Abstract
It has been shown previously that homozygous and compound-heterozygous variants affecting protein function in the human NLRP genes impact reproduction and/or fetal imprinting patterns. These variants represent so-called ‘maternal effect mutations’, that is, although female variant carriers are healthy, they are at risk of reproductive failure, and their offspring may develop aberrant methylation and imprinting disorders. In contrast, the relevance to reproductive failure of maternal heterozygous NLRP7 variants remains unclear. The present report describes the identification of a heterozygous NLRP7 variant in a healthy 28-year-old woman with a history of recurrent reproductive failure, and the molecular findings in two of the deceased offspring. Next-generation sequencing (NGS) for NLRP variants was performed. In the tissues of two offspring (one fetus; one deceased premature neonate) methylation of imprinted loci was tested using methylation-specific assays. Both pregnancies had been characterized by the presence of elevated human chorionic gonadotropin (hCG) levels and ovarian cysts. In the mother, a heterozygous nonsense 2-bp deletion in exon 5 of the NLRP7 gene was identified (NM_001127255.1:c.2010_2011del, p.(Phe671Glnfs*18)). In the two investigated offspring, heterogeneous aberrant methylation patterns were detected at imprinted loci. The present data support the hypothesis that heterozygous NLRP7 variants contribute to reproductive wastage, and that these variants represent autosomal dominant maternal effect variants which lead to aberrant imprinting marks in the offspring. Specific screening and close prenatal monitoring of NLRP7 variant carriers is proposed. Egg donation might facilitate successful pregnancy in heterozygous NLRP7 variant carriers.
Introduction
The NLRP (NOD-like receptor PYD, NALP) protein family comprises 14 members, and is a subgroup of the NLR (NOD-like receptor) protein family which has a key role in the innate immune system.1 In view of their high expression in the ovaries and in oocytes, previous authors have proposed that NLRP proteins have key functions in reproduction and early embryonic development.2 This hypothesis is supported by studies showing that genomic variants in the human genes NLRP2, NLRP5 and NLRP7 impact reproduction and/or fetal imprinting patterns.3, 4, 5, 6, 7, 8, 9
The mode of inheritance for NLRP variants differs from that observed in monogenetic disorders, as NLRP variants are maternal effect variants, that is, although female carriers are healthy, they are at risk of reproductive failure, whereas their offspring are at risk of aberrant methylation and imprinting disorders (IDs).3, 4
IDs are a group of rare clinically heterogeneous congenital disorders characterized by the same types of molecular alterations.10 These alterations (epimutations, uniparental disomies (UPDs); copy number variations (CNVs); point mutations) affect the balanced and parent-of-origin-specific expression of imprinted chromosomal regions and genes. Although UPDs, CNVs and point mutations represent genomic disturbances, epimutations involve no change in the DNA sequence, and the mechanisms through which they lead to a gain or loss in methylation remain largely unknown. However, in previous investigations of single cases, it could be shown that transacting variants in ZFP57 and NLRP genes resulted in aberrant methylation marks. The respective epimutations were observed in the patient (in the case of ZFP57), and in the offspring of a carrier (in the case of NLRP variants). In all the cases reported to date, aberrant methylation was detected at multiple imprinted loci (Multilocus Imprinting Disturbances (MLID)).3, 4
Maternal effect mutations in NLRP7 were first identified in women with a history of recurrent hydatidiform moles (HM), that is, human pregnancies characterized by failed embryonic development and excessive trophoblastic proliferation.5 Subsequently authors reported abnormal DNA methylation of imprinted DMRs in embryonic tissues obtained from the pregnancies of NLRP7 variant carriers.11, 12, 13 Recently, Sanchez-Delgado et al.14 demonstrated that the reproductive failure of NLRP7 variant carriers is associated with a defective placenta-specific imprinting and over-expression of maternally methylated transcripts. Together, these data indicate that NLRP7 is implicated in the establishment and/or maintenance of the maternal imprint.15 In 2014, Mahadevan et al.16 demonstrated that NLRP7 interacts with DMRs in a methylation-dependent manner.
Available studies have identified homozygous or compound-heterozygous variants in NLRP7 in a total of more than 130 women. Nearly all these women experienced reproductive failure, but single live-births from donated ova and even from spontaneous conceptions have also been reported (for review, Akoury et al.17). Besides HM, some of these aberrant pregnancies comprised mosaic aneuploidies.18 In pregnant heterozygous NLRP7 variant carriers and in in vitro experiments in human embryonic stem cells, increased levels of human chorionic gonadotropin (hCG) have been reported.6, 16 The contribution of homozygous or compound-heterozygous variants in the NLRP7 gene to the etiology of HM and other severe forms of reproductive failure is therefore well documented. In contrast, few data are available concerning the relevance to reproductive failure of heterozygous NLRP7 variants. To date, only single cases have been reported. However, these reports suggest that female heterozygous NLRP7 variant carriers are at increased risk of reproductive failure without necessarily the manifestation of a molar phenotype.19, 20 In previous studies, it has therefore been postulated that although heterozygous NLRP7 variant carriers are at risk of reproductive failure they have a better reproductive outcome compared with women with two defective alleles.7, 20
The present report describes the identification of a heterozygous frameshift variant in NLRP7 in a woman presenting with a history of reproductive failure and fetal MLID. Our data support the hypothesis that maternal heterozygous NLRP7 variants are associated with reproductive problems.
Materials and methods
Informed consent was obtained from the patient and her family, the study was approved by the ethical committee of the University Hospital Aachen.
DNA was extracted from lymphocytes using a conventional salting-out procedure. For other tissues (tendon, skin, heart muscle, liver, spleen, kidney), DNA was extracted using the Qiagen Mini-kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s instructions.
A quantitative methylation-specific Multiplex Ligation Dependent Probe Amplification assay (MS-MLPA; Kit ME030, MRC Holland, Amsterdam, The Netherlands) was used to identify alterations in methylation status of the following regions of chromosome 11p15.5: (i) telomeric imprinting control region 1 (ICR1, H19/IGF2: IG DMR); and (ii) the centromeric ICR2 (KCNQ1OT1: TSS DMR). Imprinted loci on chromosomes 6, 7, 14, 15 and 20 were analyzed using a methylation-specific single-nucleotide primer extension (MS-SNuPE) assay.21 Further information is available on request.
Next-generation sequencing (NGS) of six NLRP genes (NLRP1, NLRP2, NLRP3, NLRP7, NLRP12, NLRP14) was performed using the Illumina TruSight One assay and a MiSeq platform (paired end, 2 × 300 cycles)(Illumina, California/USA). Sequencing data were aligned to the hg19 reference genome (Burrows-Wheeler Aligner, BWA, v 0.7.5a), and processed using Picard (v. 1.95) to remove PCR duplicates. The Genome Analysis Toolkit (GATK v.3.0-0) was applied to perform local realignment, base quality recalibration, and SNP and indel identification (output in variant call format (vcf) files). Annotation of the raw vcf files was performed using the Illumina Variant Studio (v.2.2). All coding exons and exon/intron boundaries were covered with at least × 20.
To confirm variants detected by NGS, Sanger Sequencing was performed for exons 4 and 5 of the NLRP7 gene (NM_001127255.1). For variant classification, the bioinformatics prediction tools Alamut and PolyPhen-2 were used. The presence of second defective allele consisting of a deletion or a duplication in the genomic sequence of NLRP7 was excluded by SNP array analysis (CytoScan, Affymetrix, Wycombe, UK; Supplementary Figure 1).
Case report and molecular findings
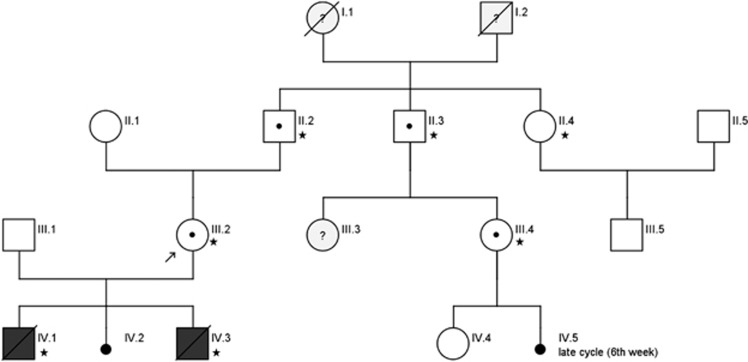
A healthy 28-year-old German woman was referred for genetic counseling having experienced one failed pregnancy due to preeclampsia and the suspicion of BWS. This event was followed by two other unsuccessful pregnancies within 2 years (Figure 1).
Figure 1.
Family pedigree. (* these family members were sequenced for the variant c.2010_2011del, p.(Phe671Glnfs*18)).
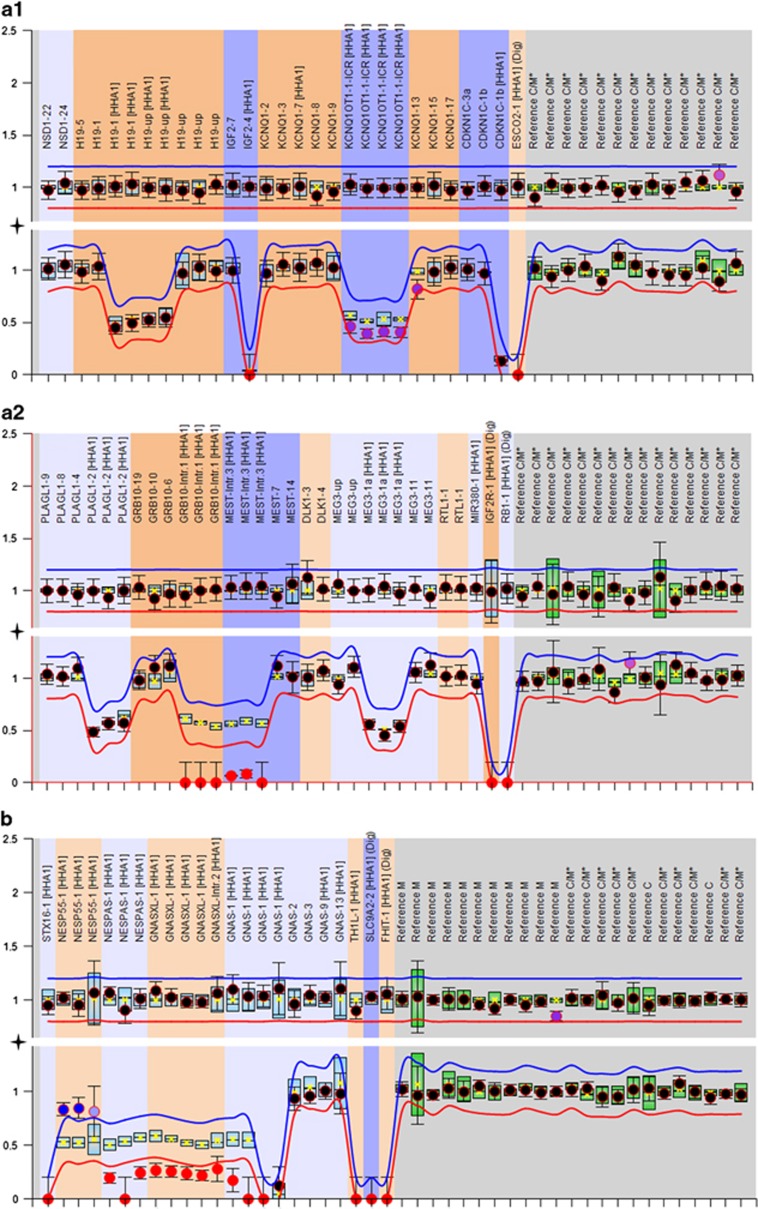
The first pregnancy had been complicated by severe preeclampsia, elevated hCG (>200 000 IU/L), and polyhydramnios at 21+5 weeks gestation. Fetal ultrasound had revealed mild macroglossia with otherwise normal biometry. In the mother, ultrasound examination had shown placental cysts of up to 9 mm in diameter, suggestive of mesenchymal dysplasia, and enlarged multicystic ovaries. Due to severe, life-threatening preeclampsia, the pregnancy was terminated at 22 weeks gestation. Fetal weight, head circumference and femur length were all within the normal range. In view of the ultrasonographic findings in the fetus, molecular testing for Beckwith–Wiedemann syndrome (BWS) was performed. MS-MLPA in cord blood revealed hypomethylation of KCNQ1OT1:TSS DMR (ICR2)in 11p15, which is consistent with a diagnosis of BWS (Figure 2). To investigate additional loci, molecular analysis of fetal tendon DNA was performed. This revealed hypomethylation of GRB10 and MEST on chromosome 7. These findings were also detected in the cord blood. In contrast, hypomethylation of KCNQ1OT1:TSS DMR was restricted to cord blood DNA only (Figure 2). Cytogenetic analysis of cord blood revealed a mosaic status for Turner Syndrome in the fetus (mos45,X[18]/46,XY[14]). Parental karyotypes were normal.
Figure 2.
Examples of copy number (upper tracks) and methylation profiles (lower tracks) from MS-MLPA of DNA of fetus IV.1 (a1 and a2) and fetus IV.3 (b). Each probe is represented by a dot and targets either an imprinted, a non-imprinted or a control sequence. Normal copy number results (upper tracks) are shown as black dots, hypomethylation is represented by purple/red dots, hypermethylation by blue dots. The arbitrary ratio border (red and blue line) indicates a 20% signal increase/loss. (a) MS-MLPA with a chromosome 11 specific kit (ME030, MRC Holland) in DNA from blood of fetus IV.1 shows a mild KCNQ1OT1 hypomethylation (a1) and a complete loss of methylation for GRB10 and MEST (red dots) on chromosome 7 (kit ME032) in DNA from tendon (a2). (b) For fetus IV.3, aberrant methylation was detected for chromosome 20 imprinted loci (kit ME031). Copy number analyses did not show any abnormalities.
The second pregnancy ended in a spontaneous abortion at 6 weeks with no further medical documentation.
In the third pregnancy, elevated βHCG (>200 000 IU/ml) levels and large multicystic ovaries were detected at 18+3 weeks. Placental and fetal ultrasound investigations were normal. However, amniotic fluid volume was in the upper normal range. In view of the similarities to the first pregnancy, amniocentesis was performed. MS-MLPA for chromosome 11 in amniotic fluid was unremarkable. However, further methylation studies revealed aberrant imprinting marks for MEST in 7q32, and for the GNAS locus in 20q13 (Figure 2). MEST hypomethylation corresponded to a paternal UPD of chromosome 7. This constitution is not associated with a clinical phenotype. However, the aberrant GNAS methylation is consistent with pseudohypoparathyroidism type 1b (PHP1B). Spontaneous preterm labor occurred at 24 weeks. The neonate died 16 h post delivery due to ruptures of the lungs. Postmortem macroscopic examination was unremarkable. However, subsequent molecular investigations of six different tissues from the deceased neonate (tendon, skin, heart muscle, liver, spleen and kidney) confirmed aberrant methylation at the 7q32 and 20q13 loci in all tissues.
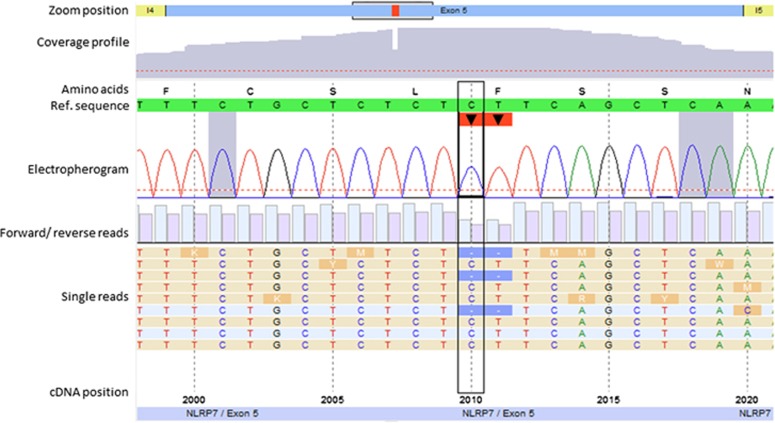
As research data suggest that recurrent MLIDs are caused by maternal effect variants in NLRP genes, sequencing of NLRP1, NLRP2, NLRP3, NLRP7, NLRP12 and NLRP14 was performed using a NGS approach (Illumina TruSight One assay). For five of the NLRP genes, no disease causing variant was detected. However, in NLRP7, a heterozygous 2 bp deletion variant in exon 5 was identified (NM_001127255.1:c.2010_2011del, p.(Phe671Glnfs*18); submitted to LOVD: www.lovd.nl/NLRP7 (Patient ID 00094948; variant #0000095349; Figure 3)). This variant leads to a premature stop codon 17 amino acids downstream, and is therefore classified to affect the protein function.
Figure 3.
Identification of the 2 bp deletion in exon 5 of the NLRP7 gene by clinical exome next-generation sequencing (c.2010_2011del, p.(Phe671Glnfs*18)) with a total coverage of 183 reads. The minimum coverage threshold was set to 30 reads (indicated by dashed red lines).The number of forward and reverse reads is represented by light and dark blue bars, respectively.
In addition, a rare heterozygous variant was identified in exon 4 (NM_001127255.1:c.574A>C, p.(Met192Leu), rs104895529; minor allele frequency (MAF) <1% LOVD entry: #0000168218). In a previous study,22 this variant was found in a patient with reproductive wastage in heterozygous state and in 2 out of 38 controls. Therefore, its pathogenicity remains unclear. However, in the present analysis, bioinformatic prediction tools generated ambiguous results for the above-mentioned variant, as small physicochemical differences between methionine and leucine might influence the interaction characteristics, due to the fact that the substitution occurs in the protein’s highly conserved NACHT domain.
Segregation analysis revealed that the patient had inherited both variants on the same allele, and that this allele had been inherited from her father (Figure 1). DNA from two female relatives of the index patient was also analyzed. A paternal aunt with one healthy son (II.4) did not carry the variant. A paternal cousin (III.4) with one healthy daughter (IV.4) carried the variant. This cousin reported that she had experienced a 2 weeks prolonged menstrual cycle once, and assumed this might have been a very early spontaneous abortion. (IV.5). No further data on this presumed pregnancy were available. In the fetal DNA samples of the first and the third pregnancy of the proposita (IV.1, IV.3), the variation could not be detected. No history of reproductive problems were reported in the maternal side of the family.
Discussion
Previous research has established an association between homozygous or compound-heterozygous maternal effect variants in the NLRP7 gene and hydatidiform moles/reproductive wastage. The present findings support the hypothesis that although heterozygous NLRP7 variant carriers are at increased risk of reproductive wastage, they do not develop the HM phenotype.19, 20 Our data also support the hypothesis that heterozygous maternal effect variants in NLRP7 lead to reduced survival and methylation pattern alterations in the offspring.6, 23 Additional evidence for this conclusion was generated by segregation analysis (Figure 1), as the variant was not detected in the paternal aunt who had no history of pregnancy-related complications. If the delayed menstrual bleeding reported by the paternal cousin did indeed represent a spontaneous abortion, our findings would provide further support for the hypothesis that female heterozygous NLRP7 variant carriers are at an increased risk of miscarriage. However, as no medical data are available to confirm the pregnancy, this remains speculative. Furthermore, the fact that the cousin had a healthy daughter indicates that heterozygous NLRP7 variant carriers are able to have healthy offspring, as documented in previous reports.7
Our index patient carried a two basepair deletion (c.2010_2011del, p.(Phe671Glnfs*18), NM_001127255.1; Figure 3) resulting in a frameshift with a stop codon 17 amino acids downstream. Bioinformatic analysis predicted a nonsense-mediated mRNA decay (NMD) for the translated mRNA product. In a previous study by Qian et al.,19 heterozygosity for a variant leading to a premature stop in NLRP7, p.(Glu99*)(c.295G>T), was detected in a woman with a history of one stillbirth and three normal pregnancies. Assuming that both mutated transcripts undergo NMD, a similar aetiopathology and clinical course might be expected in the two families. But the two women with functionally comparable genomic alterations show differences in reproductive success: In contrast to the p.(Glu99*) carrier reported by Qian et al., none of the three pregnancies experienced by our index patient resulted in a healthy live-birth. We therefore hypothesize that NMD is not the underlying pathomechanism, at least in the present family. We rather suggest that the frameshift variant has immediate effect on two functionally relevant domains of the NLRP7 protein. The variant p.(Glu99*)19 results in the absence of most of the binding domains, including the NACHT domain and the leucine-rich repeat domains (LRR). The NACHT domain and the LRRs are involved in the ATP-dependent formation of homodimers and in mediating protein–protein interactions, respectively. Thus, as a result of the p.(Glu99*) variant, only 50% of the translated proteins are able to form homodimers and interact with binding partners through their LRR domain. In contrast, the mutated protein caused by the variant c.2010_2011del (p.(Phe671Glnfs*18)) includes the NACHT domain but lacks the LRR. We postulate that although the corresponding truncated protein is expressed in the oocytes, it captures intact proteins through self-oligomerization, and thereby reduces the dosage of functional NLRP7 homodimers. In consequence, this dominant negative effect leads to a more serious phenotype during early fetal development – even in heterozygous cases – than that observed for the p.(Glu99*) variant, whereas the latter has a more severe but recessive impact on protein function.
The aberrant imprinting marks detected in the present offspring are consistent with those reported in a recent investigation by Caliebe et al.6 of a heterozygous NLRP7 variant affecting the methylation status of imprinted loci in the offspring. In the present study, the methylation patterns differed both inter-individually and intra-individually. As demonstrated in previous studies on genome-wide methylation of sporadic HMs, the methylation profile of imprinted loci is paternalized, which results in general loss of methylation at maternally derived loci, whereas paternal DMRs showed complete methylation14 However, these observations were made for sporadic HMs only. In offsprings of NLRP7 variant carriers, methylation aberrations in the sense of loss of methylation were restricted to the maternal DMRs.14 This observation is supported by the findings of the present study. The aberrant methylation changes detected in the offspring of the heterozygous NLRP7 carrier were limited to maternally methylated loci (loss of maternal methylation), thus indicating that NLRP7 has a key role in setting proper maternal methylation marks during early embryonic development.
This observation is consistent with findings for the recently reported variant NM_206828.3:c.2156C>T (p.(Ala719Val)). Here, heterozygosity is also associated with aberrant methylation in the offspring.6, 7, 18 Interestingly, although the women described in these three reports also experienced early pregnancy loss and/or partial hydatidiform moles, two individuals had a successful pregnancy.
The present case demonstrates the difficulties to predict the resulting phenotype in case of prenatally detected aberrant methylation patterns, as the clinical and molecular findings in the three offspring were inconsistent (ie, prenatal screening is already performed in clinical practice). The first offspring carried a 11p15 epimutation, which is associated with BWS, but convincing macroscopic features of BWS were not apparent. In the third pregnancy, aberrant methylation at the GNAS locus was detected, which is consistent with PHPIb. However, PHPIb normalizes post delivery and cannot be diagnosed prenatally. As the first pregnancy was terminated and the third offspring died shortly after birth, the question of whether PHPIb would have arisen in later life remains unanswered. The uncertainties associated with the prenatal prediction of an ID phenotype on the basis of the molecular analysis of single loci in single tissues limit the predictive value of prenatal testing results for these disorders,24 due to the lack of information of the mosaic distribution of epimutations and the restriction of testing to specific imprinted loci.
In consequence, the prenatal identification of aberrant imprinting marks for specific IDs represents a dilemma for genetic counselors. Furthermore, it remains unclear – at least in the present case – whether the aberrant methylation is of genuine clinical relevance, or whether it simply represents a biomarker for disturbances in imprinting secondary to NLRP7 variants. In the latter case, it remains unclear which of the loci that exhibit aberrant imprinting marks will be dominant in terms of phenotypic expression.
In addition to aberrations in methylation, the first fetus of the patient displayed a mosaic status for Turner Syndrome in more than 50% of the investigated cells (mos45,X[18]/46, XY[14]). This observation is consistent with the study of Deveault et al.18 demonstrating early cleavage abnormalities in two patients with NLRP7 variants. An important issue, therefore, is whether egg donation or pre-implantation genetic diagnosis (PGD) might increase the chances of a successful pregnancy in female NLRP7 variant carriers. In the oocyte, the NLRP7 protein is transcribed from the maternal genome, and is necessary for normal fetal development until the embryonic genome is activated.25 In recent studies, donor eggs containing intact ooplasm led to successful pregnancies in women with maternal effect variants in NLRP7.17 Although aberrant NLRP7 protein may be present in the maternal oocyte and ooplasm, PGD is focused on variants in the embryonic genome. Thus, egg donation and PGD are potential facilitators of successful pregnancy.17 However, further data are warranted before firm conclusions can be drawn.
In summary, the present data support the hypothesis that maternal heterozygous NLRP7 variants contribute to reproductive failure, and that these variants represent autosomal dominant maternal effect variants resulting in MLID. Further studies are warranted to determine whether preeclampsia, elevated hCG and/or ovarian cysts are frequent phenotypic manifestations of maternal heterozygous NLRP7 variants, and whether specific screening (eg, for fetal MLID) and intensive prenatal monitoring should be recommended for affected women. Egg donation might facilitate successful pregnancy in NLRP7 variant carriers.
Acknowledgments
We thank the patient and her family for their cooperation. LS, MB and TE are supported by the German Federal Ministry of Education and Research (Network ‘Imprinting Diseases’ 01GM1513B). They are members of the COST Action BM1208 and EUCID.net (European Congenital Imprinting Disorders Network; www.imprinting-disorders.eu). The NGS facility is funded by the Deutsche Forschungsgemeinschaft DFG (INST948/32-1FUGG).
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Broz P, Monack DM: Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol 2013; 13: 551–565. [DOI] [PubMed] [Google Scholar]
- Van Gorp H, Kuchmiy A, Van Hauwermeiren F, Lamkanfi M: NOD-like receptors interfacing the immune and reproductive systems. FEBS J 2014; 281: 4568–4582. [DOI] [PubMed] [Google Scholar]
- Meyer E, Lim D, Pasha S et al: Germline mutation in NLRP2 (NALP2) in a familial imprinting disorder (Beckwith-Wiedemann Syndrome). PLoS Genet 2009; 5: e1000423–e1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty LE, Rezwan FI, Poole RL et al: Mutations in NLRP5 are associated with reproductive wastage and multilocus imprinting disorders in humans. Nat Commun 2015; 6: 8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch S, Djuric U, Mazhar B et al: Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet 2006; 38: 300–302. [DOI] [PubMed] [Google Scholar]
- Caliebe A, Richter J, Ammerpohl O et al: A familial disorder of altered DNA-methylation. J Med Genet 2014; 51: 407–412. [DOI] [PubMed] [Google Scholar]
- Messaed C, Chebaro W, Roberto RBD et al: NLRP7 in the spectrum of reproductive wastage: rare non-synonymous variants confer genetic susceptibility to recurrent reproductive wastage. J Med Genet 2011; 48: 540–548. [DOI] [PubMed] [Google Scholar]
- Landolsi H, Rittore C, Philibert L et al: Screening for NLRP7 mutations in familial and sporadic recurrent hydatidiform moles: report of 2 Tunisian families. Int J Gynecol Pathol Off J Int Soc Gynecol Pathol 2011; 30: 348–353. [DOI] [PubMed] [Google Scholar]
- Landolsi H, Rittore C, Philibert L et al: NLRP7 mutation analysis in sporadic hydatidiform moles in Tunisian patients: NLRP7 and sporadic mole. Arch Pathol Lab Med 2012; 136: 646–651. [DOI] [PubMed] [Google Scholar]
- Eggermann T, Perez de Nanclares G, Maher ER et al: Imprinting disorders: a group of congenital disorders with overlapping patterns of molecular changes affecting imprinted loci. Clin Epigenetics 2015; 7: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward BE, De Vos M, Talati N et al: Genetic and epigenetic analysis of recurrent hydatidiform mole. Hum Mutat 2009; 30: E629–E639. [DOI] [PubMed] [Google Scholar]
- Kou YC, Shao L, Peng HH et al: A recurrent intragenic genomic duplication, other novel mutations in NLRP7 and imprinting defects in recurrent biparental hydatidiform moles. Mol Hum Reprod 2008; 14: 33–40. [DOI] [PubMed] [Google Scholar]
- Ito Y, Maehara K, Kaneki E et al: Novel nonsense mutation in the NLRP7 gene associated with recurrent hydatidiform mole. Gynecol Obstet Invest 2015; 81: 353–358. [DOI] [PubMed] [Google Scholar]
- Sanchez-Delgado M, Martin-Trujillo A, Tayama C et al: Absence of maternal methylation in biparental hydatidiform moles from women with NLRP7 maternal-effect mutations reveals widespread placenta-specific imprinting. PLoS Genet 2015; 11: e1005644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallahian M, Sebire NJ, Savage PM, Seckl MJ, Fisher RA: Mutations in NLRP7 and KHDC3L confer a complete hydatidiform mole phenotype on digynic triploid conceptions. Hum Mutat 2013; 34: 301–308. [DOI] [PubMed] [Google Scholar]
- Mahadevan S, Wen S, Wan Y-W et al: NLRP7 affects trophoblast lineage differentiation, binds to overexpressed YY1 and alters CpG methylation. Hum Mol Genet 2014; 23: 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akoury E, Gupta N, Bagga R et al: Live births in women with recurrent hydatidiform mole and two NLRP7 mutations. Reprod Biomed Online 2015; 31: 120–124. [DOI] [PubMed] [Google Scholar]
- Deveault C, Qian JH, Chebaro W et al: NLRP7 mutations in women with diploid androgenetic and triploid moles: a proposed mechanism for mole formation. Hum Mol Genet 2009; 18: 888–897. [DOI] [PubMed] [Google Scholar]
- Qian J, Deveault C, Bagga R, Xie X, Slim R: Women heterozygous for NALP7/NLRP7 mutations are at risk for reproductive wastage: report of two novel mutations. Hum Mutat 2007; 28: 741–741. [DOI] [PubMed] [Google Scholar]
- Slim R, Wallace EP: NLRP7 and the genetics of hydatidiform moles: recent advances and new challenges. Front Immunol 2013; 4: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann M, Leisten I, Soellner L, Zerres K, Eggermann T, Spengler S: Use of multilocus methylation-specific single nucleotide primer extension (MS-SNuPE) technology in diagnostic testing for human imprinted loci. Epigenetics 2012; 7: 473–481. [DOI] [PubMed] [Google Scholar]
- Slim R, Bagga R, Chebaro W, Srinivasan R, Agarwal N: A strong founder effect for two NLRP7 mutations in the Indian population: an intriguing observation. Clin Genet 2009; 76: 292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beygo J, Ammerpohl O, Gritzan D et al: Deep bisulfite sequencing of aberrantly methylated loci in a patient with multiple methylation defects. PLoS One 2013; 8: e76953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermann T, Brioude F, Russo S et al: Prenatal molecular testing for Beckwith-Wiedemann and Silver-Russell syndromes: a challenge for molecular analysis and genetic counseling. Eur J Hum Genet 2015; 24: 784–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akoury E, Zhang L, Ao A, Slim R: NLRP7 and KHDC3L, the two maternal-effect proteins responsible for recurrent hydatidiform moles, co-localize to the oocyte cytoskeleton. Hum Reprod 2015; 30: 159–169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.