are associated with MMA, not only in East Asian but also in Caucasian patients.
Interestingly, the Caucasian MMA variants are significantly grouped in a C-terminal region that includes the RING finger domain of RNF213. Case variants located in this hotspot present a number of characteristics supporting that they might be causal, eventhough no firm conclusion can be made at the level of an individual variant: (i) these variants show higher pathogenicity prediction scores, and, except for the p.(R4019C) variant, are absent or extremely rare in FREX and ExAC controls; (ii) 8 out of the 11 case variants located in the ‘hotspot’ have been identified in early-onset and/or familial MMA cases, who are most likely to be of genetic origin; (iii) for two of these variants, a recurrence was observed across multiplex families from distinct studies. Indeed, p.(D4013N), identified in M007 family, was previously shown to segregate with MMD in two large families of European ancestry.6, 13 As well, the p.(V3933M) variant was identified both in M007 family and in a previously published Taiwanese family.30 (iv) another case variant located within the hotspot, p.(H4014N), occurred de novo in a sporadic case, which is a strong argument for causality. Of note, this variant is the fifth de novo RNF213 variant to be reported in the context of sporadic early-onset MMD, the previous four being p.(C4017S), p.(K4115del), p.(S4118F) and p.(V4146A).13, 16, 17, 31 Two of these de novo variants, p.(H4014N) and p.(C4017S), affect key cysteine or histidine residues of the RING-finger domain that are directly involved in zinc coordination (Figure 3). The analysis of RING finger conservation across species reveals that the RNF213 RING-finger is most likely a C3HC4 type (Supplementary Figure S2). Indeed, the cysteine and histidine residues of the C3HC4 motif are well conserved in the RNF213 orthologs, whereas the C4009 residue is not, arguing against the hypothesis of a C4C4 structure. In addition, all best supports for modeling the RING-finger 3D structure, as proposed by Phyre2, belong to the C3HC4 subgroup (Supplementary Figure S2). To date, 9 rare variants located in the RING-finger domain of RNF213 have been detected in MMA patients. According to our 3D model of RNF213 RING finger domain, all of them are predicted to alter its structure or impair its binding to partners (Figure 3d). This finding suggests that alteration of the RNF213 RING-finger structure or function most likely plays an important role in moyamoya pathogenesis. Many data support that RING-finger proteins act as E3 ubiquitin ligases.27, 32, 33 In addition, auto-ubiquitination assays conducted in transfected cells and co-immunoprecipitation data suggest that the RING-finger domain is possibly involved in the ubiquitin ligase activity of RNF213.6, 34 However, substrates and partners of RNF213 are so far largely unknown. Their identification, as well as the identification of the biological pathways involving RNF213, is needed to characterize the functional consequences of RNF213 variants.
In our study, the penetrance of MMA in family members carrying a RNF213 variant is close to 25%, in line with what is observed in Cecchi study.13 This 25% penetrance in a family context appears much higher than the penetrance reported for the p.(R4810K) variant in East Asian population. Indeed, although heavily associated with MMA in Japan (OR>200, P<10-100 in a meta-analysis),9 the penetrance of MMA in p.(R4810K) carriers is estimated to be 1/150 in the Japanese population.35 This very low penetrance, on a population basis, contrasts with the incomplete but much higher penetrance observed in a family context. One possible explanation could be that the development of MMA requires, in addition to RNF213 pathogenic variants, at least one additional genetic factor which is most likely shared in families. This “oligogenic hypothesis” might also explain the very limited size of most MMD family trees reported in literature.
Several studies suggest that RNF213 variants might also be involved in other vascular phenotypes, such as premature coronary artery disease, renal artery or aortic disease, high blood pressure, fibro-muscular dysplasia, and recently with cerebral aneurysms.13, 36, 37, 38 In our study, several mutated probands have a personal or familial history of systemic vascular disease. Among them, the M003_4 proband had a severe form of pulmonary arterial hypertension (PAH) in addition to MMA. The association of MMA and PAH had also been described in two unrelated Japanese cases homozygous for the p.(R4810K) variant.39 Interestingly, these two patients and ours showed a peculiar PAH angiographic pattern characterized by peripheral pulmonary artery stenoses. The M003_4 proband developed in the second year of life, a transitory but marked elevation of liver enzymes of undetermined origin. A similar phenomenon was previously reported in a sporadic childhood-onset MMD patient who carried the p.(S4118F) de novo mutation, very close to the amino-acid D4122 mutated in the proband M003_4.31 This association of MMD and hepatic cytolysis has also been reported in a case-series of infant-onset MMD, but unfortunately they had not been sequenced for RNF213.40 RNF213 mutations were also found to be associated with intracranial arterial steno-occlusive lesions without moyamoya network.14, 41 In the present study, this phenotype was observed in the M019_5 relative, who carried the c.11779G>A (p.(A3927T)) variant and who presented a unilateral and asymptomatic stenosis of anterior cerebral artery without neovessels on MRA. Further studies will be required to firmly establish the role of RNF213 variants in these phenotypes distinct to MMA.
In summary, we provided herein significant evidence for the role of rare, non p.(R4810K), variants of RNF213 in the development of MMA in Caucasian patients, especially when located in a C-terminal hotspot encompassing the RING finger domain of the protein. In a diagnostic context, this study raises the question of the putative benefit of a systematic screening of RNF213 in MMA patients and their relatives. However, the difficulty to draw a clear conclusion about causality when RNF213 variants are considered individually, and the low penetrance observed in most families renders genetic counseling quite difficult at this point. The investigation of biochemical functions, substrates and partners of RNF213 will help to unravel the pathogenic mechanisms linking those variants to MMA, and facilitate variants interpretation in a diagnostic context in the future.
Acknowledgments
The authors express their gratitude to the family members for participating to this study and are indebted to Professor Dr med. Andreas Ferbert, Kassel, Germany; Dr med. Christian Roth, Kassel, Germany; Dr K. Spengos, Athens, Greece; Dr JM Pedespan, Bordeaux, France; Dr C Bellesme, Le Kremlin Bicêtre, France; Dr JM Cuisset, Lille, France; Pr C. Cordonnier, Lille, France; Dr C Francannet, Clermont-Ferrand, France for providing clinical information and samples of some patients, Lou Grangeon for stimulating discussions, Audrey Delaforge and Jessica Hadjadj for their help in samples preparation. They also thank Florence Marchelli for excellent figures editing. We acknowledge support from INSERM and from Fondation pour la Recherche Médicale (S Guey, PhD student grant, FDM40678). This work was also supported by France Génomique National infrastructure, funded as part of ‘Investissement d’avenir’ program managed by Agence Nationale pour la Recherche (contrat ANR-10-INBS-09) https://www.france-genomique.org/spip/spip.php?article158.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Contributor Information
the FREX consortium:
Génin E, Campion D, Dartigues J-F, Deleuze J-F, Lambert J-C, Redon R, Ludwig T, Grenier-Boley B, Letort S, Lindenbaum P, Meyer V, Quenez O, Dina C, Bellenguez C, Charbonnier-Le Clézio C, Giemza J, Chatel S, Férec C, Hervé Le Marec, Letenneur L, Nicolas G, Rouault K, Bacq D, Boland A, and Lechner D
Supplementary Material
References
- Hashimoto N, Tominaga T, Miyamoto S et al: Research Committee on the Pathology and Treatment of Spontaneous Occlusion of the Circle of Willis. Guidelines for diagnosis and treatment of moyamoya disease. Neurol Med Chir (Tokyo) 2012; 52: 245–266. [DOI] [PubMed] [Google Scholar]
- Kuroda S, Houkin K: Moyamoya disease: current concepts and future perspectives. Lancet Neurol 2008; 7: 1056–1066. [DOI] [PubMed] [Google Scholar]
- Scott RM, Smith ER: Moyamoya disease and moyamoya syndrome. N Engl J Med 2009; 360: 1226–1237. [DOI] [PubMed] [Google Scholar]
- Guey S, Tournier-Lasserve E, Hervé D, Kossorotoff M: Moyamoya disease and syndromes: from genetics to clinical management. Appl Clin Genet 2015; 8: 49–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineharu Y, Takenaka K, Yamakawa H et al: Inheritance pattern of familial moyamoya disease: autosomal dominant mode and genomic imprinting. J Neurol Neurosurg Psychiatry 2006; 77: 1025–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Morito D, Takashima S et al: Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE 2011; 6: e22542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada F, Aoki Y, Narisawa A et al: A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J Hum Genet 2011; 56: 34–40. [DOI] [PubMed] [Google Scholar]
- Miyatake S, Miyake N, Touho H et al: Homozygous c.14576G>A variant of RNF213 predicts early-onset and severe form of moyamoya disease. Neurology 2012; 78: 803–810. [DOI] [PubMed] [Google Scholar]
- Moteki Y, Onda H, Kasuya H et al: Systematic validation of RNF213 coding variants in japanese patients with Moyamoya disease. J Am Heart Assoc 2015; 4: pii: e001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phi JH, Choi JW, Seong MW et al: Association between moyamoya syndrome and the RNF213 c.14576G>A variant in patients with neurofibromatosis Type 1. J Neurosurg Pediatr 2016; 17: 717–722. [DOI] [PubMed] [Google Scholar]
- Morimoto T, Mineharu Y, Kobayashi H et al: Significant association of the RNF213 p.R4810K polymorphism with quasi-Moyamoya disease. J Stroke Cerebrovasc Dis 2016; 25: 2632–2636. [DOI] [PubMed] [Google Scholar]
- Liu W, Senevirathna ST, Hitomi T et al: Genomewide association study identifies no major founder variant in Caucasian moyamoya disease. J Genet 2013; 92: 605–609. [DOI] [PubMed] [Google Scholar]
- Cecchi AC, Guo D, Ren Z et al: RNF213 rare variants in an ethnically diverse population with Moyamoya disease. Stroke 2014; 45: 3200–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KR, Leventer RJ, Mackay MT et al: Identification of a novel RNF213 variant in a family with heterogeneous intracerebral vasculopathy. Int J Stroke 2014; 9: E26–E27. [DOI] [PubMed] [Google Scholar]
- Shoemaker LD, Clark MJ, Patwardhan A et al: Disease variant landscape of a large multiethnic population of moyamoya patients by exome sequencing. G3 (Bethesda) 2015; 6: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Brozman M, Kyselová K et al: RNF213 rare variants in Slovakian and Czech Moyamoya disease patients. PLoS ONE 2016; 11: e0164759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raso A, Biassoni R, Mascelli S et al: Moyamoya vasculopathy shows a genetic mutational gradient decreasing from East to West. J Neurosurg Sci 2016; e-pub ahead of print 27 October 2016. [DOI] [PubMed]
- Kiezun A, Garimella K, Do R et al: Exome sequencing and the genetic basis of complex traits. Nat Genet 2012; 44: 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Abecasis GR, Boehnke M, Lin X: Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet 2014; 95: 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E et al: The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM: Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet 2008; 83: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, Makarov V: ARRA Autism Sequencing Consortium, Buxbaum JD. Scan-statistic approach identifies clusters of rare disease variants in LRP2, a gene linked and associated with autism spectrum disorders, in three datasets. Am J Hum Genet 2012; 90: 1002–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc Callum KJ, Ionita-Laza I: Empirical Bayes scan statistics for detecting clusters of disease risk variants in genetic studies. Biometrics 2015; 71: 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ: The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015; 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, Sali A: Protein structure modeling with MODELLER. Methods Mol Biol 2014; 1137: 1–15. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC et al: UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 2004; 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA: RING domain E3 ubiquitin ligases. Ann Rev Biochem 2009; 78: 399–434. [DOI] [PubMed] [Google Scholar]
- Wu Z, Jiang H, Zhang L et al: Molecular analysis of RNF213 gene for moyamoya disease in the Chinese Han population. PLoS One 2012; 7: e48179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer PL, Reiner AP, Wang G et al: Guidelines for Large-Scale Sequence-Based Complex Trait Association Studies: Lessons Learned from the NHLBI Exome Sequencing Project. Am J Hum Genet 2016; 99: 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Chen YF, Fan PC et al: Mutation genotypes of RNF213 gene from moyamoya patients in Taiwan. J Neurol Sci 2015; 353: 161–165. [DOI] [PubMed] [Google Scholar]
- Harel T, Posey JE, Graham BH et al: Atypical presentation of moyamoya disease in an infant with a de novo RNF213 variant. Am J Med Genet A 2015; 167A: 2742–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont PS: RING for destruction? Curr Biol 2000; 10: R84–R87. [DOI] [PubMed] [Google Scholar]
- Metzger MB, Pruneda JN, Klevit RE, Weissman AM: RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta 2014; 1843: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz B, Korn C, Wojtarowicz J et al: Endothelial RSPO3 controls vascular stability and pruning through non-canonical WNT/Ca(2+)/NFAT Signaling. Dev Cell 2016; 36: 79–93. [DOI] [PubMed] [Google Scholar]
- Koizumi A, Kobayashi H, Hitomi T, Harada KH, Habu T, Youssefian S: A new horizon of moyamoya disease and associated health risks explored through RNF213. Environ Health Prev Med 2016; 21: 55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi A, Kobayashi H, Liu W et al: p.R4810K, a polymorphism of RNF213, the susceptibility gene for moyamoya disease, is associated with blood pressure. Environ Health Prev Med 2013; 18: 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiando SR, Barlassina C, Cusi D et al: Exome sequencing in seven families and gene-based association studies indicate genetic heterogeneity and suggest possible candidates for fibromuscular dysplasia. J Hypertens 2015; 33: 1802–1810. [DOI] [PubMed] [Google Scholar]
- Zhou S, Ambalavanan A, Rochefort D et al: RNF213 Is Associated with Intracranial Aneurysms in the French-Canadian Population. Am J Hum Genet 2016; 99: 1072–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima H, Takenouchi T, Kosaki K: Homozygosity for moyamoya disease risk allele leads to moyamoya disease with extracranial systemic and pulmonary vasculopathy. Am J Med Genet A 2016; 170: 2453–2456. [DOI] [PubMed] [Google Scholar]
- Law-Ye B, Saliou G, Toulgoat F et al: Early-onset stroke with moyamoya-like syndrome and extraneurological signs: a first reported paediatric series. Eur Radiol 2016; 26: 2853–2862. [DOI] [PubMed] [Google Scholar]
- Miyawaki S, Imai H, Shimizu M et al: Genetic variant RNF213 c.14576G>A in various phenotypes of intracranial major artery stenosis/occlusion. Stroke 2013; 44: 2894–2897. [DOI] [PubMed] [Google Scholar]
- Morito D, Nishikawa K, Hoseki J et al: Moyamoya disease-associated protein mysterin/RNF213 is a novel AAA+ ATPase, which dynamically changes its oligomeric state. Sci Rep 2014; 4: 4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaRosa PA, Wang Z, Jiang X et al: Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature 2015; 517: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.