Abstract
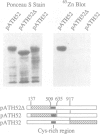
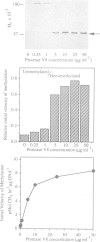
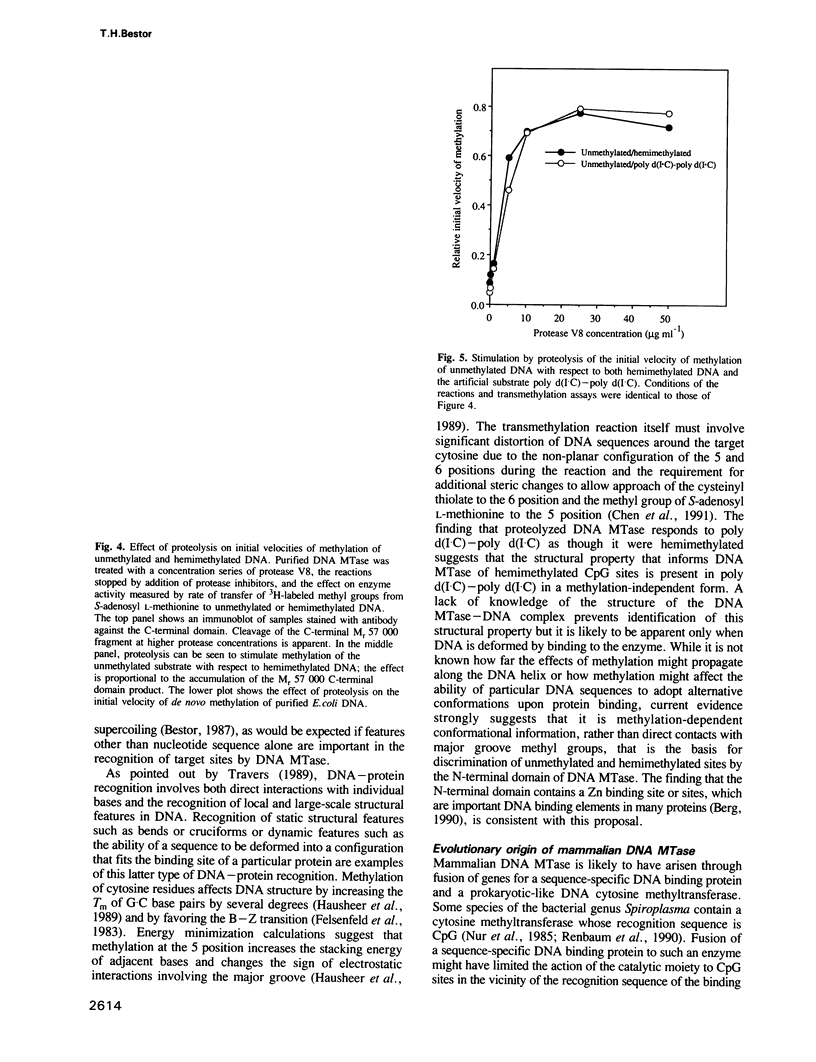
Mammalian DNA (cytosine-5) methyltransferase contains a C-terminal domain that is closely related to bacterial cytosine-5 restriction methyltransferase. This methyltransferase domain is linked to a large N-terminal domain. It is shown here that the N-terminal domain contains a Zn binding site and that the N- and C-terminal domains can be separated by cleavage with trypsin or Staphylococcus aureus protease V8; the protease V8 cleavage site was determined by Edman degradation to lie 10 residues C-terminal of the run of alternating lysyl and glycyl residues which joins the two domains and six residues N-terminal of the first sequence motif conserved between the mammalian and bacterial cytosine methyltransferases. While the intact enzyme had little activity on unmethylated DNA substrates, cleavage between the domains caused a large stimulation of the initial velocity of methylation of unmethylated DNA without substantial change in the rate of methylation of hemimethylated DNA. These findings indicate that the N-terminal domain of DNA methyltransferase ensures the clonal propagation of methylation patterns through inhibition of the de novo activity of the C-terminal domain. Mammalian DNA methyltransferase is likely to have arisen via fusion of a prokaryotic-like restriction methyltransferase and an unrelated DNA binding protein. Stimulation of the de novo activity of DNA methyltransferase by proteolytic cleavage in vivo may contribute to the process of ectopic methylation observed in the DNA of aging animals, tumors and in lines of cultured cells.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., Burdon R. H., McKinnon K., Rinaldi A. Stimulation of de novo methylation following limited proteolysis of mouse ascites DNA methylase. FEBS Lett. 1983 Nov 14;163(2):194–198. doi: 10.1016/0014-5793(83)80817-5. [DOI] [PubMed] [Google Scholar]
- Antequera F., Boyes J., Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990 Aug 10;62(3):503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Antequera F., Macleod D., Bird A. P. Specific protection of methylated CpGs in mammalian nuclei. Cell. 1989 Aug 11;58(3):509–517. doi: 10.1016/0092-8674(89)90431-5. [DOI] [PubMed] [Google Scholar]
- Baylin S. B., Fearon E. R., Vogelstein B., de Bustros A., Sharkis S. J., Burke P. J., Staal S. P., Nelkin B. D. Hypermethylation of the 5' region of the calcitonin gene is a property of human lymphoid and acute myeloid malignancies. Blood. 1987 Aug;70(2):412–417. [PubMed] [Google Scholar]
- Baylin S. B., Höppener J. W., de Bustros A., Steenbergh P. H., Lips C. J., Nelkin B. D. DNA methylation patterns of the calcitonin gene in human lung cancers and lymphomas. Cancer Res. 1986 Jun;46(6):2917–2922. [PubMed] [Google Scholar]
- Berg J. M. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J Biol Chem. 1990 Apr 25;265(12):6513–6516. [PubMed] [Google Scholar]
- Bestor T. H. DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1235):179–187. doi: 10.1098/rstb.1990.0002. [DOI] [PubMed] [Google Scholar]
- Bestor T. H., Ingram V. M. Growth-dependent expression of multiple species of DNA methyltransferase in murine erythroleukemia cells. Proc Natl Acad Sci U S A. 1985 May;82(9):2674–2678. doi: 10.1073/pnas.82.9.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T. H., Ingram V. M. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T., Ingram V. M. The DNA-methylating system of murine erythroleukemia cells. Prog Clin Biol Res. 1985;198:95–104. [PubMed] [Google Scholar]
- Bestor T., Laudano A., Mattaliano R., Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988 Oct 20;203(4):971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Bestor T. Supercoiling-dependent sequence specificity of mammalian DNA methyltransferase. Nucleic Acids Res. 1987 May 11;15(9):3835–3843. doi: 10.1093/nar/15.9.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991 Nov 19;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J Biol Chem. 1985 Feb 10;260(3):1513–1520. [PubMed] [Google Scholar]
- Felsenfeld G., Nickol J., Behe M., McGhee J., Jackson D. Methylation and chromatin structure. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):577–584. doi: 10.1101/sqb.1983.047.01.068. [DOI] [PubMed] [Google Scholar]
- Friedman S., Som S., Yang L. F. The core element of the EcoRII methylase as defined by protease digestion and deletion analysis. Nucleic Acids Res. 1991 Oct 11;19(19):5403–5408. doi: 10.1093/nar/19.19.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982 Feb 18;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
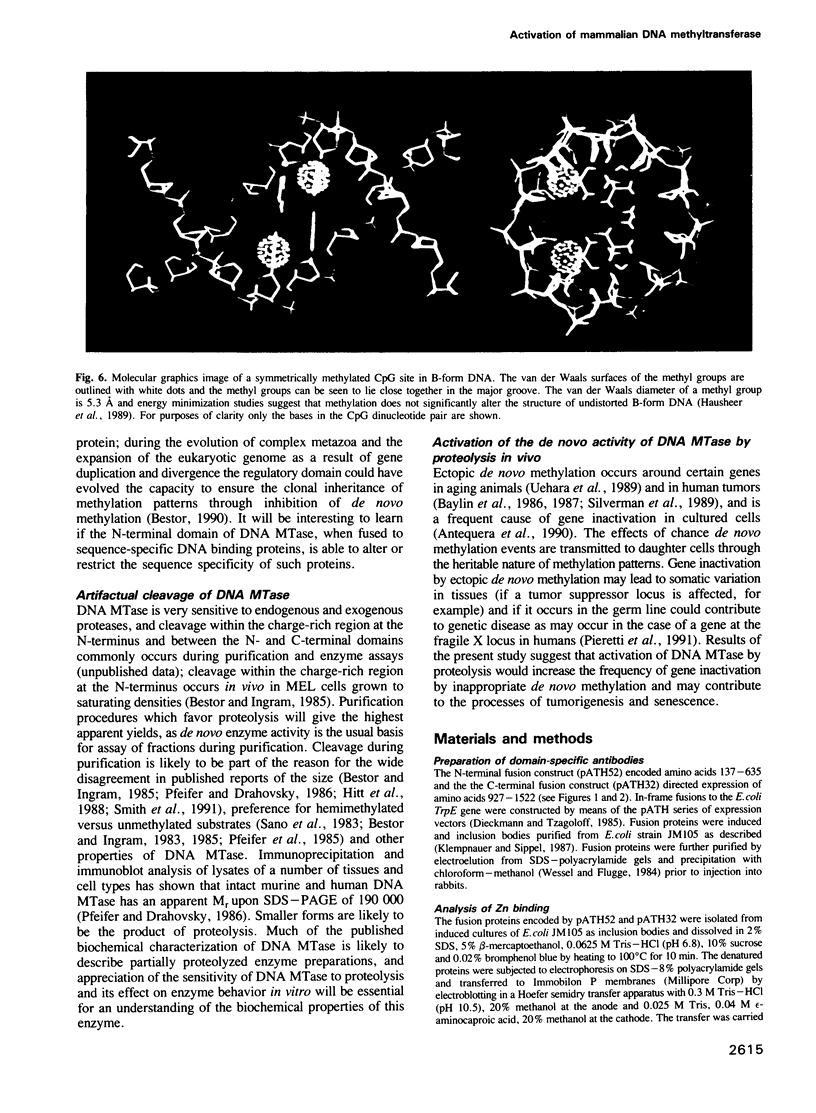
- Hausheer F. H., Rao S. N., Gamcsik M. P., Kollman P. A., Colvin O. M., Saxe J. D., Nelkin B. D., McLennan I. J., Barnett G., Baylin S. B. Computational analysis of structural and energetic consequences of DNA methylation. Carcinogenesis. 1989 Jun;10(6):1131–1137. doi: 10.1093/carcin/10.6.1131. [DOI] [PubMed] [Google Scholar]
- Hitt M. M., Wu T. L., Cohen G., Linn S. De novo and maintenance DNA methylation by a mouse plasmacytoma cell DNA methyltransferase. J Biol Chem. 1988 Mar 25;263(9):4392–4399. [PubMed] [Google Scholar]
- Ingrosso D., Fowler A. V., Bleibaum J., Clarke S. Sequence of the D-aspartyl/L-isoaspartyl protein methyltransferase from human erythrocytes. Common sequence motifs for protein, DNA, RNA, and small molecule S-adenosylmethionine-dependent methyltransferases. J Biol Chem. 1989 Nov 25;264(33):20131–20139. [PubMed] [Google Scholar]
- Kelleher J. E., Daniel A. S., Murray N. E. Mutations that confer de novo activity upon a maintenance methyltransferase. J Mol Biol. 1991 Sep 20;221(2):431–440. doi: 10.1016/0022-2836(91)80064-2. [DOI] [PubMed] [Google Scholar]
- Klempnauer K. H., Sippel A. E. The highly conserved amino-terminal region of the protein encoded by the v-myb oncogene functions as a DNA-binding domain. EMBO J. 1987 Sep;6(9):2719–2725. doi: 10.1002/j.1460-2075.1987.tb02565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S., Nelson J. L., Roberts R. J. The sequence specificity domain of cytosine-C5 methylases. Nucleic Acids Res. 1991 Nov 25;19(22):6183–6190. doi: 10.1093/nar/19.22.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauster R., Trautner T. A., Noyer-Weidner M. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J Mol Biol. 1989 Mar 20;206(2):305–312. doi: 10.1016/0022-2836(89)90480-4. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991 Aug 8;352(6335):497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Nelkin B. D., Przepiorka D., Burke P. J., Thomas E. D., Baylin S. B. Abnormal methylation of the calcitonin gene marks progression of chronic myelogenous leukemia. Blood. 1991 Jun 1;77(11):2431–2434. [PubMed] [Google Scholar]
- Nur I., Szyf M., Razin A., Glaser G., Rottem S., Razin S. Procaryotic and eucaryotic traits of DNA methylation in spiroplasmas (mycoplasmas). J Bacteriol. 1985 Oct;164(1):19–24. doi: 10.1128/jb.164.1.19-24.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Drahovsky D. DNA methyltransferase polypeptides in mouse and human cells. Biochim Biophys Acta. 1986 Dec 18;868(4):238–242. doi: 10.1016/0167-4781(86)90059-x. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Grünwald S., Palitti F., Kaul S., Boehm T. L., Hirth H. P., Drahovsky D. Purification and characterization of mammalian DNA methyltransferases by use of monoclonal antibodies. J Biol Chem. 1985 Nov 5;260(25):13787–13793. [PubMed] [Google Scholar]
- Pieretti M., Zhang F. P., Fu Y. H., Warren S. T., Oostra B. A., Caskey C. T., Nelson D. L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991 Aug 23;66(4):817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renbaum P., Abrahamove D., Fainsod A., Wilson G. G., Rottem S., Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res. 1990 Mar 11;18(5):1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Noguchi H., Sager R. Characterization of DNA methyltransferase from bovine thymus cells. Eur J Biochem. 1983 Sep 15;135(2):181–185. doi: 10.1111/j.1432-1033.1983.tb07635.x. [DOI] [PubMed] [Google Scholar]
- Schiff L. A., Nibert M. L., Fields B. N. Characterization of a zinc blotting technique: evidence that a retroviral gag protein binds zinc. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4195–4199. doi: 10.1073/pnas.85.12.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman A. L., Park J. G., Hamilton S. R., Gazdar A. F., Luk G. D., Baylin S. B. Abnormal methylation of the calcitonin gene in human colonic neoplasms. Cancer Res. 1989 Jul 1;49(13):3468–3473. [PubMed] [Google Scholar]
- Smith S. S., Kan J. L., Baker D. J., Kaplan B. E., Dembek P. Recognition of unusual DNA structures by human DNA (cytosine-5)methyltransferase. J Mol Biol. 1991 Jan 5;217(1):39–51. doi: 10.1016/0022-2836(91)90609-a. [DOI] [PubMed] [Google Scholar]
- Stein R., Gruenbaum Y., Pollack Y., Razin A., Cedar H. Clonal inheritance of the pattern of DNA methylation in mouse cells. Proc Natl Acad Sci U S A. 1982 Jan;79(1):61–65. doi: 10.1073/pnas.79.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Treich I., Riva M., Sentenac A. Zinc-binding subunits of yeast RNA polymerases. J Biol Chem. 1991 Nov 15;266(32):21971–21976. [PubMed] [Google Scholar]
- Uehara Y., Ono T., Kurishita A., Kokuryu H., Okada S. Age-dependent and tissue-specific changes of DNA methylation within and around the c-fos gene in mice. Oncogene. 1989 Aug;4(8):1023–1028. [PubMed] [Google Scholar]
- Wessel D., Flügge U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984 Apr;138(1):141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Wigler M., Levy D., Perucho M. The somatic replication of DNA methylation. Cell. 1981 Apr;24(1):33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]
- de Bustros A., Nelkin B. D., Silverman A., Ehrlich G., Poiesz B., Baylin S. B. The short arm of chromosome 11 is a "hot spot" for hypermethylation in human neoplasia. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5693–5697. doi: 10.1073/pnas.85.15.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]