Abstract
Circular RNAs (circRNAs) are implicated in a variety of cancers. However, the roles of circRNAs in gastric cancer (GC) remain largely unknown. In the current study, circRNAs expression profiles were screened in GC, using 5 pairs of GC and matched non-GC tissues with circRNA chip. Preliminary results were verified with quantitative PCR (qRT-PCR). Briefly, total of 713 circRNAs were differentially expressed in GC tissues vs. non-GC tissues (fold change ≥ 2.0, p < 0.05): 191 were upregulated, whereas 522 were downregulated in GC tissues. qRT-PCR analysis of randomly selected 7 circRNAs from the 713 circRNAs in 50 paired of GC vs. non-GC control tissues confirmed the microarray data. Gene ontology (GO) and KEGG pathway analyses showed that many circRNAs are implicated in carcinogenesis. Among differentially expressed circRNAs, hsa_circ_0076304, hsa_circ_0035431, and hsa_circ_0076305 had the highest magnitude of change. These results provided a preliminary landscape of circRNAs expression profile in GC.
Introduction
Gastric cancer (GC) is one of the most common cancers worldwide1. Diagnosis and treatment have improved over the last decades, but the 5-year survival rate remains low in patients with advanced GC2. A lack of reliable and efficient early diagnostic biomarkers, as well as, poorly understood molecular mechanisms of this disease is a major factor. To improve patient outcome, identifying effective biomarkers with early diagnostic value is essential. Novel biomarkers may also reflect the characteristics of cancer and clarify the molecular mechanisms of GC.
Over the past decade, the roles of non-coding RNA in cancer have been under intense investigation, encompassing miRNAs to long non-coding RNAs (lncRNA) and recently identified circular RNAs (circRNAs)3, 4. Accumulating evidence has demonstrated that both miRNAs and lncRNAs are closely associated with human cancers; many play crucial roles in cancer progression. Recent studies have implicated circRNAs in cancer development5. However, there have been relatively few reports describing circRNAs in GC.
CircRNAs are novel circular non-coding RNAs that are covalently closed6. CircRNAs could mediate the activity of microRNAs through binding and functioning as their sponges. Increasing evidence has suggested that circRNAs are often abnormally expressed in human cancers, and contribute to oncogenesis through miRNAs7. CircRNAs regulate cancer-related pathways and linear RNA transcription as well as protein expression8, 9. However, the expression levels and potential roles of circRNAs in GC are still poorly understood. In the present study, we investigated the alteration of circRNA expression profiles in GC tissues.
Materials and Methods
Tissue samples
A total of 55 patients (44 men and 11 women; mean age 59.8 years with a range of 23–81) with GC who underwent radical resection of the primary lesions between June 2014 and May 2015 at the Fuzhou General Hospital were included in this study. All tissues were histologically identified, diagnosed as gastric adenocarcinoma, and graded according to the guidelines of modified American Joint Committee on Cancer (AJCC). The initial screening step (Table 1) was conducted with microarray chip assay in 5 pairs of GC vs. non-GC tissue sample; the remaining 50 pairs were used for verification with quantitative reverse transcription PCR (qRT-PCR). Prior to analysis, all tissue samples were processed using a previously published method10 and stored at −80 °C.
Table 1.
The information of patients with gastric cancer subjected to circRNA expression profile chip assay.
| NO | Gender (male or female) | Age (years) | Histological type | Histologic differentiation | TNM stage |
|---|---|---|---|---|---|
| 286 | M | 74 | Ulcerative | Moderately | T2N0Mx |
| 287 | M | 74 | Ulcerative | Poorly | T4aN1Mx |
| 292 | M | 78 | Ulcerative | Moderately | T4aN2Mx |
| 313 | M | 58 | Ulcerative | Moderately-poorly | T2N0Mx |
| 326 | M | 61 | Ulcerative | Moderately | T4aN0Mx |
RNA preparation for chip assay
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) were used to isolate and purify total RNA. Sample quality (purity) was verified using standard spectrophotometer (ND-1000). The RNA integrity was assessed by electrophoresis with denaturing agarose gel.
Labeling and hybridization
After removing linear RNAs with ribonuclease R, RNA (5 μg from each sample) was reverse transcribed into cDNA using random primers containing T7 promoter by First Strand Enzyme Mix Kit. The DNA-RNA mixture was transformed to the second strand DNA by Second Strand Enzyme Mix. This DNA was used as a template to synthesize cRNA by the T7 enzyme mix. Subsequently, the cRNA was used as a template to obtain cDNA by reverse transcription through CbcScript II enzyme combined with random primers. This cDNA, in turn, was used as a template to synthesize the complementary strand DNA labeled fluorescently by Klenow Fragment enzyme combined with random primers and dNTP with fluorescent tags such as Cy3-dCTP and Cy5-dCTP. The samples were then hybridized with a CapitalBio Technology Human CircRNA Array v1 (Agilent, USA).
Signals were scanned by Agilent G2565CA Microarray Scanner. Images were introduced into Agilent Feature Extraction to obtain raw data (v10.7). Differential expression was analyzed with Agilent GeneSpring software, and the processing included raw data quantile normalization and data analysis. Post quantile normalization by log2-ratio, low-intensity filtering was conducted, and circRNAs with at least 60 percent samples flagged as “Detected” were selected for further analysis: differentially expressed circRNAs were analyzed with Independent samples t-test. CircRNAs with ≥2.0 fold-changes (FC) and p < 0.05 were selected as circRNAs with significant differential expression11.
Bioinformatics analysis
circRNA targets identified with profiling data were subjected to gene ontology (GO) and KEGG pathway analyses based on their correlated mRNAs using Gene Ontology (http://www.geneongoloty.org/) and KOBAS software (KEGG Orthology-Based Annotation System). The differentially expressed circRNAs-targeted miRNAs were sought and predicted by miRanda software coupled with statistical analysis. In order to understand the association between circRNAs and their related miRNAs, 3 most significantly altered circRNAs were used to draw the circRNA-miRNA network using miRanda combined with patterning software. The circRNAs expression profile microarray chip assay and data and bioinformatics analysis were carried out by Capitalbio Corporation (Beijing, China).
qRT-PCR assay
Total RNA was extracted by TRIzol reagent as described previously10. The expression levels of 7 randomly selected differentially expressing circRNAs (Fold changes ≥ 2, p < 0.05) were measured by qRT-PCR; among them, 2 were upregulated and 5 were downregulated in the GC tissues: (upregulated: hsa_circ_0081146, hsa_circ_0084720), (downregulated: hsa_circ_0060108, hsa_circ_0057104, hsa_circ_0054971, hsa_circ_0063561, and hsa_circ_0058766). GAPDH expression was used as an internal reference. The primers used for these amplifications are listed in Table S1. PCRs were a relative estimation in triplicate as per the following temperature profile:denaturation 95 °C for 10 min followed by amplification by 40 cycles of 95 °C for 10 s and 60 °C for 1 min10.
Statistical analysis
For comparisons involving multiple groups, data were analyzed by analysis of variance (ANOVA); For analysis involving only two groups, data were analyzed with Student’s t-test. Results are expressed as the mean ± SEM. p < 0.05 was regarded as statistically significant. Data analysis was performed by Statistical Program for Social Sciences (SPSS) 22.0 software (SPSS, Chicago, IL, USA).
Compliance with ethical standards
The tissue samples used in this study were obtained with patients informed consent. All the methods were performed in compliance with the permitted or institutional protocols.This study was approved by the Fuzhou General Hospital Ethics Committee (No. 2014CXTD04). This article does not contain any studies with animals performed by any of the authors.
Results
CircRNAs expression profiles in GC
The microarray screening detected a total of 62,998 circRNAs, in GC, non-GC or both tissues (such information could be accessed with GSE100170 at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE100170). As illustrated in Fig. 1, 713 of these exhibited differential expressions between GC and non-GC tissues (FC ≥ 2.0, p < 0.05) (Table S2), among which 191 were upregulated and the remaining 522 were downregulated in cancer tissues. A total of 207 circRNAs were differentially expressed between GC and non-GC tissues by both long and short probes (the two kinds of probe were named CBC1 and CBC2, respectively), among which 57 were upregulated and 150 were downregulated. The magnitude of fold change was highest for hsa_circ_0044516 in upregulated circRNAs (fold changes = 6.28, p = 0.036), and hsa_circ_0076305 for downregulated circRNAs (fold changes = −125.95, p = 0.030). Hierarchical clustering (Fig. 1A), volcano plots (Fig. 1B), and scatter plots (Fig. 1C) revealed that the expression profiles of circRNAs between GC and non-GC tissues were diverse. The top up- and down-regulated circRNAs are displayed in Table 2.
Figure 1.
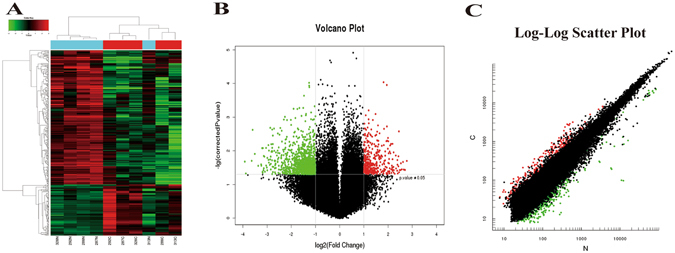
Hierarchical clustering, volcano plots, and scatter plots exhibited the differentially expressed circRNAs in gastric cancer tissues compared to paired non-gastric cancer tissues. (A) Hierarchical clustering, numbers were the samples used for the microarray assay. C: cancer tissues, N: non-cancerous tissues. (B) Differentially expressed circRNAs were displayed by volcano plots. The green and red parts indicated >2 fold-decreased and -increased expression of the dysregulated circRNAs in GC tissues, respectively (p < 0.05). (C) Differentially expressed circRNAs were displayed by scatter plots. The green and red parts indicated >2 fold-decreased and -increased expression of the dysregulated circRNAs in GC tissues (p < 0.05).
Table 2.
The top up- and down-regulated differentially expressed circRNAs in GC tissues compared to those in non-cancerous tissues by both probes.
| Name | Probe CBC1 | Probe CBC2 | chr | gene symbol | ||||
|---|---|---|---|---|---|---|---|---|
| p | FC (abs) | Regulation | p | FC (abs) | Regulation | |||
| hsa_circ_0076305 | 0.029925665 | 125.95259 | down | 0.03022709 | 109.61993 | down | chr6 | PGC |
| hsa_circ_0076304 | 0.014676455 | 31.56073 | down | 0.011826442 | 28.374382 | down | chr6 | PGC |
| hsa_circ_0035431 | 0.00300389 | 20.631256 | down | 0.003938972 | 21.39568 | down | chr15 | CGNL1 |
| hsa_circ_0000390 | 0.01983777 | 12.444183 | down | 0.026148466 | 7.417858 | down | chr12 | FGD4 |
| hsa_circ_0037362 | 0.027888432 | 10.314745 | down | 0.014241457 | 8.81237 | down | chr16 | C16orf73 |
| hsa_circ_0076307 | 0.025423396 | 9.498694 | down | 0.020947674 | 15.158471 | down | chr6 | PGC |
| hsa_circ_0037361 | 0.027664987 | 9.436992 | down | 0.002388634 | 12.108136 | down | chr16 | C16orf73 |
| hsa_circ_0007315 | 0.042465463 | 9.14959 | down | 0.03788745 | 8.887227 | down | chr3 | PVRL3 |
| hsa_circ_0027969 | 0.014839446 | 8.641936 | down | 0.03794105 | 7.963011 | down | chr12 | SLC41A2 |
| hsa_circ_0001679 | 0.031237341 | 8.059054 | down | 0.04918231 | 6.926098 | down | chr7 | GLCCI1 |
| hsa_circ_0035435 | 0.014802514 | 7.2164187 | down | 0.045140408 | 5.798211 | down | chr15 | CGNL1 |
| hsa_circ_0027971 | 0.03575782 | 7.201395 | down | 0.008316714 | 9.217224 | down | chr12 | SLC41A2 |
| hsa_circ_0073770 | 0.008143293 | 6.911025 | down | 0.006314759 | 6.7912025 | down | chr5 | SLC12A2 |
| hsa_circ_0074239 | 0.002184121 | 6.5593286 | down | 0.003311638 | 5.6043277 | down | chr5 | C5orf32 |
| hsa_circ_0051995 | 0.020385692 | 6.4125986 | down | 0.027742783 | 5.7211037 | down | chr19 | VRK3 |
| hsa_circ_0025842 | 0.024139885 | 6.305559 | down | 0.019139148 | 6.9465156 | down | chr12 | FGD4 |
| hsa_circ_0077736 | 0.048207846 | 6.2890162 | down | 0.033191722 | 6.435054 | down | chr6 | CEP85L |
| hsa_circ_0006034 | 0.01381651 | 6.038208 | down | 0.003529549 | 6.7874093 | down | chr5 | SLC12A2 |
| hsa_circ_0066971 | 0.011570533 | 5.8554688 | down | 0.036805384 | 5.452018 | down | chr3 | EAF2 |
| hsa_circ_0035432 | 0.011778097 | 5.5990286 | down | 0.011175622 | 6.2109637 | down | chr15 | CGNL1 |
| hsa_circ_0044516 | 0.036079686 | 6.276136 | up | 0.04336383 | 5.479236 | up | chr17 | COL1A1 |
| hsa_circ_0044518 | 0.037375137 | 5.72242 | up | 0.04342119 | 3.3064938 | up | chr17 | COL1A1 |
| hsa_circ_0077033 | 0.018847013 | 5.221709 | up | 0.023423905 | 5.7803392 | up | chr6 | COL12A1 |
| hsa_circ_0006401 | 0.013833045 | 4.9873238 | up | 0.017758103 | 4.545301 | up | chr2 | COL6A3 |
| hsa_circ_0081090 | 0.036175337 | 4.5390043 | up | 0.021447672 | 6.136692 | up | chr7 | COL1A2 |
| hsa_circ_0058132 | 0.045273677 | 4.3983254 | up | 0.022904716 | 4.0176225 | up | chr2 | FN1 |
| hsa_circ_0081146 | 0.010543266 | 4.187957 | up | 0.011812083 | 4.0625715 | up | chr7 | COL1A2 |
| hsa_circ_0058100 | 0.047036752 | 4.166924 | up | 0.04413781 | 4.6881175 | up | chr2 | FN1 |
| hsa_circ_0081091 | 0.008671624 | 3.9734251 | up | 0.014162468 | 3.2844515 | up | chr7 | COL1A2 |
| hsa_circ_0091742 | 0.005013032 | 3.8982337 | up | 0.014553196 | 5.383641 | up | chrX | BGN |
| hsa_circ_0058097 | 0.049916536 | 3.8704908 | up | 0.033016972 | 4.187093 | up | chr2 | FN1 |
| hsa_circ_0081136 | 0.013954006 | 3.6851623 | up | 0.021678727 | 3.2075522 | up | chr7 | COL1A2 |
| hsa_circ_0044519 | 0.03606566 | 3.6805367 | up | 0.03702461 | 6.5064387 | up | chr17 | COL1A1 |
| hsa_circ_0016294 | 0.034532506 | 3.5717776 | up | 0.038935255 | 3.4514186 | up | chr1 | CD55 |
| hsa_circ_0081143 | 0.002223194 | 3.5639381 | up | 0.03638 | 3.226277 | up | chr7 | COL1A2 |
| hsa_circ_0044515 | 0.048010923 | 3.5043917 | up | 0.047074426 | 5.2711334 | up | chr17 | COL1A1 |
| hsa_circ_0081066 | 0.040320095 | 3.4538522 | up | 0.006778345 | 3.5394926 | up | chr7 | COL1A2 |
| hsa_circ_0016292 | 0.016833305 | 3.2611954 | up | 0.017872557 | 3.1238286 | up | chr1 | CD55 |
| hsa_circ_0091743 | 0.015532353 | 3.1929755 | up | 0.006474511 | 3.045686 | up | chrX | BGN |
| hsa_circ_0020788 | 0.005037079 | 3.030931 | up | 0.005184689 | 3.8050945 | up | chr11 | TCONS_00063837_H19 |
FC: Fold changes. abs: absolute value.
The results of qRT-PCR verification of the differentially expressed circRNAs
Seven differentially expressed circRNAs were randomly selected for qRT-PCR verification by using 50 paired of samples. The results confirmed the upregulation of hsa_circ_0081146 and hsa_circ_0084720 in GC, and downregulation of hsa_circ_0060108, hsa_circ_0057104, hsa_circ_0054971, hsa_circ_0063561, and hsa_circ_0058766 in GC (Fig. 2).
Figure 2.
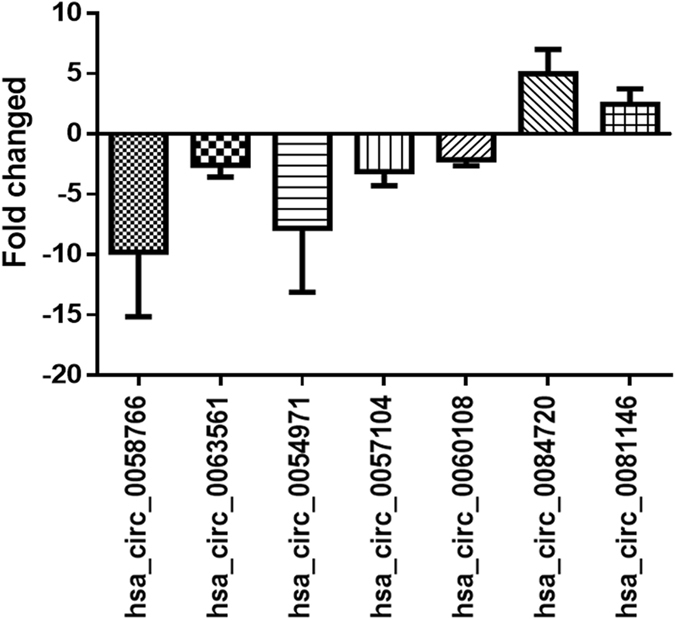
Verification of the differentially expressed circRNAs by qRT-PCR. The expression of 7 lncRNAs in 50 paired GC tissues was detected by qRT-PCR, which were shown by the expression fold changes. Comparison of the results obtained from qPCR and microarray assay revealed satisfactory consistency.
The results of bioinformatics analysis
Differentially expressed circRNAs could be mapped to all chromosomes, except for chromosome 21 and Y. A lot of miRNAs were predicted to be their targets (Table 3). 1026 miRNAs were predicted to be the targets of hsa_circ_0001210, which is an intragenic circRNA, located on chromosome 22 with a length of 25285 nt and downregulated in GC. 116 mRNAs were shown to be the potential corresponding linear transcripts of these dysregulated circRNAs (Table S3). GO, KEGG, and enrichment (Table S4) analyses suggest that these differentially expressed circRNAs are relevant to several vital physiological processes, cellular components, molecular functions, and critical signaling pathways such as growth factor binding, cell adhesion molecule binding, and response to transforming growth factor beta (TGF-β). Many of the known pathways associated with carcinogenesis, such as focal adhesion pathway, PI3K–Akt signaling pathway, and degradation of the extracellular matrix pathway were also implicated. Figure 3A–C illustrated the top 30 significantly enriched GO terms, pathway terms, and disease terms.
Table 3.
The numbers of potential targeted miRNAs of the differentially expressed circRNAs.
| ProbeName | p | FC (abs) | Regulation | chr | gene symbol | No. miRNA targets |
|---|---|---|---|---|---|---|
| hsa_circ_0006401 | 0.013833045 | 4.9873238 | up | chr2 | COL6A3 | 17 |
| hsa_circ_0014202 | 0.008794556 | 2.5519805 | up | chr1 | S100A10 | 4 |
| hsa_circ_0016292 | 0.016833305 | 3.2611954 | up | chr1 | CD55 | 0 |
| hsa_circ_0016294 | 0.034532506 | 3.5717776 | up | chr1 | CD55 | 0 |
| hsa_circ_0018424 | 0.004929639 | 2.2052047 | up | chr10 | BICC1 | 1 |
| hsa_circ_0020788 | 0.005037079 | 3.030931 | up | chr11 | TCONS_00063837_H19 | 0 |
| hsa_circ_0020790 | 0.002316416 | 2.4726825 | up | chr11 | TCONS_00063837_H19 | 0 |
| hsa_circ_0034428 | 0.03272182 | 2.6071007 | up | chr15 | THBS1 | 87 |
| hsa_circ_0034475 | 0.028014906 | 2.2003295 | up | chr15 | THBS1 | 23 |
| hsa_circ_0034495 | 0.018290553 | 2.2112164 | up | chr15 | THBS1 | 3 |
| hsa_circ_0034496 | 0.023052732 | 2.150319 | up | chr15 | THBS1 | 8 |
| hsa_circ_0035137 | 0.026556138 | 2.3628845 | up | chr15 | FBN1 | 1 |
| hsa_circ_0044513 | 0.044563152 | 2.5272765 | up | chr17 | COL1A1 | 46 |
| hsa_circ_0044515 | 0.048010923 | 3.5043917 | up | chr17 | COL1A1 | 180 |
| hsa_circ_0044516 | 0.036079686 | 6.276136 | up | chr17 | COL1A1 | 191 |
| hsa_circ_0044517 | 0.03509533 | 2.7368069 | up | chr17 | COL1A1 | 230 |
| hsa_circ_0044518 | 0.037375137 | 5.72242 | up | chr17 | COL1A1 | 245 |
| hsa_circ_0044519 | 0.03606566 | 3.6805367 | up | chr17 | COL1A1 | 267 |
| hsa_circ_0044521 | 0.03614439 | 2.4449718 | up | chr17 | COL1A1 | 176 |
| hsa_circ_0046707 | 0.033796836 | 2.1029172 | up | chr18 | SMCHD1 | 5 |
| hsa_circ_0057391 | 0.04772664 | 2.256704 | up | chr2 | COL3A1 | 193 |
| hsa_circ_0057403 | 0.03509203 | 2.146545 | up | chr2 | COL3A1 | 46 |
| hsa_circ_0058097 | 0.049916536 | 3.8704908 | up | chr2 | FN1 | 159 |
| hsa_circ_0058100 | 0.047036752 | 4.166924 | up | chr2 | FN1 | 56 |
| hsa_circ_0058132 | 0.045273677 | 4.3983254 | up | chr2 | FN1 | 9 |
| hsa_circ_0077033 | 0.018847013 | 5.221709 | up | chr6 | COL12A1 | 13 |
| hsa_circ_0077055 | 0.038407695 | 2.5744588 | up | chr6 | COL12A1 | 0 |
| hsa_circ_0077056 | 0.04552718 | 2.9007647 | up | chr6 | COL12A1 | 1 |
| hsa_circ_0077057 | 0.04089677 | 2.6701639 | up | chr6 | COL12A1 | 0 |
| hsa_circ_0080229 | 0.03741656 | 2.027931 | up | chr7 | EGFR | 0 |
| hsa_circ_0081066 | 0.040320095 | 3.4538522 | up | chr7 | COL1A2 | 243 |
| hsa_circ_0081084 | 0.032507733 | 2.7151222 | up | chr7 | COL1A2 | 136 |
| hsa_circ_0081089 | 0.03845984 | 2.0764067 | up | chr7 | COL1A2 | 163 |
| hsa_circ_0081090 | 0.036175337 | 4.5390043 | up | chr7 | COL1A2 | 227 |
| hsa_circ_0081091 | 0.008671624 | 3.9734251 | up | chr7 | COL1A2 | 260 |
| hsa_circ_0081092 | 0.044405155 | 2.7602344 | up | chr7 | COL1A2 | 4 |
| hsa_circ_0081111 | 0.005212786 | 4.0334992 | up | chr7 | COL1A2 | 237 |
| hsa_circ_0081125 | 0.013989674 | 2.8943481 | up | chr7 | COL1A2 | 182 |
| hsa_circ_0081136 | 0.013954006 | 3.6851623 | up | chr7 | COL1A2 | 156 |
| hsa_circ_0081137 | 0.039436605 | 2.9486153 | up | chr7 | COL1A2 | 112 |
| hsa_circ_0081138 | 0.047790088 | 2.2264013 | up | chr7 | COL1A2 | 121 |
| hsa_circ_0081143 | 0.002223194 | 3.5639381 | up | chr7 | COL1A2 | 89 |
| hsa_circ_0081146 | 0.010543266 | 4.187957 | up | chr7 | COL1A2 | 121 |
| hsa_circ_0081149 | 0.045875933 | 2.6191776 | up | chr7 | COL1A2 | 69 |
| hsa_circ_0081152 | 0.011682078 | 2.843584 | up | chr7 | COL1A2 | 99 |
| hsa_circ_0081155 | 0.00731891 | 2.8246753 | up | chr7 | COL1A2 | 94 |
| hsa_circ_0081159 | 0.006505117 | 2.5019772 | up | chr7 | COL1A2 | 76 |
| hsa_circ_0081160 | 0.04243376 | 2.0236626 | up | chr7 | COL1A2 | 4 |
| hsa_circ_0081163 | 0.044002496 | 2.5647683 | up | chr7 | COL1A2 | 10 |
| hsa_circ_0081167 | 0.0097287 | 3.1536498 | up | chr7 | COL1A2 | 24 |
| hsa_circ_0084720 | 0.02980772 | 3.712714 | up | chr8 | SULF1 | 4 |
| hsa_circ_0087215 | 0.023945637 | 2.500071 | up | chr9 | ANXA1 | |
| hsa_circ_0089433 | 0.008899449 | 2.1055155 | up | chr9 | COL5A1 | 364 |
| hsa_circ_0090450 | 1.11E-04 | 3.8579054 | up | chrX | TIMP1 | 11 |
| hsa_circ_0090452 | 0.003296774 | 2.235864 | up | chrX | TIMP1 | 5 |
| hsa_circ_0091742 | 0.005013032 | 3.8982337 | up | chrX | BGN | 192 |
| hsa_circ_0091743 | 0.015532353 | 3.1929755 | up | chrX | BGN | 181 |
| hsa_circ_0000019 | 0.020847283 | 2.5991628 | down | chr1 | DDI2 | 0 |
| hsa_circ_0000258 | 0.042388447 | 2.687611 | down | chr10 | PDCD11 | 0 |
| hsa_circ_0000390 | 0.01983777 | 12.444183 | down | chr12 | FGD4 | 0 |
| hsa_circ_0000580 | 0.04103441 | 4.64374 | down | chr14 | None | 0 |
| hsa_circ_0000615 | 0.047749862 | 5.541818 | down | chr15 | ZNF609 | 17 |
| hsa_circ_0000642 | 0.040490992 | 2.6040976 | down | chr15 | ZFAND6 | 2 |
| hsa_circ_0001074 | 0.030960364 | 3.0543585 | down | chr2 | ORC4 | 0 |
| hsa_circ_0001112 | 0.018396724 | 2.6625469 | down | chr2 | DGKD | 0 |
| hsa_circ_0001114 | 5.34E-04 | 3.171953 | down | chr2 | DGKD | 0 |
| hsa_circ_0001210 | 0.03488208 | 2.5489702 | down | chr22 | None | 1026 |
| hsa_circ_0001216 | 0.026226409 | 3.7022111 | down | chr22 | XBP1 | 2 |
| hsa_circ_0001438 | 0.029758973 | 6.8401494 | down | chr4 | LARP1B | 0 |
| hsa_circ_0001679 | 0.031237341 | 8.059054 | down | chr7 | GLCCI1 | 2 |
| hsa_circ_0001998 | 0.025869323 | 2.7446353 | down | chr14 | FUT8 | 0 |
| hsa_circ_0002110 | 0.018834386 | 2.806639 | down | chr12 | AMN1 | 0 |
| hsa_circ_0002138 | 0.032208133 | 3.2452636 | down | chr15 | USP3 | 0 |
| hsa_circ_0002190 | 0.030215895 | 2.5628076 | down | chr7 | KLHDC10 | 1 |
| hsa_circ_0002422 | 0.037262026 | 4.9764647 | down | chr3 | FNDC3B | 7 |
| hsa_circ_0002449 | 0.005389997 | 4.227186 | down | chr5 | C5orf32 | 3 |
| hsa_circ_0002504 | 0.03395744 | 2.3386385 | down | chr17 | ENGASE | 4 |
| hsa_circ_0003012 | 0.0350584 | 5.214465 | down | chr12 | SLC41A2 | 0 |
| hsa_circ_0003201 | 0.017809883 | 2.6226783 | down | chr4 | TBC1D14 | 0 |
| hsa_circ_0003787 | 0.009270906 | 5.055101 | down | chr5 | RGNEF | 2 |
| hsa_circ_0003911 | 0.0193295 | 3.0724642 | down | chr16 | CNOT1 | 8 |
| hsa_circ_0004689 | 0.043355685 | 6.3922377 | down | chr1 | SWT1 | 0 |
| hsa_circ_0005028 | 0.004585072 | 2.5328124 | down | chr3 | TSEN2 | 0 |
| hsa_circ_0005135 | 0.025206376 | 2.680219 | down | chr19 | LOC100506033 | 1 |
| hsa_circ_0006034 | 0.01381651 | 6.038208 | down | chr5 | SLC12A2 | 1 |
| hsa_circ_0006511 | 0.008116972 | 2.9554853 | down | chr2 | FARP2 | 2 |
| hsa_circ_0007315 | 0.042465463 | 9.14959 | down | chr3 | PVRL3 | 0 |
| hsa_circ_0007538 | 0.021333938 | 2.2191985 | down | chr1 | C1orf27 | 0 |
| hsa_circ_0007619 | 0.024274996 | 6.2581415 | down | chr4 | LARP1B | 0 |
| hsa_circ_0007715 | 0.02931771 | 3.3169591 | down | chr19 | CIRBP | 4 |
| hsa_circ_0007754 | 0.007596167 | 2.5428722 | down | chr13 | PCCA | 0 |
| hsa_circ_0007840 | 0.01876142 | 2.3082905 | down | chr15 | COX5A | 0 |
| hsa_circ_0008962 | 0.023675451 | 2.5180075 | down | chr5 | ELL2 | 0 |
| hsa_circ_0011092 | 0.027778216 | 2.8495584 | down | chr1 | STX12 | 0 |
| hsa_circ_0014614 | 0.046407204 | 3.509566 | down | chr1 | DAP3 | 1 |
| hsa_circ_0015948 | 0.036354423 | 2.187048 | down | chr1 | IPO9 | 4 |
| hsa_circ_0017445 | 0.032844365 | 4.300893 | down | chr10 | WDR37 | 0 |
| hsa_circ_0017974 | 0.003326846 | 2.0536025 | down | chr10 | KIAA1217 | 3 |
| hsa_circ_0020752 | 0.038835317 | 3.8155284 | down | chr11 | None | 18 |
| hsa_circ_0020757 | 0.045137018 | 3.7709012 | down | chr11 | None | 481 |
| hsa_circ_0020762 | 0.049438722 | 3.4821498 | down | chr11 | None | 729 |
| hsa_circ_0020763 | 0.024364103 | 7.188659 | down | chr11 | None | 623 |
| hsa_circ_0022351 | 0.048745744 | 2.1941059 | down | chr11 | C11orf9 | 22 |
| hsa_circ_0023597 | 0.025558302 | 2.48522 | down | chr11 | XRRA1 | 2 |
| hsa_circ_0025842 | 0.024139885 | 6.305559 | down | chr12 | FGD4 | 14 |
| hsa_circ_0025847 | 0.01065637 | 6.9226236 | down | chr12 | FGD4 | 0 |
| hsa_circ_0027969 | 0.014839446 | 8.641936 | down | chr12 | SLC41A2 | 1 |
| hsa_circ_0027971 | 0.03575782 | 7.201395 | down | chr12 | SLC41A2 | 1 |
| hsa_circ_0028323 | 0.03792535 | 4.157252 | down | chr12 | TMEM116 | 0 |
| hsa_circ_0029235 | 0.04259122 | 2.0699773 | down | chr12 | DDX55 | 0 |
| hsa_circ_0031281 | 0.016793832 | 2.7682478 | down | chr14 | SLC7A8 | 0 |
| hsa_circ_0031423 | 0.020782415 | 2.8116136 | down | chr14 | SCFD1 | 2 |
| hsa_circ_0035431 | 0.00300389 | 20.631256 | down | chr15 | CGNL1 | 46 |
| hsa_circ_0035432 | 0.011778097 | 5.5990286 | down | chr15 | CGNL1 | 74 |
| hsa_circ_0035435 | 0.014802514 | 7.2164187 | down | chr15 | CGNL1 | 9 |
| hsa_circ_0035875 | 0.039918724 | 2.113102 | down | chr15 | SPG21 | 0 |
| hsa_circ_0036510 | 0.028307231 | 2.6944883 | down | chr15 | ZFAND6 | 1 |
| hsa_circ_0037361 | 0.027664987 | 9.436992 | down | chr16 | C16orf73 | 0 |
| hsa_circ_0037362 | 0.027888432 | 10.314745 | down | chr16 | C16orf73 | 0 |
| hsa_circ_0037861 | 0.017640278 | 3.1812923 | down | chr16 | TXNDC11 | 0 |
| hsa_circ_0037862 | 0.02676344 | 5.1893096 | down | chr16 | TXNDC11 | 3 |
| hsa_circ_0037863 | 0.014628965 | 3.431537 | down | chr16 | TXNDC11 | 11 |
| hsa_circ_0038521 | 0.012550134 | 3.4246309 | down | chr16 | CDR2 | 41 |
| hsa_circ_0039090 | 0.008517502 | 2.1161213 | down | chr16 | SRCAP | 467 |
| hsa_circ_0039216 | 0.032217234 | 2.741417 | down | chr16 | GPT2 | 1 |
| hsa_circ_0039218 | 0.028291393 | 3.9539077 | down | chr16 | GPT2 | 15 |
| hsa_circ_0039271 | 0.047586497 | 2.770235 | down | chr16 | PHKB | 4 |
| hsa_circ_0039658 | 0.04334005 | 3.0598512 | down | chr16 | CNOT1 | 14 |
| hsa_circ_0039940 | 0.016690876 | 2.131468 | down | chr16 | SLC7A6 | 0 |
| hsa_circ_0040081 | 0.013927639 | 2.3884165 | down | chr16 | NQO1 | 0 |
| hsa_circ_0040373 | 0.021966241 | 3.0635164 | down | chr16 | AP1G1 | 11 |
| hsa_circ_0040388 | 0.018369772 | 2.0574872 | down | chr16 | AP1G1 | 3 |
| hsa_circ_0041440 | 0.023598213 | 2.3573241 | down | chr17 | RAP1GAP2 | 53 |
| hsa_circ_0042853 | 0.049323324 | 2.8299448 | down | chr17 | TCONS_00025103 | 0 |
| hsa_circ_0042968 | 0.027372789 | 2.9627814 | down | chr17 | SUZ12 | 1 |
| hsa_circ_0045272 | 0.041974243 | 2.0751245 | down | chr17 | ERN1 | 7 |
| hsa_circ_0047700 | 0.02186925 | 5.2570233 | down | chr18 | ME2 | 0 |
| hsa_circ_0047785 | 0.049457386 | 2.5616474 | down | chr18 | ATP8B1 | 6 |
| hsa_circ_0047975 | 0.044554852 | 2.6958208 | down | chr18 | ZNF236 | 40 |
| hsa_circ_0048201 | 0.024692174 | 2.7309535 | down | chr19 | STK11 | 110 |
| hsa_circ_0048536 | 0.049628958 | 2.0178084 | down | chr19 | EEF2 | 63 |
| hsa_circ_0049289 | 0.034041822 | 4.4006753 | down | chr19 | SLC44A2 | 226 |
| hsa_circ_0051047 | 0.04167667 | 2.575785 | down | chr19 | FCGBP | 432 |
| hsa_circ_0051050 | 0.043033753 | 3.2892206 | down | chr19 | FCGBP | 3 |
| hsa_circ_0051995 | 0.020385692 | 6.4125986 | down | chr19 | VRK3 | 14 |
| hsa_circ_0054086 | 0.032298878 | 3.289804 | down | chr2 | HEATR5B | 6 |
| hsa_circ_0054186 | 0.044439603 | 3.2368143 | down | chr2 | MAP4K3 | 0 |
| hsa_circ_0054970 | 0.030601952 | 5.295868 | down | chr2 | SLC1A4 | 0 |
| hsa_circ_0054971 | 0.02345602 | 3.2384007 | down | chr2 | SLC1A4 | 0 |
| hsa_circ_0056240 | 0.008702193 | 3.180426 | down | chr2 | PTPN4 | 0 |
| hsa_circ_0057104 | 0.047582004 | 4.0253515 | down | chr2 | PDK1 | 1 |
| hsa_circ_0057105 | 0.03679815 | 3.6738498 | down | chr2 | PDK1 | 2 |
| hsa_circ_0057106 | 0.047164652 | 4.9790606 | down | chr2 | PDK1 | 1 |
| hsa_circ_0057480 | 0.030882323 | 3.0643332 | down | chr2 | PMS1 | 1 |
| hsa_circ_0058443 | 0.008067183 | 4.725038 | down | chr2 | ACSL3 | 1 |
| hsa_circ_0058762 | 0.012233879 | 4.721563 | down | chr2 | DGKD | 0 |
| hsa_circ_0058766 | 0.002712246 | 3.6002114 | down | chr2 | DGKD | 1 |
| hsa_circ_0058767 | 0.020062199 | 2.9678054 | down | chr2 | DGKD | 18 |
| hsa_circ_0058768 | 0.001202278 | 2.1620224 | down | chr2 | DGKD | 75 |
| hsa_circ_0058769 | 0.001321101 | 2.6733668 | down | chr2 | DGKD | 32 |
| hsa_circ_0058770 | 0.005304494 | 5.227343 | down | chr2 | DGKD | 0 |
| hsa_circ_0060108 | 0.011829588 | 2.895927 | down | chr20 | FER1L4 | 217 |
| hsa_circ_0062721 | 0.03316098 | 3.6204636 | down | chr22 | XBP1 | 26 |
| hsa_circ_0063555 | 0.038891237 | 2.0088708 | down | chr22 | ACO2 | 23 |
| hsa_circ_0063561 | 0.016673693 | 2.77844 | down | chr22 | ACO2 | 35 |
| hsa_circ_0063562 | 0.03148357 | 4.083042 | down | chr22 | ACO2 | 9 |
| hsa_circ_0063563 | 0.015282579 | 3.6569278 | down | chr22 | ACO2 | 9 |
| hsa_circ_0063567 | 0.019898534 | 2.1735334 | down | chr22 | ACO2 | 0 |
| hsa_circ_0065143 | 0.040677425 | 2.0057971 | down | chr3 | SETD2 | 57 |
| hsa_circ_0066873 | 0.048354574 | 3.8894832 | down | chr3 | TIMMDC1 | 0 |
| hsa_circ_0066877 | 0.034795098 | 3.1437237 | down | chr3 | TIMMDC1 | 0 |
| hsa_circ_0066971 | 0.011570533 | 5.8554688 | down | chr3 | EAF2 | 0 |
| hsa_circ_0067450 | 0.01625185 | 7.951924 | down | chr3 | PPP2R3A | 6 |
| hsa_circ_0068032 | 0.035557568 | 4.645977 | down | chr3 | NAALADL2 | 5 |
| hsa_circ_0069113 | 0.027285358 | 2.006047 | down | chr4 | TBC1D14 | 0 |
| hsa_circ_0069114 | 0.002569319 | 2.1330538 | down | chr4 | TBC1D14 | 0 |
| hsa_circ_0070936 | 0.04316312 | 2.6107051 | down | chr4 | LARP1B | 7 |
| hsa_circ_0071107 | 0.006979433 | 3.2499113 | down | chr4 | ARHGAP10 | 2 |
| hsa_circ_0071321 | 9.78E-04 | 4.8015165 | down | chr4 | FGA | 3 |
| hsa_circ_0072309 | 0.002072746 | 7.3090854 | down | chr5 | LIFR | 0 |
| hsa_circ_0072789 | 0.009060388 | 3.3077662 | down | chr5 | MARVELD2 | 9 |
| hsa_circ_0072997 | 0.048613824 | 2.7817066 | down | chr5 | RGNEF | 1 |
| hsa_circ_0072998 | 0.015955605 | 5.7427354 | down | chr5 | RGNEF | 0 |
| hsa_circ_0073006 | 0.014696714 | 2.9636796 | down | chr5 | RGNEF | 35 |
| hsa_circ_0073035 | 0.013912499 | 2.19241 | down | chr5 | HMGCR | 0 |
| hsa_circ_0073244 | 0.004625377 | 4.716427 | down | chr5 | EDIL3 | 0 |
| hsa_circ_0073582 | 0.019003673 | 3.4118779 | down | chr5 | EPB41L4A | 0 |
| hsa_circ_0073763 | 0.029358098 | 2.9471374 | down | chr5 | SLC12A2 | 2 |
| hsa_circ_0073768 | 0.029927656 | 4.6921773 | down | chr5 | SLC12A2 | 1 |
| hsa_circ_0073769 | 0.005703302 | 3.6955311 | down | chr5 | SLC12A2 | 1 |
| hsa_circ_0073770 | 0.008143293 | 6.911025 | down | chr5 | SLC12A2 | 1 |
| hsa_circ_0073771 | 0.031154156 | 4.1421623 | down | chr5 | SLC12A2 | 1 |
| hsa_circ_0073772 | 0.031598363 | 3.3706727 | down | chr5 | SLC12A2 | 1 |
| hsa_circ_0073955 | 0.03778204 | 4.6350393 | down | chr5 | SEC. 24 A | 21 |
| hsa_circ_0074239 | 0.002184121 | 6.5593286 | down | chr5 | C5orf32 | 13 |
| hsa_circ_0075447 | 0.043269653 | 4.7036705 | down | chr6 | GMDS | 0 |
| hsa_circ_0075538 | 0.018290881 | 4.5513425 | down | chr6 | F13A1 | 19 |
| hsa_circ_0076303 | 0.0419991 | 2.85898 | down | chr6 | PGC | 16 |
| hsa_circ_0076304 | 0.014676455 | 31.56073 | down | chr6 | PGC | 35 |
| hsa_circ_0076305 | 0.029925665 | 125.95259 | down | chr6 | PGC | 49 |
| hsa_circ_0076307 | 0.025423396 | 9.498694 | down | chr6 | PGC | 1 |
| hsa_circ_0077168 | 0.04191364 | 2.536258 | down | chr6 | BCKDHB | 1 |
| hsa_circ_0077736 | 0.048207846 | 6.2890162 | down | chr6 | CEP85L | 2 |
| hsa_circ_0082915 | 0.022677844 | 2.0698295 | down | chr7 | SLC4A2 | 2 |
| hsa_circ_0083027 | 0.02473872 | 3.3468018 | down | chr7 | MLL3 | 8 |
| hsa_circ_0084925 | 0.017582932 | 5.8715267 | down | chr8 | KIAA1429 | 0 |
| hsa_circ_0088633 | 0.002749995 | 2.1568172 | down | chr9 | GARNL3 | 0 |
Figure 3.
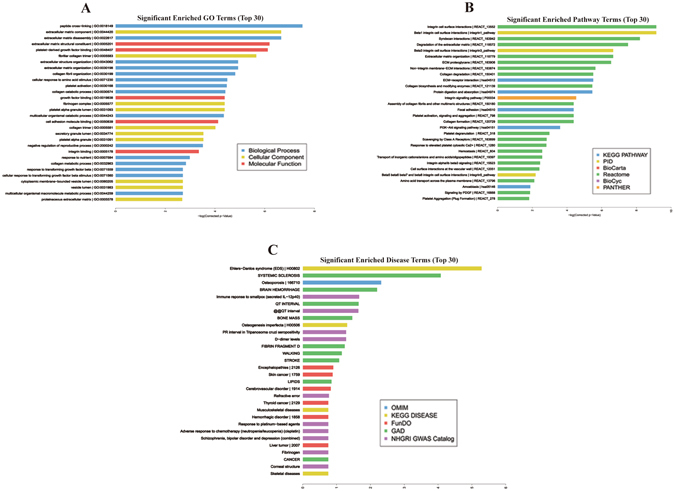
Results of Gene Ontology, KEGG pathway, and disease enrichment analysis. (A) Top 30 classes of GO enrichment terms. (B) Top 30 classes of KEGG pathway enrichment terms. (C) Top 30 disease enrichment terms.
CircRNA-miRNA network
The 3 circRNAs with most robust differential expression were used to construct a represent circRNA-miRNA network. The CBC1 and CBC2 probes identified a total of 207 differentially expressed circRNAs; among these circRNAs, hsa_circ_0076304, hsa_circ_0035431, and hsa_circ_0076305 had the highest magnitude of difference. Figure 4 illustrates the interaction of the 3 circRNAs with miRNA.
Figure 4.
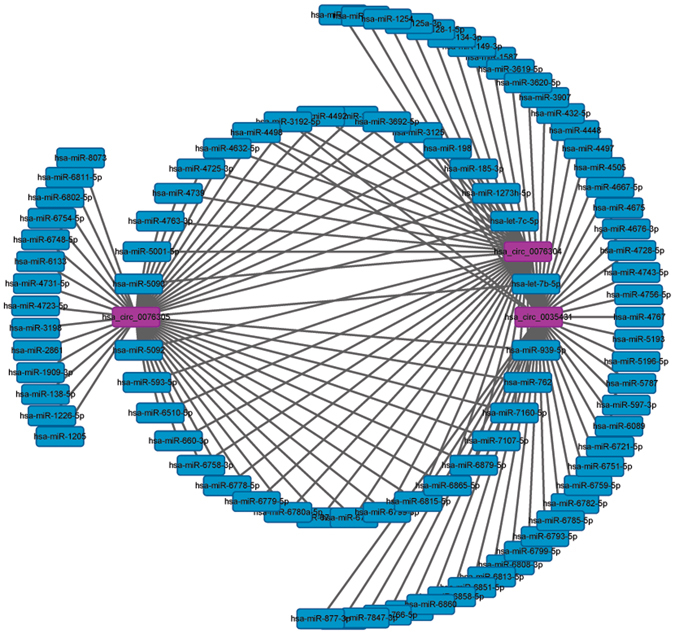
Represent circRNA-miRNA network. This network was based on the expression profile results and the related software. The 3 dysregulated circRNAs, hsa_circ_0076304, hsa_circ_0035431, and hsa_circ_0076305 (purple red nodes) having the highest magnitude of change, were predicted to be functionally connected with their targeted miRNAs in the network.
Discussion
CircRNAs are recently identified as disease-related and ubiquitously expressed noncoding RNAs, that can act as sponges of miRNAs and affect the expression of parent gene11–14. During the past several years, increasing evidence suggested that circRNAs play a vital role in cancer development and may be used as novel biomarkers15–18. By comparing circRNAs expression profiles in parental cell line and established cell line with radioresistant effects, Su et al. found that dysregulated circRNAs are related to the progression of radiation resistance in esophageal cancer cells19. Huang et al.20 reported that dysregulated lncRNAs and circRNAs are linked to the development of bladder cancer. They identified that several hundreds of circRNAs showed altered expression in bladder cancer tissues as analyzed by the expression profiles of 4 paired cancer and para-carcinoma tissues. They postulated that several of the dysregulated circRNAs are functional molecules and contribute to bladder cancer tumorigenesis. In the present study, 207 circRNAs were found to be differentially expressed between GC and non-cancerous tissues by both CBC1 and CBC2 probes in the microarray chip. Hsa_circ_0044516 had the highest magnitude of upregulation, whereas hsa_circ_0076305 had the highest magnitude of downregulation. The randomly selected 7 circRNAs that were significantly altered were further verified by qRT-PCR. These results conformed the validity of the microarray findings.
Some of the previously identified circRNAs are implicated to be associated with tumorigenesis and malignant behavior of cancer cells, such as uncontrolled growth, proliferation, migration, and invasion. For example, Hsa_circ_0067934 has been shown to be upregulated in esophageal squamous cell carcinoma (ESCC)21, and associated with poor tumor differentiation. In their findings, hsa_circ_0067934 was able to increase ESCC cell proliferation, migration, and cell cycle progression21. Xu et al.22 showed that patients with hepatocellular carcinoma (HCC) with higher expression level of circular RNA ciRS-7 (Cdr1as) in cancerous tissues had shorter median recurrent time than those with lower circRNA expression. Additionally, Cdr1as was related to the high hepatic microvascular invasion (MVI) in HCC, and the mechanism may be associated with its potential activity as the sponge of miR-7. Therefore, the study concluded that Cdr1as might be a novel biomarker and treatment target for MVI.
CircRNAs can regulate the transcription of parent genes. In the present study, we identified 116 corresponding linear mRNAs. GO and pathway enrichment analysis showed that these mRNAs are involved in critical pathways associated with cancer, including the PI3K-AKT pathway. Previously studies have shown that activation of the PI3K-AKT pathway promote cancer cell growth and proliferation23, 24. One of the potential targets of hsa_circ_0039090, hsa-let-7c-5p is associated with stage I endometrioid endometrial carcinoma progression potentially through regulation of cell cycle pathway25. Hsa-miR-107, one of the targets of several dysregulated circRNAs identified in the present study, is widely confirmed to be associated with cancers26–30, which is the downstream target of circTCF25, and the interaction between this circRNA with miR-107 and miR-103a-3p leads to increased proliferation and migration of bladder cancer cells31.
CircRNA-miRNA network is a widely accepted approach for exploring the function of dysregulated circRNAs and the interaction between these two types of non-coding RNAs. Hence, this network was constructed based on the microarray data. Among altered circRNAs, hsa_circ_0076304, hsa_circ_0035431, and hsa_circ_0076305 had the highest magnitude of difference. Concurrently, the potential links between them and the most important targeted miRNAs were established.
In summary, this study provided a preliminary landscape of circRNA differential expression in GC vs. non-GC. Further studies are required to explore their potential as biomarkers for GC as well as their pathologic role.
Electronic supplementary material
Acknowledgements
This work was supported by the Innovation Team Foundation of Fuzhou General Hospital (No. 2014CXTD04; to Lie Wang), the National Natural Science Foundation of China (No. 81372788; to Qiaojia Huang), the Medical Scientific Research Key Foundation of Nanjing Command (No. 11Z032; Qiaojia Huang), and the Natural Science Foundation of Fujian Province (No. 2014J01427; to Qiaojia Huang).
Author Contributions
Y.D. performed the qRT-PCR and collected data. X.O., K.W., F.Z., Y.L. and B.S. participated in samples collected. L.W. and Y.W. guided major fresh samples collected. L.W. also contributed to the major funding support. Q.H. designed the study. Q.H. and Y.D. wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-09076-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lie Wang, Email: fzptwk@xmu.edu.cn.
Qiaojia Huang, Email: huangqj100@126.com.
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Dang Y, Wang YC, Huang QJ. Microarray and next-generation sequencing to analyse gastric cancer. Asian Pacific journal of cancer prevention: APJCP. 2014;15:8033–8039. [PubMed] [Google Scholar]
- 3.He Y, et al. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer letters. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 4.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer research. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, et al. Circular RNAs in human cancer. Molecular cancer. 2017;16 doi: 10.1186/s12943-017-0598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu S, et al. Circular RNA: A new star of noncoding RNAs. Cancer letters. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 8.Li J, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. American journal of cancer research. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- 9.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang Y, et al. Expression and clinical significance of long non-coding RNA HNF1A-AS1 in human gastric cancer. World journal of surgical oncology. 2015;13 doi: 10.1186/s12957-015-0706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng HH, et al. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PloS one. 2016;11 doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Scientific reports. 2015;5 doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Q, et al. Circular RNA Related to the Chondrocyte ECM Regulates MMP13 Expression by Functioning as a MiR-136 ‘Sponge’ in Human Cartilage Degradation. Scientific reports. 2016;6 doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed I, et al. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7:36366–36381. doi: 10.18632/oncotarget.8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li F, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. International journal of clinical and experimental pathology. 2015;8:16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 18.Qu S, et al. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genomics data. 2015;5:385–387. doi: 10.1016/j.gdata.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su H, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. Journal of translational medicine. 2016;14 doi: 10.1186/s12967-016-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang M, et al. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7:47186–47200. doi: 10.18632/oncotarget.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia W, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Scientific reports. 2016;6 doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L, et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. Journal of cancer research and clinical oncology. 2017;143:17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PubMed] [Google Scholar]
- 23.Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis: an international journal on programmed cell death. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- 24.Qu JL, et al. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Digestive and liver disease: official journal of the Italian Society of Gastroenterology and the Italian Association for the Study of the Liver. 2009;41:875–880. doi: 10.1016/j.dld.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Xiong H, et al. Integrated microRNA and mRNA transcriptome sequencing reveals the potential roles of miRNAs in stage I endometrioid endometrial carcinoma. PloS one. 2014;9 doi: 10.1371/journal.pone.0110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma P, Saraya A, Gupta P, Sharma R. Decreased levels of circulating and tissue miR-107 in human esophageal cancer. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals. 2013;18:322–330. doi: 10.3109/1354750X.2013.781677. [DOI] [PubMed] [Google Scholar]
- 27.Wang S, et al. miR-107 regulates tumor progression by targeting NF1 in gastric cancer. Scientific reports. 2016;6 doi: 10.1038/srep36531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi Y, et al. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PloS one. 2009;4 doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen HY, et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer research. 2012;72:3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- 30.Feng L, Xie Y, Zhang H, Wu Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Medical oncology. 2012;29:856–863. doi: 10.1007/s12032-011-9823-1. [DOI] [PubMed] [Google Scholar]
- 31.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Scientific reports. 2016;6 doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


