Abstract
Sex chromosome aneuploidies (SCA) is a group of conditions in which individuals have an abnormal number of sex chromosomes. SCA, such as Klinefelter’s syndrome, XYY syndrome, and Triple X syndrome are associated with a large range of neurological outcome. Another genetic event such as another cytogenetic abnormality may explain a part of this variable expressivity. In this study, we have recruited fourteen patients with intellectual disability or developmental delay carrying SCA associated with a copy-number variant (CNV). In our cohort (four patients 47,XXY, four patients 47,XXX, and six patients 47,XYY), seven patients were carrying a pathogenic CNV, two a likely pathogenic CNV and five a variant of uncertain significance. Our analysis suggests that CNV might be considered as an additional independent genetic factor for intellectual disability and developmental delay for patients with SCA and neurodevelopmental disorder.
Introduction
Sex chromosome aneuploidies (SCA) are defined by an abnormal number of sex chromosomes. SCA are the most frequent aneuploidy in humans with an estimated prevalence of over 1 per 500. Drawing an exact description of the phenotype of SCA is still challenging, especially given that >50% of individuals with Klinefelter’s syndrome and XYY syndrome would not be identified.1 However, it is well established that SCA are associated with very variable cognitive, psychiatric and neurological outcome. For example, it has been documented that the intellectual capacities of individuals with Klinefelter’s syndrome (47,XXY) range from below average to gifted intelligence quotient (IQ).1 Numerous individuals with 47,XXY display no cognitive or behavioral deficits, highlighting the variability of this phenotype. It is very likely that other factors (environmental, hormonal, parental genetic background…) contribute to this adverse neurodevelopmental outcome in this population.1 Genetic mechanisms such as X chromosome gene excess dosage, the parental origin of the supernumerary X chromosome, the androgen receptor (AR) CAGn repeat length and the pattern of X chromosome inactivation could be related to the variable neurological outcome.1 This variability is still very problematic especially when SCA are discovered antenatally, by chance. Overall, intellectual ability for individuals with SCA is usually in the average to low-average range.1, 2 Therefore, as in the general population, a causative event should be looked for individuals with SCA showing developmental delay (DD) or intellectual disability (ID).
In this study, we report a subgroup of 14 patients with developmental delay carrying a sex chromosome aneuploidy and a copy-number variant (CNV).
Methods
Patient recruitment
Venous blood samples were obtained from the patients and their parents using standard procedures. Written informed consent was obtained from all individuals or parents. Samples were ascertained from geneticists through a multi-center collaboration. Patients recruited, retrospectively and prospectively (December 2014 to October 2015), were:
Patients known to carry SCA for whom another genetic factor was suspected due to developmental delay. Karyotype was performed in antenatal period for three patients, because of abnormal first trimester screening.
Patients with developmental delay for whom array CGH was indicated.
Only patients carrying CNV considered as pathogenic, likely pathogenic or of uncertain significance were recruited.
Clinical analysis
To compare the phenotype of the patients, we used description from a meta-analysis of SCA patients.2 Patients with Klinefelter syndrome tended to have a verbal IQ depressed in relation to the level expected from social background, but generally within low normal limits, performance IQ is relatively unimpaired, they also presented delays in the earliest stages of language development.2 Patients with 47,XXX presented reduction in IQ of ~20 points affecting both verbal and performance, they also presented difficulties in speech and language.2 Patients with 47,XYY syndrome presented deficits in verbal IQ relative to comparison group, less marked in performance IQ. Nevertheless, IQ scores were not impaired in relation to general population norms.2
Karyotype
Karyotyping based on RH G-banding was performed using standard methods on metaphase spreads from peripheral blood of the patient.
Microarray-based comparative genomic hybridization
Microarray-based comparative genomic hybridization (array-CGH). experiments were performed on genomic DNA, extracted from peripheral blood by conventional methods, using Agilent Human Genome CGH 60 K, 105 K or 180 K oligonucleotides arrays (Agilent, Santa Clara, CA, USA; www.agilent.com) following manufacter’s protocols. Genomic positions are relative to human genome Build NCBI37/hg19. The median probe spacing of this microarray is about 41 kb, 22 kb, 13 kb (www.genomics.agilent.com) for 60 K, 105 K and 180 K arrays, respectively. The arrays were analyzed with the CytoGenomics3.0.4.1 software. CNV were submitted in Decipher database (https://decipher.sanger.ac.uk/) or LOVD database (https://databases.lovd.nl).
CNV validation
Chromosomal rearrangements were confirmed by fluorescence in situ hybridization (FISH) with various specific probes on chromosome preparations from leukocyte cultures or by quantitative PCR using standard protocols.
CNVs were classified as pathogenic, likely pathogenic or of uncertain significance (VOUS) according to the recommendations of the American College of Medical Genetics.3 A pathogenic CNV is documented as clinically significant in multiple peer-reviewed publications, even if penetrance and expressivity of the CNV are known to be variable. Likely pathogenic CNV is a variant of uncertain clinical significance but assessed as potentially pathogenic depending on some evidences such as gene content. VOUS is a variant of uncertain significance with insufficient evidence, preventing unequivocal pathogenic or benign classification.
Results
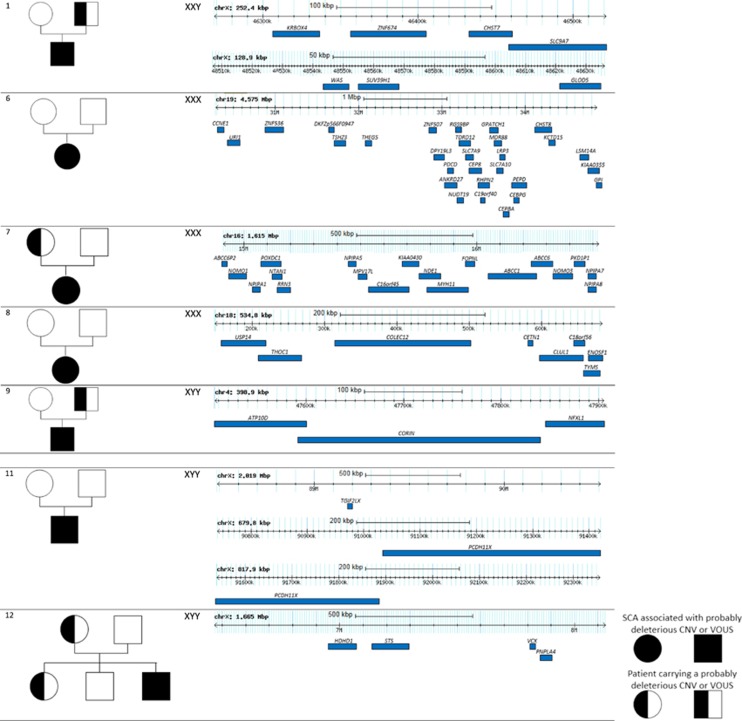
Patients were referred in genetic consultation when neurodevelopmental assessments were noticed. The main clinical and molecular data are listed in Table 1, more informations about these patients are noted in Supplementary Information. In summary, the most prevalent clinical features were mild or moderate developmental delay (100%), speech delay (71%), psychiatric/behavioral features (50%) and congenital malformations (43%). The Figure 1 shows gene content concerning likely pathogenic CNVs and VOUS identified in patients with SCA.
Table 1. Phenotypic and genetics features in individuals with sex chromosome aneuploidy.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 | Patient 10 | Patient 11 | Patient 12 | Patient 13 | Patient 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | Male | Male | Male | Male | Female | Female | Female | Female | Male | Male | Male | Male | Male | Male |
| Consanguinity | no | no | no | no | no | no | no | no | no | unknown | unknown | no | no | no |
| Karyotype | 47,XXY | 47,XXY | 47,XXY | 47,XXY | 47,XXX | 47,XXX | 47,XXX | 47,XXX | 47,XYY | 47,XYY | 47,XYY | 47,XYY | 47,XYY | 47,XYY |
| Karyotype performing period | antenatal | Postnatal | Postnatal | Antenatal | Antenatal | Postnatal | Postnatal | Postnatal | Postnatal | Postnatal | Postnatal | Antenatal | Postnatal | Postnatal |
| Developmental delay | +, mild | +, mild | +, mild | + | +, mild | +, mild | +, mild | +, mild | +, mild | + | + | +, mild | +, mild | +, mild |
| Speech delay | unknown | + | + | + | + | + | + | unknown | + | - | + | + | unknown | + |
| Learning difficulties | + | + | unknown | + | unknown | + | + | unknown | + | unknown | + | + | unknown | + |
| Psychiatric/Behavioral Features | − | + | + | − | − | − | + | − | + | + | − | + | + | − |
| Dysmorphic features | + | + | + | + | − | + | + | + | + | − | − | + | + | + |
| Congenital malformation | + | − | − | − | − | − | − | + | + | + | − | − | + | + |
| CNV | Xp11.3p11.23 duplication Xq11.23 duplication | 7q11.23 duplication | 17q12 duplication | 7q11.23 deletion | 16p11.2 duplication | 19q12q13.1 duplication | 16p13.11 duplication | 18p11.32 deletion | 4p12 duplication | 16p11.2 duplication | Xq21.31 duplication Xq21.31 duplication Xq21.31q21.32 duplication | Xp22.31 duplication | 15q13.2q13.3 deletion | 16p11.2 duplication |
| Chromosomal rearrangment boundaries (Hg 19) | chrX:g.(?_46,268,725)_(46,521,166_?)dup chrX:g.(?_48,507,137)_(48,636,027_ ?)dup | chr7:g.(?_72,726,578)_(74,139,390_?)dup | chr17:g.(34,585,698_34,611,352)_(36,248,910_36,290,301)dup | chr7:g.(?_73,428,837)_(73,643,684_?)del | chr16:g.(?_29,673,954)_(30,198,600_?)dup | chr19:g.(?_30,286,892)_(34,862,143_?)dup | chr16:g.(?_14,910,005)_(16,525,489_?)dup | chr18:g.(?_148,963)_(683,794_776,907)del | chr4:g.(?_47,506,373)_(47,905,283_?)dup | chr16:g.(?_29,258,477)_(28,492,125_?)dup | chrX:g.(?_88,489,310)_(90,507,890_?)dup; chrX:g.(?_90,740,429)_(91,420,193_?)dup; chrX:g.(?_91,539,487)_(92,357,367_?)dup | chrX:g.(?_6,467,006)_(8,131,810_?)dup | chr15:g.(?_31,014,508)_(32,914,140_?)del | ch16:g.(?_29,652,999)_ (30,197,290_?)dup |
| Inherited/de novo | Inherited (father) | Inherited (mother) | De novo | De novo | De novo | De novo | Inherited (mother) | De novo | Inherited (father) | Inherited (mother) | De novo | Inherited (mother) | Inherited (father) | Inherited (mother) |
| Parental phenotype | Father asymptomatic | Learning difficulties | Mother: epilepsy, no degree | Asymptomatic | Unknown | Mother asymptomatic, present in sister (asymptomatic) | Speech delay | Psychiatric disorder Congenital coronary artery anomalies | ||||||
| CNV classification | VOUS | CNV affecting function | CNV affecting function | CNV affecting function | CNV affecting function | Likely pathogenic | Likely pathogenic | VOUS | VOUS | CNV affecting function | VOUS | VOUS | CNV affecting function | CNV affecting function |
| Public database | DECIPHER: 337300 | DECIPHER: 263470 | DECIPHER: 314486 | LOVD database: #00103955 | DECIPHER: 253372 | DECIPHER: 293419 | DECIPHER: 261417 | DECIPHER: 307605 | DECIPHER: 337301 | DECIPHER: 269663 | DECIPHER: 258856 | DECIPHER: 337302 | DECIPHER: 337303 | DECIPHER: 317706 |
Figure 1.
Likely pathogenic and of uncertain significance variants in patients with SCA. Large CNVs, in individuals with likely pathogenic CNVs associated with SCA are shown. The CNV regions with genes are indicated on the last column and the cytogenetic extent and size are labeled. Also shown, on the left column, the pedigrees of individuals with known available parental information. Circles indicate females and squares indicate males. Genomic positions are relative to human genome Build NCBI37/hg19.
We identified four patients (patients 1, 2, 3 and 4) with a 47,XXY karyotype and a CNV (three with a pathogenic CNV and one with a VOUS). Patient 1 carried a 253 kb Xp11.3p11.23 duplication associated with a 130 kb Xp11.23 duplication, both inherited from his healthy father. The first region (Xp11.3p11.23) contains the ZNF674 gene, which has been involved in non-syndromic X-linked intellectual disability in one female patient.4, 5 Patient 2 carried a 1.4 Mb 7q11.23 duplication, responsible for the Williams–Beuren region duplication syndrome, and inherited from his mother who had some learning difficulties (OMIM 609757). Patient 3 was carrying a de novo 1.65 Mb 17q12 duplication, previously involved in the chromosomal 17q12 duplication syndrome (OMIM 614526). Patient 4 carried the recurrent de novo 7q11.23 deletion, identified by FISH, responsible for Williams–Beuren syndrome (OMIM 194050).
We identified four patients with a 47,XXX karyotype (patients 5, 6, 7 and 8) and a CNV (one with a pathogenic CNV, two with a likely pathogenic CNV and one with a VOUS). Patient 5 was carrying a de novo 525 kb 16p11.2 duplication, corresponding to the 16p11.2 duplication syndrome (OMIM 614671). Patient 6 was carrying a de novo 4.5 Mb 19q12q13.1 duplication. Children with dysmorphic features and developmental delay associated with a 19q12q13.1 duplication have already been described in several reports.6, 7 For patient 7, we identified a duplication of 1.6 Mb in 16p13.1, inherited from her mother who showed learning difficulties and epilepsy. Chromosomal deletions or reciprocal duplications at the 16p13.1 locus have been involved in a wide range of neuropsychiatric disorders such as autism, schizophrenia, epilepsy and attention deficit hyperactivity disorder.8, 9
Patient 8 was carrying a de novo terminal 535 kb 18p11.32 deletion. This deletion encompassed eight genes (USP14, THOC1, COLEC12, CETN1, CLUL1, C18orf56, TYM and ENOSF1). Monosomy 18p (OMIM 146390) refers to a chromosomal disorder resulting from the deletion of all or part of the short arm of chromosome 18. The main clinical features are short stature, facial dysmorphism and mild to moderate intellectual deficiency. USP14 (ubiquitin-specific protease 14) is a good candidate gene for short stature phenotype considering that mice with a loss of function Usp14 mutation had growth failure. USP14 would also have a role in synaptic development and plasticity.10, 11
We identified six patients with a 47,XYY karyotype (patients 9, 10, 11, 12, 13 and 14) and a CNV. Patient 9 was carrying a 4p12 duplication, encompassing the NFXL1 gene, and inherited from his healthy father. A recent report indicates that variants within NFXL1 may contribute to language impairment with incomplete penetrance.12 Patient 10 harbored a 766 kb 16p11.2 duplication corresponding to the 16p11.2 duplication syndrome, inherited from his mother (OMIM 614671). Patient 11 was carrying three de novo Xq21.31 duplications (2 Mb, 680 kb and 820 kb). This patient presented developmental delay and severe obesity diagnosed at 4 years of age. The diagnosis of Prader–Willi syndrome (PWS) was ruled out as a DNA probe for the 15q11q12 PWS region and methylation studies failed to detect any abnormalities. These Xq21.31 duplications have already been suggested as a locus for PWS-like phenotype.13 The main candidate gene is PCDH11X, which encodes a protocadherin, highly expressed in brain, implicated in cell–cell communication and dendritic synaptic plasticity. Duplication at this locus might be also responsible for dyslexia.14 Patient 12 was carrying an inherited 1.6 Mb Xp22.31 duplication. The pathogenicity of this CNV is not definitively established. Several studies have suggested that the Xp22.31 duplications may contribute to developmental delay, intellectual disability, seizures and autism spectrum disorder.15, 16, 17 Patient 13 was carrying a 1.9 Mb 15q13.2q13.3 deletion corresponding to the 15q13.3 deletion syndrome (OMIM 612001). This deletion was inherited from the father who had speech delay in childhood. Patient 14 was carrying a 544 kb 16p11.2 duplication corresponding to the 16p11.2 duplication syndrome (OMIM 614671). This duplication was inherited from the mother who presented psychiatric disorders and congenital coronary artery anomalies.
We did not recruit any patient with Turner Syndrome carrying an additional CNV.
Discussion
Here we present 14 patients with SCA and developmental delay harboring another cytogenetic event. These patients had more severe or distinct phenotypes than the typical features associated with SCA, including mild to moderate speech and motor disability, facial dysmorphism, hypotonia, seizures or congenital malformations.
We can define two groups of patients: a first subgroup with patients carrying a well-established pathogenic CNV explaining the full phenotype; a second subgroup with patients carrying a variant of uncertain clinical significance possibly involved in the severity of the cognitive phenotype. The first subgroup includes seven patients: one patient with Williams–Beuren syndrome, one with a 7q11.23 duplication, one with a 17q12 duplication, three with 16p11.2 duplication and one with a 15q11.3 deletion. The second subgroup includes seven patients: two with a likely pathogenic CNV (one with a 19q12q13.1 duplication and one with a 16p13.11 duplication) and five with VOUS (one with a Xp11.3p11.23 duplication associated with a Xq11.23 duplication, one with a 18p11.32 deletion, one with a 4p12 duplication, one with a Xq21.31 duplication and one with a Xq22.31 duplication).
Surprisingly, no individual with Turner syndrome was recruited in our study. It is difficult at the moment to draw any conclusions considering this is not a prospective study. Another CNV associated with Turner syndrome might lead to more frequent spontaneous abortion or other mechanisms could be involved for girls with Turner syndrome explaining an unusual severe delay.
Owing to the high incidence of SCA (one in 375 male livebirths and one in 660 female livebirths)1 and CNV of 1 Mb or larger (2%),18 it is not surprising that some individuals carry two independent genetics event. Our study is biased as we only considered patients known to carry SCA and CNV. French laboratories have different procedures for genetic analysis in patients with DD/ID. Therefore, this study does not allow us to estimate the frequency of additional CNV in patients with SCA and developmental delay. For example, in the cytogenetic laboratory of Nantes, we use to perform array CGH in first place for patients with developmental delay. We identified 27 patients with SCA (11 with 47,XXY, 8 with 47,XXX, 4 with 47,XYY and 4 with 45,X) from 2008 to 2015. In this population presenting with cognitive defect, four patients harboured another cytogenetic event (4/27, 14.8%). We did not obtain these data for the other centers that participated in this study. Further prospective studies comparing the frequency of CNV in two groups of patients with SCA (with or without severe neurological symptoms) would allow answering this question.
These results strengthen that SCA should not be always considered as causative for patients presenting with ID/DD. For those patients, physicians should be aware to look for an additional environmental or genetic event for individuals with SCA presenting with an unusual or severe phenotype. These data encourage performing array CGH for any further studies evaluating the cognitive profile of patients with SCA. Finally, determining additional genetics factor leading to cognitive impairment for patients with SCA is crucial especially for antenatal diagnosis. Indeed, genetic counselling for SCA during pregnancy is still a matter of debate. If we consider the high frequency of CNV in general population, these results raise the question whether or not to perform array on a fetal sample, which are available after karyotyping. Absence of additional CNV would not provide complete reassurance about the outcome but the discovery of an additional pathogenic or likely pathogenic CNV would probably worsen the prognosis. In these cases, array CGH results could complete the antenatal decision-making process.
In conclusion, additional (pathogenic, likely pathogenic or VOUS) CNV might be a significant, independent, genetic factor for intellectual disability and developmental delay in patients with SCA. The poor neurological prognosis of some of those patients could be due to additional CNV rather than SCA itself. This additional genetic independent factor might be another cytogenetic or molecular variant involving a gene of interest or an environmental event that modifies the phenotype. These results encourage us to look for another genetic event such as a CNV for patients with SCA and cognitive, psychiatrics or neurological symptoms. Performing array-CGH when a SCA is discovered during pregnancy could lead to more accurate genetic counselling.
Acknowledgments
We are grateful to the patients and their families for participating in this study.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Hong DS, Reiss AL: Cognitive and neurological aspects of sex chromosome aneuploidies. Lancet Neurol 2014; 13: 306–318. [DOI] [PubMed] [Google Scholar]
- Leggett V, Jacobs P, Nation K, Scerif G, Bishop D: Neurocognitive outcomes of individuals with a sex chromosome trisomy: XXX, XXY, or XYY: a systematic review. Dev Med Child Neurol 2010; 52: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee: American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med 2011; 13: 680–685. [DOI] [PubMed] [Google Scholar]
- Lugtenberg D, Yntema HG, Banning MJG et al: ZNF674: a new kruppel-associated box-containing zinc-finger gene involved in nonsyndromic X-linked mental retardation. Am J Hum Genet 2006; 78: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy V, Castillo M, Bolduc FV: Developmental disability: duplication of zinc finger transcription factors 673 and 674. Pediatr Neurol 2010; 43: 209–212. [DOI] [PubMed] [Google Scholar]
- Davidsson J, Jahnke K, Forsgren M, Collin A, Soller M: dup(19)(q12q13.2): array-based genotype-phenotype correlation of a new possibly obesity-related syndrome. Obesity (Silver Spring) 2010; 18: 580–587. [DOI] [PubMed] [Google Scholar]
- Lugli L, Malacarne M, Cavani S, Pierluigi M, Ferrari F, Percesepe A: A 12.4 Mb direct duplication in 19q12-q13 in a boy with cardiac and CNS malformations and developmental delay. J Appl Genet 2011; 52: 335–339. [DOI] [PubMed] [Google Scholar]
- Ingason A, Rujescu D, Cichon S et al: Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry 2011; 16: 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann R, Turner G, Kirchhoff M et al: Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat 2007; 28: 674–682. [DOI] [PubMed] [Google Scholar]
- Anderson C, Crimmins S, Wilson JA et al: Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem 2005; 95: 724–731. [DOI] [PubMed] [Google Scholar]
- Wilson SM, Bhattacharyya B, Rachel RA et al: Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet 2002; 32: 420–425. [DOI] [PubMed] [Google Scholar]
- Villanueva P, Nudel R, Hoischen A et al: Exome sequencing in an admixed isolated population indicates NFXL1 variants confer a risk for specific language impairment. PLoS Genet 2015; 11: 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramyothin P, Pithukpakorn M, Arakaki RF: A 47, XXY patient and Xq21.31 duplication with features of Prader-Willi syndrome: results of array-based comparative genomic hybridization. Endocrine 2010; 37: 379–382. [DOI] [PubMed] [Google Scholar]
- Veerappa AM, Saldanha M, Padakannaya P, Ramachandra NB: Genome-wide copy number scan identifies disruption of PCDH11X in developmental dyslexia. Am J Med Genet B Neuropsychiatr Genet 2013; 162B: 889–897. [DOI] [PubMed] [Google Scholar]
- Esplin ED, Li B, Slavotinek A et al: Nine patients with Xp22.31 microduplication, cognitive deficits, seizures, and talipes anomalies. Am J Med Genet A 2014; 164A: 2097–2103. [DOI] [PubMed] [Google Scholar]
- Faletra F, D’Adamo AP, Santa Rocca M et al: Does the 1.5 Mb microduplication in chromosome band Xp22.31 have a pathogenetic role? New contribution and a review of the literature. Am J Med Genet A 2012; 158A: 461–464. [DOI] [PubMed] [Google Scholar]
- Li F, Shen Y, Köhler U et al: Interstitial microduplication of Xp22.31: Causative of intellectual disability or benign copy number variant? Eur J Med Genet 2010; 53: 93–99. [DOI] [PubMed] [Google Scholar]
- Männik K, Mägi R, Macé A et al: Copy number variations and cognitive phenotypes in unselected populations. JAMA 2015; 313: 2044–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.