Abstract
RNA-Seq is a powerful tool in transcriptomic profiling of cells and tissues. We recently identified many more taste buds than previously appreciated in chickens using molecular markers to stain oral epithelial sheets of the palate, base of oral cavity, and posterior tongue. In this study, RNA-Seq was performed to understand the transcriptomic architecture of chicken gustatory tissues. Interestingly, taste sensation related genes and many more differentially expressed genes (DEGs) were found between the epithelium and mesenchyme in the base of oral cavity as compared to the palate and posterior tongue. Further RNA-Seq using specifically defined tissues of the base of oral cavity demonstrated that DEGs between gustatory (GE) and non-gustatory epithelium (NGE), and between GE and the underlying mesenchyme (GM) were enriched in multiple GO terms and KEGG pathways, including many biological processes. Well-known genes for taste sensation were highly expressed in the GE. Moreover, genes of signaling components important in organogenesis (Wnt, TGFβ/ BMP, FGF, Notch, SHH, Erbb) were differentially expressed between GE and GM. Combined with other features of chicken taste buds, e.g., uniquely patterned array and short turnover cycle, our data suggest that chicken gustatory tissue provides an ideal system for multidisciplinary studies, including organogenesis and regenerative medicine.
Introduction
RNA sequencing (RNA-Seq) technology has emerged as a powerful and revolutionary approach to quantify gene expression levels and survey detailed transcriptomic profiling at unprecedented resolution and sensitivity1, 2. Additionally, it is an invaluable tool for gene discovery3, 4. In comparison to microarray platforms, RNA-Seq has several advantages, including a wider dynamic range of expression levels, higher accuracy and reproducibility, and lower noise-to-signal ratio, resulting in an enhanced ability to detect novel transcripts2, 5. Consequently, RNA-Seq has attracted broad interest and led to significant breakthroughs in our understanding of the genetic and molecular basis of living organisms, including traits of economic interest in livestock species6–16.
Chickens (Gallus sp.) are widely used as a research model in multidisciplinary studies including developmental biology17, 18, molecular biology19–22, and food science23. Their popularity stems from their comparative advantages over other animal models, including the convenience of in ovo embryo manipulation, rapid development, high availability and low costs. Similarly to mammals, chickens have many taste buds in the oral cavity and respond to taste stimuli24–27. Our recent studies using molecular markers to label chicken taste buds in oral epithelial sheets, i.e., palate, base of the oral cavity and posterior region of the tongue, demonstrated that chicken taste buds, like those of mammals, are distributed in a unique pattern28. In peeled chicken oral epithelial sheets, taste buds labeled with Vimentin and α-Gustducin were easily identified. Many more taste buds, patterned in rosette-like clusters, were found than previously reported28 suggesting that chickens possess a more advanced taste system than previously believed. Moreover, the clustered taste bud patterning in the oral cavity of chickens is reminiscent of the mammalian soft palate29. Taken together, these data suggest that taste sensory organs in chickens can potentially provide a system for organogenesis studies, including pattern formation.
To better understand the transcriptomic architecture of gustatory tissue in the oral cavity of chickens, RNA-Seq analysis was carried out with the following specific objectives: i) to demonstrate the validity of chicken taste organs as an ideal system for organogenesis studies, ii) to provide new insights into the underlying mechanisms implicated in the development of taste buds. Such information will facilitate studies on mechanisms underlying chicken taste bud formation which will be beneficial for understanding taste organ development in birds and potentially mammals, including humans.
Materials and Methods
Animal and tissue collection
The use of animals throughout the study was approved by The University of Georgia Institutional Animal Care and Use Committee and was in compliance with the National Institutes of Health Guidelines for the care and use of animals in research.
Newly hatched Cobb 500 (P0) broiler-type male chickens were provided by Cobb-Vantress Inc. from its hatchery in Cleveland, Georgia. The chicks were housed in separate cages in the animal facility at the Department of Animal and Dairy Science, University of Georgia until 3 days of age (P3). The brooder temperature was ~35°C and room temperature was maintained at 30°C with food (starter feed) and water available ad libitum under a 12/12 hr light/dark cycle.
P3 chicks (n = 3) were euthanized by decapitation. The oral tissue in the palate, base of the oral cavity, and posterior region of the tongue were dissected and processed for RNA extraction. To separate the epithelium from the underlying connective tissue, the upper and lower beak were dissected and briefly rinsed in 0.1 M phosphate buffered saline (PBS). An enzyme mixture of Collagenase A (1 mg/ml, Cat# 10103578001, Roche Diagnostics) and Dispase II (2.5 mg/ml, Cat# 04942078001, Roche Diagnostics) was injected (~6 ml in total) into the sub-epithelial space of the palate, the base of the oral cavity, and the posterior region of the tongue, followed by incubation at 37°C for 2 hr. Following enzymatic tissue digestion, the palate, the base of the oral cavity and the posterior region of the tongue were dissected in sterile PBS, and epithelial sheets were separated from underlying connective tissue.
To collect gustatory and non-gustatory tissues at the base of the oral cavity, the soft tissue of the base of oral cavity was cut off from the lower beak. Tissues with taste buds were easy to recognize under a stereomicroscope and they were separated from the surrounding region lacking taste buds. The epithelium and underlying connective tissue were further separated for these two regions (with and without taste buds) and the following tissue compartments were collected: gustatory epithelium (GE), gustatory mesenchyme/connective tissue (GM), non-gustatory epithelium (NGE), non-gustatory mesenchyme/connective tissue (NGM).
RNA isolation and quality assessment
Freshly isolated tissues were immersed in Trizol and immediately stored at −80°C until time of RNA extraction. Upon harvest of all tissues, samples were thawed on ice and homogenized with a PowerGen 700D tissue homogenizer (Fisher Scientific, Waltham, MA). The resulting homogenate was then processed via RNeasy Plus kit (Qiagen, Hilden, Germany), omitting the gDNA eliminator step, and was reconstituted in RNAse-free water. Initial RNA quantity and quality were determined on a Nanodrop 8000 spectrophotometer (Nanodrop, Thermo Scientific, Waltham, MA), and high quality samples were further assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) at the University of Georgia Genomics Facility (Athens, GA).
cDNA library preparation, pooling, and sequencing
cDNA libraries for each sample were prepared with Kapa Stranded mRNA-seq kit (KAPA Biosystems, Wilmington, MA). Library quality and quantity were assessed via Fragment Analyzer Automated CE (Advanced Analytical, Evry Cedex, France) and Qubit (Thermo Fisher) systems, respectively. Libraries were subsequently subjected to 2 × 75 bp paired-end (PE75) sequencing on a NextSeq. 500 system (Illumina). All samples were pooled and sequenced on one lane. All library preparation and sequencing were performed by the Georgia Genomics Facility (Athens, GA).
Reads alignment to the chick reference genome and annotated transcripts
To ensure high-quality data, reads with low-quality, containing adapter contamination, or at least 10 Ns from raw data (FASTQ format) were removed using in-house developed Perl scripts. Prior to downstream analysis, the overall quality of pre-processed data was further examined using FastQC v0.11.4 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The Galgal4 reference assembly (FASTA format) and annotated gene model (GTF format) were downloaded from Ensembl database (http://ftp//ftp.ensembl.org/pub/release-76/). For each library, we estimated the actual insert size distribution after indexing the reference genome using Bowtie2 v2.2.4 with default parameters30. Subsequently, paired-end clean reads were aligned to the reference genome using Tophat v2.0.14 (http://tophat.cbcb.umd.edu/)31. The detailed alignment information is presented in Supplemental Table S1, including total numbers of reads, mapped reads, and unique mapped reads.
Identification of differentially expressed genes (DEGs)
The number of RNA-Seq reads produced by a transcript is directly proportional to its abundance. Thus, the gene expression level could be easily quantified by the read count. Cuffdiff v2.2.132 and DESeq2 R packages4, 33, the main statistical methods to identify DEGs across different experimental conditions, were used.
For Cuffdiff, commonly used fragments were normalized by the relevance to the transcript’s length and the total yield of the fragments to ensure accurate quantification of each gene’s expression. The obtained values are represented by the number of fragments per kilobase of transcript per a million of mapped fragments (FPKM)34 in paired-end sequencing experiments32. TopHat’s read alignments were assembled by Cufflinks32, and then the DEGs and transcripts between two groups were detected and quantified by Cuffdiff using a rigorous sophisticated statistical analysis32. The corresponding attributes including fold changes, p-values, and q-values (false discovery rate corrected p values) of DEGs were reported in the output files from Cuffdiff. The difference in gene expression was determined as significant if the p-value was less than the false discovery rate after Benjamini-Hochberg correction for multiple testing (http://cufflinks.cbcb.umd.edu/manual.html).
For DESeq2 method, DEGs were detected using the DESeq2 R/Bioconductor package that performs independent filtering. To enhance the statistical power for identifying DEGs, we removed genes with weak expression levels using the HTS Filter package35. RNA-Seq read counts were analyzed using a generalized linear model for comparisons focusing on the GE vs NGE and GE vs GM. The resulting p-values were adjusted using Benjamini and Hochberg’s approach for controlling for false discovery rate. The fold changes, p-values and q-values (false discovery rate corrected p-values) of the DEGs were reported in the output files from DESeq2. Genes with a q-value <0.05 were assumed as differentially expressed.
GO and gene functional analysis of differentially expressed genes
To gain insight into the biological functions of DEGs, the enriched Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were analyzed using Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/). The functional groups with at least two DEGs in the background terms were selected and those with a p-value < 0.05 were considered as significantly overrepresented.
Real-time quantitative RT-PCR (qRT-PCR) assay
To confirm sequencing results, qRT-PCR on 14 randomly selected DEGs from GE vs NGE and 14 DEGs from GE vs GM was performed with the same RNA samples used for RNA-Seq. Primers were designed via Primer Express 3.0.1 software (Applied Biosystems) and are shown in Supplemental Table S2 (GE vs NGE) and Supplemental Table S3 (GE vs GM). Briefly, 1 μg of total RNA was reverse-transcribed to cDNA using the SuperScript First Strand cDNA conversion kit (Invitrogen, Carlsbad, CA). qRT-PCR was subsequently carried out in duplicate 10 μl reactions using SYBR green master mix (Genecopoeia, Rockville, MD). Reactions were run in a 96-well plate using an MX3000 P system (Agilent Technologies). The cycle conditions were as follows: 1 cycle of pre-incubation at 95°C for 10 min, 40 cycles of amplification (95°C for 30 s, 63°C for 15 s, and 72°C for 20 s). Relative gene expressions of DEGs were calculated using 2−ΔΔCt method, with the housekeeping gene GAPDH serving as internal control.
Results
More differentially expressed genes (DEGs) between epithelium and connective tissue of the base of oral cavity than palate and tongue
In the first RNA-Seq experiment, the entire epithelial sheet and underlying connective tissues of the palate, base of oral cavity, and posterior tongue were used. We demonstrated that (1) more DEGs were found between the epithelium (1783 highly expressed) and connective tissue (1906 highly expressed) in the base of the oral cavity (Supplemental Table S4) than palate or posterior tongue (1021 highly expressed in the epithelium and 1730 in the connective tissue of palate; 1512, 1880 in the posterior tongue, listed in Supplemental Tables S5 and S6, respectively); (2) a large overlap of DEGs (640 and 889) between epithelium and connective tissues of the palate, base of oral cavity, and posterior tongue were observed using DESeq2 and Cuffdiff (Fig. 1).
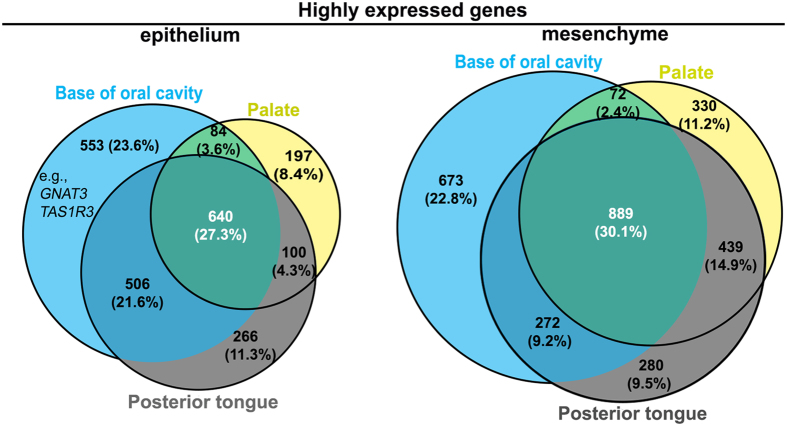
Figure 1.
Number and overlapping of DEGs between epithelium and underlying mesenchyme of the palate, base of oral cavity, and posterior tongue in P3 male chickens. The highly expressed genes in the oral epithelium (left) or the underlying mesenchyme (right) in different tissues were analyzed separately followed by further comparisons of highly DEGs in the epithelium (left) or mesenchyme (right) of different tissues. The DEGs were determined by statistical algorithms Cuffdiff and DESeq2. Notably, the base of oral cavity had a greater number of highly expressed DEGs (p < 0.05) in both the epithelium and mesenchyme compared to the palate and posterior tongue. Genes that are well known to be involved in taste perception, e.g., GNAT3, TAS1R3, were revealed as highly DEGs in the epithelium of the base of oral cavity.
More importantly, DEGs that are associated with taste perception were found in the base of oral cavity, e.g., GNAT3, TAS1R3 (Fig. 1, Supplemental Table S4), but were not detected in the palate and posterior tongue (Supplemental Tables S5, S6). In addition, our recent studies have demonstrated that taste bud clusters in the base of oral cavity are more uniformly distributed than those in the palate and tongue28. Further, the epithelium of the base of oral cavity was easy to separate from underlying tissues28, and the topography and tissue structure are less complex, e.g., no papillary spines. Based on these results, we focused our interest on the base of the oral cavity. Tissue samples from the gustatory epithelium (GE, epithelial region that contains taste buds), the non-gustatory epithelium (NGE, epithelium that surrounds the taste bud region), and the corresponding underlying connective tissue/mesenchyme (GM and NGM) of the base of the oral cavity were collected and used for additional RNA-Seq analyses.
DEGs between gustatory epithelium and surrounding tissue compartments (NGE and GM)
Comparisons were focused on GE vs NGE and vs GM, all in triplicate, resulting in nine total tissues. We acquired a total of 335.66 million raw 100-bp paired-end reads. After quality filtering, the RNA-Seq of nine samples yielded around 325.59 million of clean reads with an average of 36.18 million reads per sample. Alignment of the sequence of clean reads against Galgal4 yielded 80.26–85.65% uniquely aligned reads across the nine samples using Tophat2 aligner. Furthermore, 96.55–98.70% of the reads were aligned in a unique manner, while the remaining were multiple-mapped reads. Detailed information of data quality and mapping statistics are listed in Supplemental Table S1.
In order to identify candidate genes involved in the development of taste buds in the gustatory epithelium (GE), Cufflinks and DESeq2 were used to identify the DEGs between GE vs NGE, and GE vs GM. A total of 187 and 6047 genes were differentially expressed between the GE and NGE and between GE and GM, respectively, based on |FC| > 1, p < 0.05, and FDR q < 0.05 detected by Cuffdiff or DESeq2 or both. Volcano plots of DEGs between the different tissue compartments are presented in Fig. 2.
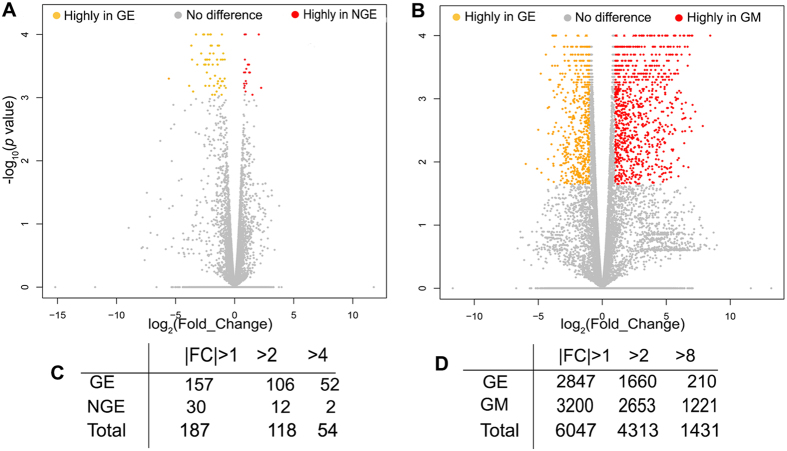
Figure 2.
Volcano plots (A,B) and number of highly expressed DEGs at different fold changes (FC) (C,D) to display the DEGs in the GE vs NGE (A,C) and GE vs GM (B,D) in the base of oral cavity of P3 male chickens. In A and B, the Y-axis corresponds to the mean expression value of log10 (p-Value), and the X-axis displays the log2 (Fold_Change). The orange and red dots represent highly expressed genes in the GE (A,B) or NGE (A) or GM (B) (p < 0.05, FDR q < 0.05); the gray dots represent the transcripts whose difference of expression levels did not reach statistical significance (p > 0.05, FDR q > 0.05). GE: gustatory epithelium; NGE: non-gustatory epithelium; GM: gustatory mesenchyme.
Between GE and NGE, 118 genes were highly differentially expressed with a |FC| > 2, p < 0.05, and FDR q < 0.05, of which 106 and 12 genes were highly expressed in GE and NGE, respectively (Fig. 2C). Additionally, 65 of these 118 highly DEGs were identified by both Cuffdiff and DESeq2 methods (only 24 and 29 DEGs were identified by DESeq2 or Cuffdiff alone respectively). A much larger number of DEGs was identified between GE and GM using the same significance criteria. In fact, 4313 genes were differentially expressed with 1660 and 2653 highly expressed in the GE and GM, respectively (Fig. 2D). Almost 60% (2584) of these 4313 genes were identified by both pieces of software used in the analyses, and only 981(23%) and 748 (17%) were detected by either DESeq2 or Cuffdiff Approach alone.
The top 10 highly expressed genes in the GE or NGE based on the GE vs NGE comparison are listed in Table 1. The top 10 highly expressed genes in the GE or GM according to the GE vs GM comparison are presented in Table 2. Detailed information (Gene ID, Gene name, Fold_Change, p_value, q_value) of all DEGs are presented in Supplemental Table S7 (GE vs NGE) and Supplemental Table S8 (GE vs GM).
Table 1.
Top 10 DEGs in GE and in NGE in comparison between GE and NGE.
| Highly expressed genes in the GE | Highly expressed genes in the NGE | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene ID | Name | Fold Change | p_value | q_value | Gene ID | Name | Fold Change | p_value | q_value |
| ENSGALG00000004083 | GRIA1 | 47.60 | 6.82E-07 | 1.50E-04 | ENSGALG00000007815 | ANKK1 | 4.87 | 3.84E-09 | 1.49E-06 |
| ENSGALG00000010163 | LGR5 | 28.59 | 2.34E-22 | 7.58E-19 | ENSGALG00000019551 | OVALY | 4.12 | 1.00E-04 | 8.16E-03 |
| ENSGALG00000011633 | SLC5A8 | 25.42 | 1.54E-10 | 7.87E-08 | ENSGALG00000008909 | USP13 | 3.51 | 1.20E-07 | 3.07E-05 |
| ENSGALG00000020084 | CAPN13 | 21.17 | 5.00E-05 | 4.43E-03 | ENSGALG00000000619 | ANGPTL4 | 3.15 | 5.00E-05 | 4.43E-03 |
| ENSGALG00000014884 | ISL1 | 20.83 | 6.68E-48 | 6.49E-44 | ENSGALG00000039830 | DHX30 | 2.86 | 5.00E-05 | 4.43E-03 |
| ENSGALG00000016325 | GSTA3 | 19.46 | 6.03E-20 | 1.46E-16 | ENSGALG00000000544 | RAB44 | 2.84 | 9.00E-04 | 4.90E-02 |
| ENSGALG00000016943 | OLFM4 | 13.13 | 5.00E-05 | 4.43E-03 | ENSGALG00000008759 | LAMP3 | 2.78 | 7.43E-07 | 1.64E-04 |
| ENSGALG00000011962 | NDNF | 12.67 | 3.53E-10 | 1.71E-07 | ENSGALG00000041114 | GOLGA4 | 2.35 | 5.00E-05 | 4.43E-03 |
| ENSGALG00000016428 | ENPP2 | 12.30 | 3.16E-09 | 1.39E-06 | ENSGALG00000013994 | TAAR1 | 2.35 | 4.00E-04 | 2.56E-02 |
| ENSGALG00000026246 | GIF | 12.17 | 2.61E-07 | 6.33E-05 | ENSGALG00000011687 | AHNAK2 | 2.17 | 3.00E-04 | 2.00E-02 |
Table 2.
Top 10 DEGs in GE and in GM in comparison between GE and GM.
| Highly expressed genes in the GE | Highly expressed genes in the GM | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene ID | Name | Fold Change | p_value | q_value | Gene ID | Name | Fold Change | p_value | q_value |
| ENSGALG00000000341 | KRTAP10-4 | 41.53 | 1.05E-04 | 2.94E-04 | ENSGALG00000033157 | ADAMTS10 | 316.56 | 5.00E-05 | 2.14E-04 |
| ENSGALG00000007815 | ANKK1 | 35.58 | 1.68E-08 | 8.02E-08 | ENSGALG00000005643 | LYVE1 | 316.14 | 1.75E-39 | 1.17E-37 |
| ENSGALG00000035166 | LYPD2 | 24.80 | 5.00E-05 | 2.14E-04 | ENSGALG00000010391 | MMRN1 | 302.00 | 1.28E-70 | 6.17E-68 |
| ENSGALG00000039572 | PADI1 | 23.01 | 5.00E-05 | 2.14E-04 | ENSGALG00000022815 | AvBD1 | 252.53 | 3.39E-18 | 4.53E-17 |
| ENSGALG00000010864 | EREG | 22.94 | 5.31E-24 | 1.14E-22 | ENSGALG00000029811 | BMPER | 234.06 | 2.26E-16 | 2.57E-15 |
| ENSGALG00000042657 | Wnt3a | 22.57 | 5.00E-05 | 2.14E-04 | ENSGALG00000028158 | FGF7 | 224.87 | 5.40E-49 | 6.51E-47 |
| ENSGALG00000002449 | DUOXA1L | 22.29 | 3.65E-16 | 4.02E-15 | ENSGALG00000010331 | MME | 185.25 | 2.15E-33 | 9.53E-32 |
| ENSGALG00000006322 | CLCA2 | 21.99 | 4.01E-11 | 2.60E-10 | ENSGALG00000026736 | OGN | 180.56 | 7.59E-64 | 2.43E-61 |
| ENSGALG00000041822 | FGF22 | 20.64 | 5.00E-05 | 2.14E-04 | ENSGALG00000001071 | ELN | 178.38 | 1.08E-93 | 2.42E-90 |
| ENSGALG00000009877 | KCNH1 | 20.36 | 1.97E-23 | 4.09E-22 | ENSGALG00000006877 | KLHL4 | 175.81 | 4.72E-29 | 1.51E-27 |
Validation of RNA-Seq data with qRT-PCR
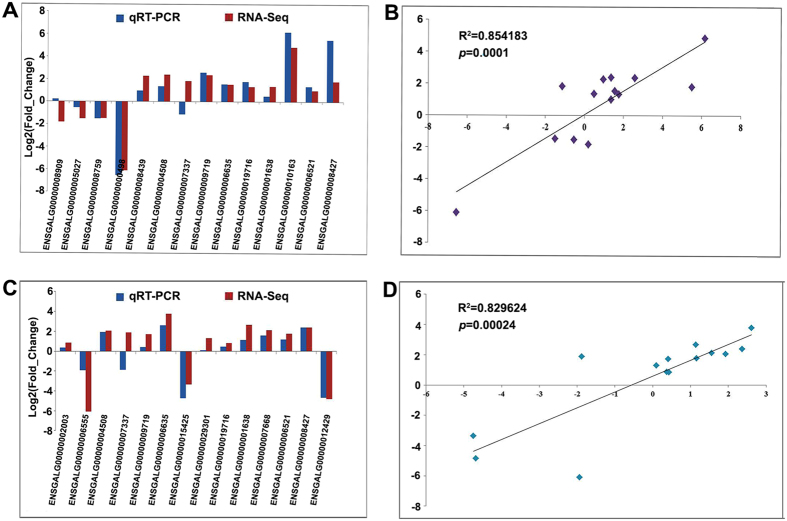
To verify the accuracy of DEGs based on RNA-Seq and the aforementioned statistical criteria, qRT-PCR analysis was performed for 20 selected DEGs (|FC| > 1.5, p < 0.05 and FDR q < 0.05) (DHX30, EYA2, LPP, PCDH10, RHCG, USP13, LAMP3, ACE, IGF2, LPL, CD36, KRT20, TAS1R3, LGR5, TRPM5, GNAT3, BMP2, BMP4, BMP7 and Bmpr1a) including highly and lowly expressed genes in the GE compared to NGE (6 DEGs) or GM (6 DEGs), and 8 DEGs (KRT20, TAS1R3, TRPM5, GNAT3, RHCG, PCDH10, LPP, EYA2) were shared DEGs by both GE vs NGE and GE vs GM comparisons. A comparison of expression of these 20 genes was made between the data from qRT-PCR normalized to GAPDH and those from RNA-Seq (Fig. 3). A high overlap is shown clearly by the concordant expression patterns between RNA-Seq and qRT-PCR and validates the adequacy of the statistical criteria used in these analyses. The computational and experimental fold changes in our study also showed a strong positive correlation (p < 0.001) with R2 = 0.854183 (p = 0.0001) in GE vs NGE (Fig. 3B) and R2 = 0.829624 (p = 0.00024) in GE vs GM (Fig. 3D) confirming the high reliability of RNA-Seq data in this study.
Figure 3.
Correlation of expression levels of DEGs (20 in total, 8 in common in A and C) between GE vs NGE (A,B) or GE vs GM (C,D) detected by qRT-PCR and RNA-Seq. A, C: Histograms of expression levels of 14 selected DEGs between GE vs NGE (A) and GE vs GM (C). Y-axis represents the log2 (Fold_Change) derived from qRT-PCR and RNA-Seq. B, D: Regression analysis of the log2 (Fold_Change) values between RNA-Seq and qRT-PCR. The X- and Y-axis represents the log2 (Fold_Change) measured by qRT-PCR (normalized by GAPDH) and RNA-Seq, respectively.
Specific DEGs in the GE compared to surrounding tissue compartments (NGE and GM)
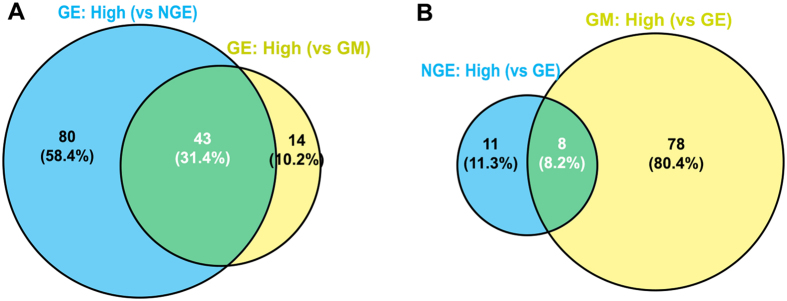
To detect genes specific to the gustatory epithelium in which taste buds reside, we analyzed genes that are differentially expressed between GE and either of the two surrounding tissues (NGE and GM). Of the 143 DEGs identified, 51 genes were differentially expressed in the GE compared to both NGE and GM (43 at a higher and 8 at a lower level, |FC| > 1.5, p < 0.05 and FDR q < 0.05) (Fig. 4). This result suggests that these 51 genes (Table 3) are likely to be promising candidates that are particularly important for taste perception or regulation of taste organ development. Out of the 43 highly DEGs in the GE, 5 genes are already well-known to be important genes involved in the taste sensation and organogenesis (GNAT3, TAS1R3, TRPM5, GNG13, SHH).
Figure 4.
Venn diagram illustrating the number of distinct and overlapping DEGs in the GE compared to NGE and GM of the base of oral cavity. (A) Highly expressed DEGs in the GE compared with NGE (blue) and GM (yellow). (B) Highly expressed DEGs in the NGE and GM compared with GE. Statistically significant genes (p < 0.05) with a fold change > 1.5 were included. GE: gustatory epithelium; NGE: non-gustatory epithelium; GM: gustatory mesenchyme.
Table 3.
Fifty-one highly (43) or lowly (8) expressed transcripts in the GE compared to both NGE and GM.
| Gene ID | Name | FC(NGE/GE) | FC(GM/GE) | p_value | q_value |
|---|---|---|---|---|---|
| ENSGALG00000016325 | GSTA3 | −19.46 | −2.78** | 6.03E-20 | 1.46E-16 |
| ENSGALG00000035166 | LYPD2 | −9.02 | −24.80** | 5.00E-05 | 4.43E-03 |
| ENSGALG00000133710 | SPINK5 | −5.68 | −4.08** | 5.00E-05 | 4.43E-03 |
| ENSGALG00000004508 | EYA2 | −5.21 | −3.87** | 7.61E-13 | 4.92E-10 |
| ENSGALG00000010901 | ABCB5 | −5.15 | −2.54** | 6.66E-05 | 6.73E-03 |
| ENSGALG00000023303 | Six2 | −4.93 | −3.64** | 1.59E-07 | 3.96E-05 |
| ENSGALG00000008185 | AOX1 | −4.30 | −6.29** | 5.00E-05 | 4.43E-03 |
| ENSGALG00000009866 | CAMK1D | −3.98 | −3.31** | 8.02E-06 | 1.17E-03 |
| ENSGALG00000003553 | ABCA12 | −3.92 | −1.94** | 5.00E-05 | 4.43E-03 |
| ENSGALG00000028598 | SERPINA12 | −3.75 | −3.69** | 1.43E-06 | 2.88E-04 |
| ENSGALG00000014987 | OTOP1 | −3.65 | −14.64** | 7.33E-15 | 7.11E-12 |
| ENSGALG00000007337 | LPP | −3.55 | −3.68** | 5.00E-05 | 4.43E-03 |
| ENSGALG00000008427 | GNAT3 | −3.48 | −5.23* | 4.78E-14 | 3.86E-11 |
| ENSGALG00000020876 | AOX2 | −3.34 | −7.63** | 2.69E-17 | 4.34E-14 |
| ENSGALG00000013033 | CMBL | −3.19 | −9.83** | 1.43E-04 | 1.19E-02 |
| ENSGALG00000006635 | RHCG | −2.90 | −12.68** | 2.48E-09 | 1.15E-06 |
| ENSGALG00000016522 | PPEF1 | −2.76 | −16.23** | 1.20E-07 | 3.07E-05 |
| ENSGALG00000021232 | TGM6 | −2.73 | −4.23** | 6.34E-04 | 4.00E-02 |
| ENSGALG00000003690 | KRT14 | −2.63 | −11.68** | 3.30E-09 | 1.39E-06 |
| ENSGALG00000001638 | TAS1R3 | −2.58 | −6.32** | 1.65E-08 | 5.17E-06 |
| ENSGALG00000007257 | TEKT5 | −2.56 | −18.49** | 3.44E-09 | 1.39E-06 |
| ENSGALG00000017295 | PTHLH | −2.53 | −2.38** | 7.31E-04 | 4.46E-02 |
| ENSGALG00000019716 | KRT20 | −2.53 | −1.78* | 9.52E-09 | 3.37E-06 |
| ENSGALG00000002024 | COMT | −2.35 | −1.95** | 1.55E-06 | 3.07E-04 |
| ENSGALG00000001617 | CAMKK1 | −2.13 | −3.81** | 2.82E-06 | 4.89E-04 |
| ENSGALG00000014730 | ELOVL7 | −2.11 | −2.42** | 1.27E-04 | 1.08E-02 |
| ENSGALG00000005293 | GNG13 | −2.03 | −8.48* | 1.31E-05 | 1.80E-03 |
| ENSGALG00000023897 | RGS21 | −2.02 | −11.64** | 2.96E-06 | 5.03E-04 |
| ENSGALG00000015593 | EPHA7 | −2.01 | −3.35** | 2.85E-05 | 3.41E-03 |
| ENSGALG00000006521 | TRPM5 | −2.00 | −3.37** | 3.22E-05 | 3.67E-03 |
| ENSGALG00000015764 | FABP5 | −1.98 | −4.07** | 6.00E-06 | 9.53E-04 |
| ENSGALG00000016128 | B3GALT5 | −1.89 | −4.78** | 3.18E-05 | 3.67E-03 |
| ENSGALG00000006379 | SHH | −1.88 | −3.82** | 1.75E-04 | 1.41E-02 |
| ENSGALG00000028946 | CSRP2 | −1.84 | −3.37** | 2.74E-06 | 4.89E-02 |
| ENSGALG00000008105 | CYP2AB4 | −1.83 | −6.08** | 1.78E-04 | 1.43E-02 |
| ENSGALG00000014372 | SLC34A2 | −1.83 | −4.92** | 3.07E-04 | 2.27E-02 |
| ENSGALG00000040607 | MED23 | −1.79 | −2.37** | 2.50E-04 | 1.76E-02 |
| ENSGALG00000003213 | LGALS2 | −1.74 | −2.42** | 5.99E-04 | 3.79E-02 |
| ENSGALG00000011654 | ACPP | −1.69 | −7.12** | 3.59E-04 | 2.58E-02 |
| ENSGALG00000024272 | S100A9 | −1.68 | −13.47** | 6.95E-04 | 4.32E-02 |
| ENSGALG00000016464 | VSNL1 | −1.64 | −5.42** | 1.41E-05 | 1.90E-03 |
| ENSGALG00000006425 | SLC6A6 | −1.57 | −2.06** | 3.68E-05 | 4.00E-03 |
| ENSGALG00000016324 | GSTA4 | −1.56 | −4.01** | 1.50E-04 | 1.24E-02 |
| ENSGALG00000013145 | EPB41L4B | 1.75 | 1.59** | 9.22E-05 | 8.61E-03 |
| ENSGALG00000001527 | VWA1 | 1.75 | 22.51** | 4.83E-04 | 3.23E-02 |
| ENSGALG00000005180 | COL11A1 | 1.77 | 14.78** | 1.05E-04 | 9.45E-03 |
| ENSGALG00000026470 | BOD1L1 | 1.81 | 2.17** | 9.00E-04 | 4.90E-02 |
| ENSGALG00000010763 | TRIP11 | 1.94 | 2.08* | 5.00E-05 | 4.43E-03 |
| ENSGALG00000008399 | ANKRD12 | 1.99 | 2.63** | 6.00E-04 | 3.60E-02 |
| ENSGALG00000009690 | CENPF | 2.15 | 2.29** | 5.00E-05 | 4.43E-03 |
| ENSGALG00000011687 | AHNAK2 | 2.17 | 11.03** | 3.00E-04 | 2.00E-02 |
*p < 0.05, **p < 0.01 for the expression of listed genes in the GE compared to both NGE and GM.
Gene Ontology enrichment and pathway analysis
To further investigate the functional associations of the DEGs, we performed gene ontology (GO) analysis at the GO database (http://www.geneontology.org/GO.database.shtml). The significance criteria was set at an unadjusted threshold at p < 0.05 and at least five DEGs in the background terms to assess potential functions. Multiple pathways and GO terms including biological processes, cellular components and molecular function were significantly enriched for these DEGs (Fig. 5 and 6). The details of the significant pathways in the two comparisons (GE vs NGE and GE vs GM) are shown in Supplemental Table S9, and the significant GO terms are shown in Supplemental Table S10.
Figure 5.

Histogram presentation of the number of DEGs between GE vs NGE of the base of oral cavity belonging to significant pathways (A) and selected gene ontology (GO) terms (B).
Figure 6.
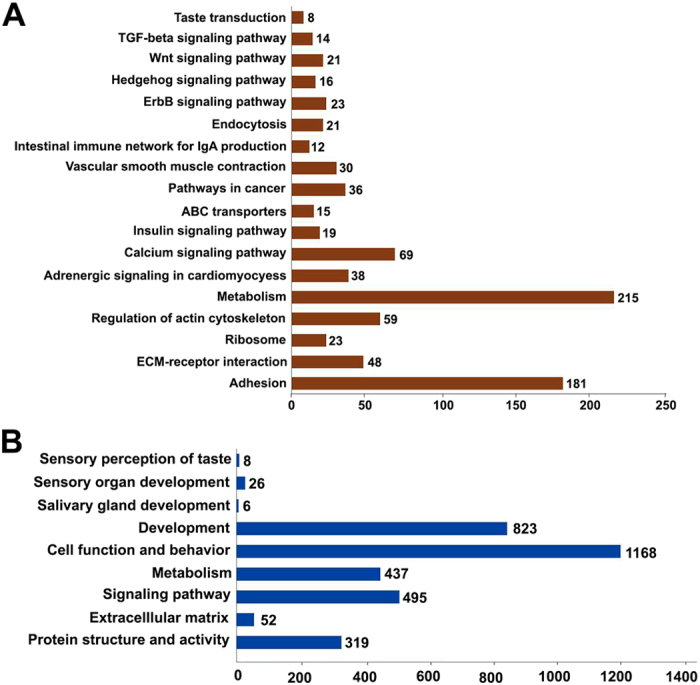
Histogram presentation of the number of DEGs between GE vs GM of the base of oral cavity belonging to significant pathways (A) and selected gene ontology (GO) terms (B).
From the comparison of GE vs NGE, 118 highly DEGs (|FC| > 2) were detected, among which 30 DEGs (Fig. 5A, Supplemental Table S9) were revealed to belong to significant pathways (8 in total) in the KEGG pathway analysis, including taste transduction, TGF-β signaling pathway, melanogenesis, dopaminergic synapse, drug metabolism-cytochrome P450, amphetamine addiction, nicotinate and nicotinamide metabolism, and tyrosine metabolism pathways (Fig. 5A, Supplemental Table S9). These 118 DEGs were classified into 32 significant GO categories (p < 0.05, Supplemental Table S10). Multiple GO terms including sensory perception of taste, development and other terms were significantly enriched for these DEGs as indicated in Fig. 5B. These DEGs include genes that are known to be involved in taste perception, e.g., TAS1R3, GNG13, GNAT3, and CD36, and genes that are important for the development of taste organs in rodents, such as LGR5, TGFβ2, SHH, and LEF1 (Supplemental Table S10). Of note, some of the DEGs were shown to be involved in multiple functions (marked in red, Supplemental Table S10).
From the comparison of GE vs GM, 4313 highly DEGs (|FC| > 2) were detected and KEGG pathway analysis revealed 18 significant pathways (p < 0.05) with 848 gene profiles involved (Fig. 6A, Supplemental Table S9). The significant pathways include taste transduction, as well as TGF-β, hedgehog, and Wnt signaling pathways (Fig. 6A, Supplemental Table S9) that are well known and have been reported for their involvement in the development of tongue, taste papillae and taste buds36–46. These 4313 DEGs were classified into 378 significant GO categories (p < 0.05, Supplemental Table S10). The majority of these DEGs (3334) belong to 9 categories as shown in Fig. 6B. Similarly to the DEGs between GE vs NGE, many of the DEGs between GE vs GM were shown in multiple GO terms (Supplemental Table S10) indicating the involvement of these DEGs in multiple functions. Multiple GO terms including sensory perception of taste, sensory organ development, salivary gland development and development of other tissues were significantly enriched for these DEGs. Among the detected DEGs were TRPM5, GNG13, TAS1R3, SCNN1G, SCNN1B, SCNN1A, PRKX and GNAT3, which are known to be involved in the sensory perception of taste, and LGR5, SHH, TGFβ2, and LEF1 (Supplemental Table S10), which are known to be involved in the development of taste organs.
Among the 51 DEGs (Table 3), which were differentially expressed in the GE at a high (43) (Fig. 4A) or low (8) (Fig. 4B) level compared to both surrounding tissue compartments (NGE and GM), 18 gene profiles were found to fall into 7 significant KEGG pathways (Fig. 7A, Supplemental Table S9) and 46 gene profiles fall into 14 significant GO terms (Supplemental Table S10). Selected terms, as shown in Fig. 7B, include taste transduction pathway and sensory perception of taste.
Figure 7.
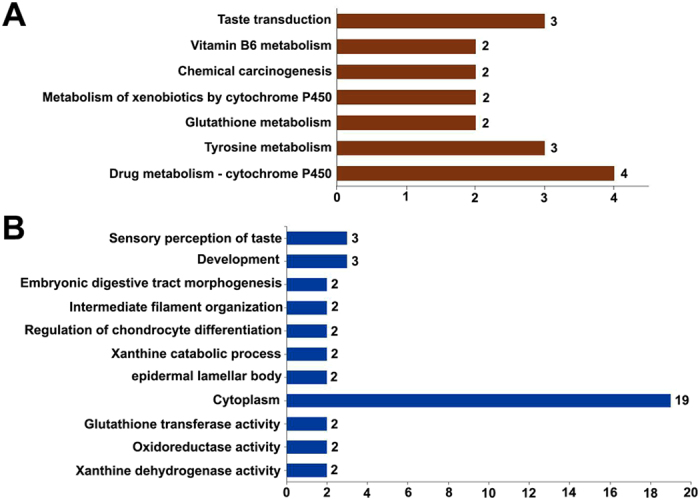
Histogram presentation of the number of shared DEGs belonging to significant pathways (A) and selected gene ontology (GO) terms (B). The shared DEGs were detected by comparisons of both GE vs NGE and GE vs GM of the base of oral cavity.
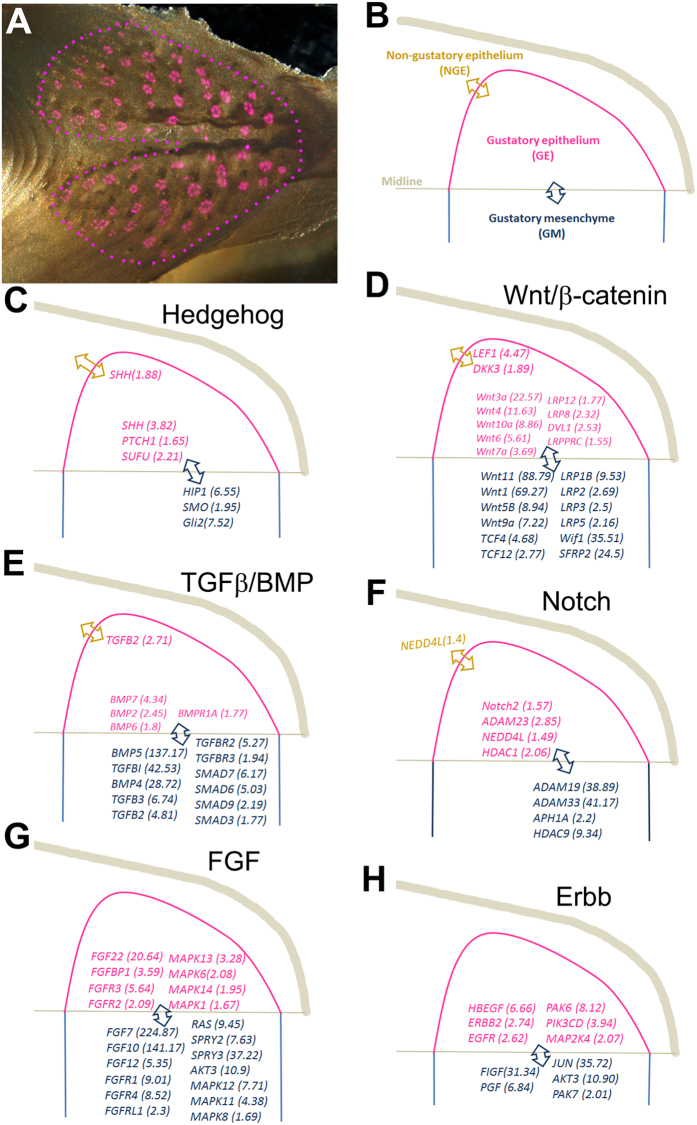
Mapping the expression of signaling components of molecular pathways in the gustatory epithelium and surrounding tissues
To better understand the potential function of molecular signaling pathways in regulating taste organ development in chickens, we selected several pathways that are known to be involved in taste organogenesis in rodents, e.g., Hedgehog37, 41–43, 47–49 (Fig. 8C), Wnt/β-catenin38, 40, 50–52 (Fig. 8D), TGF-β/BMP36, 46, 53, 54 (Fig. 8E), Notch55–57 (Fig. 8F), fibroblast growth factor (FGF) 53, 58, 59 (Fig. 8G), and Erbb60–62 (Fig. 8H), and schematically illustrated the spatial distribution of highly expressed key components in gustatory epithelium (GE), non-gustatory epithelium (NGE), and gustatory mesenchyme (GM).
Figure 8.
Distribution pattern of highly DEGs that are key components of well-known molecular signaling pathways in organogenesis (C–H). (A) An epithelial sheet of the base of the oral cavity of a P3 chicken that was immunoreacted with Vimentin for labeling taste buds (purple). Purple dots outline the gustatory epithelium that contains taste buds. (B) A diagram to illustrate the tissue compartments collected for RNA-Seq and DEG detection. (B–H) Double pointed arrows indicate the tissue compartments for comparisons of gene expressions. Values in parentheses in (C–H) represent the fold change of DEGs at a high expression level in the tissue compartment where the gene’s name is placed.
Some key components were found to be highly expressed in the GE compared to the NGE, e.g., SHH of the hedgehog signaling pathway (Fig. 8C), LEF1 and DKK3 of the Wnt/β-catenin signaling pathway (Fig. 8D), TGFβ2 of the TGFβ/BMP signaling pathway (Fig. 8E). An ubiquitin ligase neural precursor cell expressed developmentally downregulated gene 4-like (NEDD4L) was found to be expressed in the gradient of NGE > GE > GM (Fig. 8F).
From the comparison between GE and GM, a large number of signaling components in each of the aforementioned signaling pathways were identified as DEGs (Supplemental Table S8), including ligands, receptors, intracellular signaling components, and transcription factors. Some key components were selected and presented in Fig. 8C-H. The gene expression patterns suggest their potentially active involvement in the epithelial-mesenchymal interactions in the gustatory tissue.
Discussion
RNA sequencing (RNA-Seq) is currently the most powerful tool available to study the complexity of the transcriptome. It has many advantages over traditional cDNA microarray technologies, including issues related to probe design hybridization bias63 and sensitivity to detect low-abundance transcripts64. To identify candidate genes involved in the development of taste organs, we used RNA-Seq and investigated the transcriptomic profile of gustatory tissue and its surrounding compartments found in the chicken oral cavity.
In our analysis, we minimized false-positives and ensured substantial detection power and accuracy using several strategies to detect DEGs between samples by controlling critical influencing factors. Analyses were performed on deep-sequenced transcripts using two commonly used packages: DESeq2 and Cuffdiff, to ensure a higher sensitivity of algorithms that control FDR65 and provide greater inferential power in a typical RNA-Seq experiment with small replicate numbers4, 66, 67, and a rigorous statistical analysis32.
Furthermore, we confirmed the RNA-Seq results using qRT-PCR assay for a selected subset of DEGs. Overall, there was a high concordance and strong positive correlation between statistical and experimental results, revealing a high detection sensitivity and accuracy, which is similar to previous reports in animals9, 14, 16, 68, 69.
The mammalian taste organs including the tongue, taste papillae, and taste buds have been an ideal system for organogenesis studies and have attracted numerous researchers seeking to advance the field of developmental biology70–72. The topographic distribution of taste papillae and taste buds asserts taste organs as an ideal system for studying pattern formation73. Chicken taste buds, like those of mammals, are distributed in a patterned array28 and the many large clusters enables us to collect enough tissue for different analyses conveniently and efficiently. In the present study, our findings using RNA-Seq analysis provide useful information for further functional studies in organogenesis and regenerative medicine.
The functional enrichment analyses showed that multiple KEGG pathways and most GO terms are rigorously involved in sensory perception of taste, as well as the development of sensory organs and salivary glands. These results indicated that the identified DEGs may play important roles in taste function and taste organ development. Combining the significant number and expression levels of DEGs, GO and pathway results, and gene function, allows us to suggest that GNG13, SCNN1A, SCNN1B, SCNN1G and PRKX as 5 promising candidate genes for sensory perception of taste in chickens. In addition, 3 known genes (GNAT3, TRPM5 and TAS1R3) were revealed to be involved in taste perception and 4 analogous genes (SHH, TGFβ2, DKK3 and LEF1) are known to be involved in the development of taste organs in mouse. A large subset of key components of well-known molecular signaling pathways (Hedgehog, Wnt/β-catenin, TGFβ/BMP, Notch, FGF, Erbb) that are important for organogenesis in rodents were found to be highly DEGs between GE and GM. This suggests a potentially active involvement of these signaling pathways in the epithelial-mesenchymal interactions that are required for organogenesis74.
Of note, DEGs between GE and GM were also found to be enriched in salivary gland development. Chicken taste buds are often clustered in areas surrounding salivary gland openings24, 28. It is reasonable to speculate that the development of taste buds and salivary glands in chickens are likely to be associated or linked. Further studies on whether, what and how signaling molecules coordinate the organogenesis of taste buds and salivary glands openings will be beneficial for better understanding the formation of both organs.
In summary, chicken taste buds are distributed in a unique pattern in the gustatory tissue of the oral cavity. They have a much shorter lifespan (~4 days)75, 76 compared to mammals (~10–12 days in rodents)77, 78 indicating a more active progenitor cell niche and allowing for a more efficient way to study taste bud cell renewal compared to the rodent model. Moreover, the connective tissue cell marker Vimentin is expressed in a large population of taste bud cells in chickens, similar to humans79, 80, suggesting a comparable mechanism underlying the contribution of connective tissue to taste buds in both organisms. Furthermore, new taste buds continue to develop after hatching in female-line male chickens28, which provides a time window to study the regulation of taste bud development. In combination with other beneficial aspects of using chickens as a research model, e.g., convenience of in ovo embryo manipulation, high availability and rapid development81, 82, we propose that the chicken taste organ can serve as an ideal model for studies in organogenesis and regenerative medicine.
Electronic supplementary material
Acknowledgements
We give thanks to the staff at Cobb Vantress Cleaveland Hatchery, GA for providing the chickens. This study was supported by the National Institutes of Health (grant number R01 DC012308 to HXL) and University of Georgia Start-up fund to HXL.
Author Contributions
Conceived and designed the experiments: X.C., B.M., R.R., H.X.L., Performed the experiments: X.C., B.M., N.S., H.X.L., Analyzed data: X.C., B.M., R.R., H.X.L., Contributed reagents/materials/tools: S.C., R.R., H.X.L., Wrote and edited manuscript: X.C., B.M., S.C., R.R., H.X.L.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-09299-7
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nature reviews. Genetics. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews. Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ching T, Huang S, Garmire LX. Power analysis and sample size estimation for RNA-Seq differential expression. RNA. 2014;20:1684–1696. doi: 10.1261/rna.046011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biology. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nookaew I, et al. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Research. 2012;40:10084–10097. doi: 10.1093/nar/gks804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Verdal H, et al. Improving the efficiency of feed utilization in poultry by selection. 2. Genetic parameters of excretion traits and correlations with anatomy of the gastro-intestinal tract and digestive efficiency. BMC Genetics. 2011;12:71–71. doi: 10.1186/1471-2156-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saintilan R, et al. Genetics of residual feed intake in growing pigs: Relationships with production traits, and nitrogen and phosphorus excretion traits1. Journal of Animal Science. 2013;91:2542–2554. doi: 10.2527/jas.2012-5687. [DOI] [PubMed] [Google Scholar]
- 8.Ahanda M-LE, et al. Impact of the Genetic Background on the Composition of the Chicken Plasma MiRNome in Response to a Stress. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0114598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui X, et al. Transcriptional profiling of mammary gland in Holstein cows with extremely different milk protein and fat percentage using RNA sequencing. BMC Genomics. 2014;15:226–226. doi: 10.1186/1471-2164-15-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jing L, et al. Transcriptome analysis of mRNA and miRNA in skeletal muscle indicates an important network for differential Residual Feed Intake in pigs. Scientific Reports. 2015;5 doi: 10.1038/srep11953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao X, Qin QLX. Genome-wide transcriptome analysis of mRNAs and microRNAs in Dorset and Small Tail Han sheep to explore the regulation of fecundity. Molecular and Cellular Endocrinology. 2015;402:32–42. doi: 10.1016/j.mce.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 12.Sodhi SS, et al. Comparative transcriptomic analysis by RNA-seq to discern differential expression of genes in liver and muscle tissues of adult Berkshire and Jeju Native Pig. Gene. 2014;546:233–242. doi: 10.1016/j.gene.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Taylor JF, et al. Lessons for livestock genomics from genome and transcriptome sequencing in cattle and other mammals. Genetics, Selection, Evolution: GSE. 2016;48 doi: 10.1186/s12711-016-0237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi G, et al. In-Depth Duodenal Transcriptome Survey in Chickens with Divergent Feed Efficiency Using RNA-Seq. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0136765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, et al. Whole genomic prediction of growth and carcass traits in a Chinese quality chicken population1. Journal of Animal Science. 2017;95:72–80. doi: 10.2527/jas.2016.0823. [DOI] [PubMed] [Google Scholar]
- 16.Li C, et al. RNA-Seq reveals 10 novel promising candidate genes affecting milk protein concentration in the Chinese Holstein population. Scientific Reports. 2016;6 doi: 10.1038/srep26813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjørnstad S, Austdal LPE, Roald B, Glover JC, Paulsen RE. Cracking the Egg: Potential of the Developing Chicken as a Model System for Nonclinical Safety Studies of Pharmaceuticals. Journal of Pharmacology and Experimental Therapeutics. 2015;355:386–396. doi: 10.1124/jpet.115.227025. [DOI] [PubMed] [Google Scholar]
- 18.Schock EN, et al. Utilizing the chicken as an animal model for human craniofacial ciliopathies. Developmental Biology. 2016;415:326–337. doi: 10.1016/j.ydbio.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deusch S, Tilocca B, Camarinha-Silva A, Seifert J. News in livestock research — use of Omics-technologies to study the microbiota in the gastrointestinal tract of farm animals. Computational and Structural Biotechnology Journal. 2015;13:55–63. doi: 10.1016/j.csbj.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuse M. Screening of central functions of amino acids and their metabolites for sedative and hypnotic effects using chick models. European Journal of Pharmacology. 2015;762:382–393. doi: 10.1016/j.ejphar.2015.06.036. [DOI] [PubMed] [Google Scholar]
- 21.Lee HJ, Lee HC, Han JY. Germline Modification and Engineering in Avian Species. Molecules and Cells. 2015;38:743–749. doi: 10.14348/molcells.2015.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaeffel F, Feldkaemper M. Animal models in myopia research. Clinical and Experimental Optometry. 2015;98:507–517. doi: 10.1111/cxo.12312. [DOI] [PubMed] [Google Scholar]
- 23.Tufarelli V, Selvaggi M, Dario C, Laudadio V. Genetically Modified Feeds in Poultry Diet: Safety, Performance, and Product Quality. Critical Reviews in Food Science and Nutrition. 2015;55:562–569. doi: 10.1080/10408398.2012.667017. [DOI] [PubMed] [Google Scholar]
- 24.Ganchrow D, Ganchrow JR. Number and distribution of taste buds in the oral cavity of hatchling chicks. Physiology & behavior. 1985;34:889–894. doi: 10.1016/0031-9384(85)90009-5. [DOI] [PubMed] [Google Scholar]
- 25.Ganchrow JR, Ganchrow D. Taste bud development in chickens (Gallus gallus domesticus) The Anatomical Record. 1987;218:88–93. doi: 10.1002/ar.1092180113. [DOI] [PubMed] [Google Scholar]
- 26.Ganchrow JR, Steiner JE, Bartana A. Behavioral reactions to gustatory stimuli in young chicks (Gallus gallus domesticus) Developmental Psychobiology. 1990;23:103–117. doi: 10.1002/dev.420230202. [DOI] [PubMed] [Google Scholar]
- 27.Gentle MJ. Taste preference in the chicken (Gallus domesticus L.) British Poultry Science. 1972;13:141–155. doi: 10.1080/00071667208415928. [DOI] [PubMed] [Google Scholar]
- 28.Rajapaksha P, et al. Labeling and analysis of chicken taste buds using molecular markers in oral epithelial sheets. Scientific Reports. 2016;6 doi: 10.1038/srep37247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama A, et al. Expression of the basal cell markers of taste buds in the anterior tongue and soft palate of the mouse embryo. The Journal of Comparative Neurology. 2008;509:211–224. doi: 10.1002/cne.21738. [DOI] [PubMed] [Google Scholar]
- 30.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14:R36–R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, et al. Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:R106–R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Meth. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 35.Rau A, Gallopin M, Celeux G, Jaffrézic F. Data-based filtering for replicated high-throughput transcriptome sequencing experiments. Bioinformatics. 2013;29:2146–2152. doi: 10.1093/bioinformatics/btt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beites CL, et al. Follistatin modulates a BMP autoregulatory loop to control the size and patterning of sensory domains in the developing tongue. Development (Cambridge, England) 2009;136:2187–2197. doi: 10.1242/dev.030544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ermilov AN, et al. Maintenance of Taste Organs Is Strictly Dependent on Epithelial Hedgehog/GLI Signaling. PLoS Genetics. 2016;12 doi: 10.1371/journal.pgen.1006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaillard D, Barlow LA. Taste bud cells of adult mice are responsive to Wnt/β-catenin signaling: implications for the renewal of mature taste cells. Genesis (New York, N.Y.: 2000) 2011;49:295–306. doi: 10.1002/dvg.20731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han D, et al. A TGFβ-Smad4-Fgf6 signaling cascade controls myogenic differentiation and myoblast fusion during tongue development. Development (Cambridge, England) 2012;139:1640–1650. doi: 10.1242/dev.076653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwatsuki K, et al. Wnt signaling interacts with Shh to regulate taste papilla development. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2253–2258. doi: 10.1073/pnas.0607399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu HX, et al. Multiple Shh signaling centers participate in fungiform papilla and taste bud formation and maintenance. Developmental biology. 2013;382:82–97. doi: 10.1016/j.ydbio.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H-X, MacCallum DK, Edwards C, Gaffield W, Mistretta CM. Sonic hedgehog exerts distinct, stage-specific effects on tongue and taste papilla development. Developmental Biology. 2004;276:280–300. doi: 10.1016/j.ydbio.2004.07.042. [DOI] [PubMed] [Google Scholar]
- 43.Mistretta CM, Liu H-X, Gaffield W, MacCallum DK. Cyclopamine and jervine in embryonic rat tongue cultures demonstrate a role for Shh signaling in taste papilla development and patterning: fungiform papillae double in number and form in novel locations in dorsal lingual epithelium. Developmental Biology. 2003;254:1–18. doi: 10.1016/S0012-1606(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura S-i, Kawai T, Kamakura T, Ookura T. TGF-β3 is expressed in taste buds and inhibits proliferation of primary cultured taste epithelial cells. In Vitro Cellular & Developmental Biology - Animal. 2010;46:36–44. doi: 10.1007/s11626-009-9239-9. [DOI] [PubMed] [Google Scholar]
- 45.Schneider FT, et al. Sonic Hedgehog Acts as a Negative Regulator of β-Catenin Signaling in the Adult Tongue Epithelium. The American Journal of Pathology. 2010;177:404–414. doi: 10.2353/ajpath.2010.091079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y, Liu H-X, Mistretta CM. Bone morphogenetic proteins and noggin: Inhibiting and inducing fungiform taste papilla development. Developmental Biology. 2006;297:198–213. doi: 10.1016/j.ydbio.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 47.Kim JY, et al. Shh and ROCK1 modulate the dynamic epithelial morphogenesis in circumvallate papilla development. Dev Biol. 2009;325:273–280. doi: 10.1016/j.ydbio.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 48.Hall JM, Bell ML, Finger TE. Disruption of sonic hedgehog signaling alters growth and patterning of lingual taste papillae. Dev Biol. 2003;255:263–277. doi: 10.1016/S0012-1606(02)00048-9. [DOI] [PubMed] [Google Scholar]
- 49.Hall JM, Hooper JE, Finger TE. Expression of sonic hedgehog, patched, and Gli1 in developing taste papillae of the mouse. J Comp Neurol. 1999;406:143–155. doi: 10.1002/(SICI)1096-9861(19990405)406:2<143::AID-CNE1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 50.Zhu X, et al. Gpr177-mediated Wnt Signaling is Required for Fungiform Placode Initiation. J Dent Res. 2014;93:582–588. doi: 10.1177/0022034514531985. [DOI] [PubMed] [Google Scholar]
- 51.Liu F, et al. Wnt-beta-catenin signaling initiates taste papilla development. Nat Genet. 2007;39:106–112. doi: 10.1038/ng1932. [DOI] [PubMed] [Google Scholar]
- 52.Liu F, Millar SE. Wnt/beta-catenin signaling in oral tissue development and disease. J Dent Res. 2010;89:318–330. doi: 10.1177/0022034510363373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han D, et al. A TGFbeta-Smad4-Fgf6 signaling cascade controls myogenic differentiation and myoblast fusion during tongue development. Development. 2012;139:1640–1650. doi: 10.1242/dev.076653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawasaki K, et al. Bmp signalling in filiform tongue papillae development. Arch Oral Biol. 2012;57:805–813. doi: 10.1016/j.archoralbio.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren W, et al. Transcriptome analyses of taste organoids reveal multiple pathways involved in taste cell generation. Sci Rep. 2017;7 doi: 10.1038/s41598-017-04099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seta Y, Toyono T, Kataoka S, Toyoshima K. Regulation of taste bud cell differentiation by notch signaling pathway. Chem Senses. 2005;30(Suppl 1):i48–49. doi: 10.1093/chemse/bjh107. [DOI] [PubMed] [Google Scholar]
- 57.Seta Y, Seta C, Barlow LA. Notch-associated gene expression in embryonic and adult taste papillae and taste buds suggests a role in taste cell lineage decisions. J Comp Neurol. 2003;464:49–61. doi: 10.1002/cne.10787. [DOI] [PubMed] [Google Scholar]
- 58.Petersen CI, et al. FGF signaling regulates the number of posterior taste papillae by controlling progenitor field size. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ookura T, et al. Fibroblast and epidermal growth factors modulate proliferation and neural cell adhesion molecule expression in epithelial cells derived from the adult mouse tongue. In Vitro Cell Dev Biol Anim. 2002;38:365–372. doi: 10.1290/1071-2690(2002)038<0365:FAEGFM>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 60.McLaughlin SKE. and c-Kit receptors have distinctive patterns of expression in adult and developing taste papillae and taste buds. J Neurosci. 2000;20:5679–5688. doi: 10.1523/JNEUROSCI.20-15-05679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu HX, Henson BS, Zhou Y, D’Silva NJ, Mistretta CM. Fungiform papilla pattern: EGF regulates inter-papilla lingual epithelium and decreases papilla number by means of PI3K/Akt, MEK/ERK, and p38 MAPK signaling. Dev Dyn. 2008;237:2378–2393. doi: 10.1002/dvdy.21657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morris-Wiman J, Sego R, Brinkley L, Dolce C. The effects of sialoadenectomy and exogenous EGF on taste bud morphology and maintenance. Chem Senses. 2000;25:9–19. doi: 10.1093/chemse/25.1.9. [DOI] [PubMed] [Google Scholar]
- 63.Croucher NJ, Thomson NR. Studying bacterial transcriptomes using RNA-seq. Current Opinion in Microbiology. 2010;13:619–624. doi: 10.1016/j.mib.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Research. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maza E. In Papyro Comparison of TMM (edgeR), RLE (DESeq. 2), and MRN Normalization Methods for a Simple Two-Conditions-Without-Replicates RNA-Seq Experimental Design. Frontiers in Genetics. 2016;7 doi: 10.3389/fgene.2016.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burden CJ, Qureshi SE, Wilson SR. Error estimates for the analysis of differential expression from RNA-seq count data. PeerJ. 2014;2 doi: 10.7717/peerj.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seyednasrollah F, Laiho A, Elo LL. Comparison of software packages for detecting differential expression in RNA-seq studies. Briefings in Bioinformatics. 2015;16:59–70. doi: 10.1093/bib/bbt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coble DJ, et al. RNA-seq analysis of broiler liver transcriptome reveals novel responses to high ambient temperature. BMC Genomics. 2014;15 doi: 10.1186/1471-2164-15-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X, et al. Growth Hormone-Regulated mRNAs and miRNAs in Chicken Hepatocytes. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kapsimali M, Barlow LA. DEVELOPING A SENSE OF TASTE. Seminars in cell & developmental biology. 2013;24:200–209. doi: 10.1016/j.semcdb.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krimm RF. Factors that regulate embryonic gustatory development. BMC Neuroscience. 2007;8:S4–S4. doi: 10.1186/1471-2202-8-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krimm, R. F., Thirumangalathu, S., Barlow, L. A. Development of the Taste System. Handbook of Olfaction and Gustation, 3rd Edition. pp. 727–747 (2015).
- 73.Mistretta CM, Liu H-X. Development of fungiform papillae: Patterned lingual gustatory organs. Archives of Histology and Cytology. 2006;69:199–208. doi: 10.1679/aohc.69.199. [DOI] [PubMed] [Google Scholar]
- 74.Ribatti D, Santoiemma M. Epithelial-mesenchymal interactions: a fundamental Developmental Biology mechanism. Int J Dev Biol. 2014;58:303–306. doi: 10.1387/ijdb.140143dr. [DOI] [PubMed] [Google Scholar]
- 75.Ganchrow D, Ganchrow JR, Romano R, Kinnamon JC. Ontogenesis and taste bud cell turnover in the chicken. I. Gemmal cell renewal in the hatchling. J Comp Neurol. 1994;345:105–114. doi: 10.1002/cne.903450108. [DOI] [PubMed] [Google Scholar]
- 76.Ganchrow JR, Ganchrow D, Royer SM, Kinnamon JC. Aspects of vertebrate gustatory phylogeny: morphology and turnover of chick taste bud cells. Microsc Res Tech. 1993;26:106–119. doi: 10.1002/jemt.1070260204. [DOI] [PubMed] [Google Scholar]
- 77.Perea-Martinez I, Nagai T, Chaudhari N. Functional cell types in taste buds have distinct longevities. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Farbman AI. Renewal of taste bud cells in rat circumvallate papillae. Cell Tissue Kinet. 1980;13:349–357. doi: 10.1111/j.1365-2184.1980.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 79.Venkatesan N, et al. Distribution of alpha-Gustducin and Vimentin in premature and mature taste buds in chickens. Biochem Biophys Res Commun. 2016 doi: 10.1016/j.bbrc.2016.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Witt M, Reutter K, Ganchrow D, Ganchrow JR. Fingerprinting taste buds: intermediate filaments and their implication for taste bud formation. Philos Trans R Soc Lond B Biol Sci. 2000;355:1233–1237. doi: 10.1098/rstb.2000.0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hughes A. The development of the neural tube of the chick embryo. A study with the ultraviolet microscope. Journal of Embryology and Experimental Morphology. 1955;3:305–325. [Google Scholar]
- 82.Odani, N., Hou, X. & Nakamura, H. In Electroporation and Sonoporation in Developmental Biology 9–16 (Springer, 2009).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.