Abstract
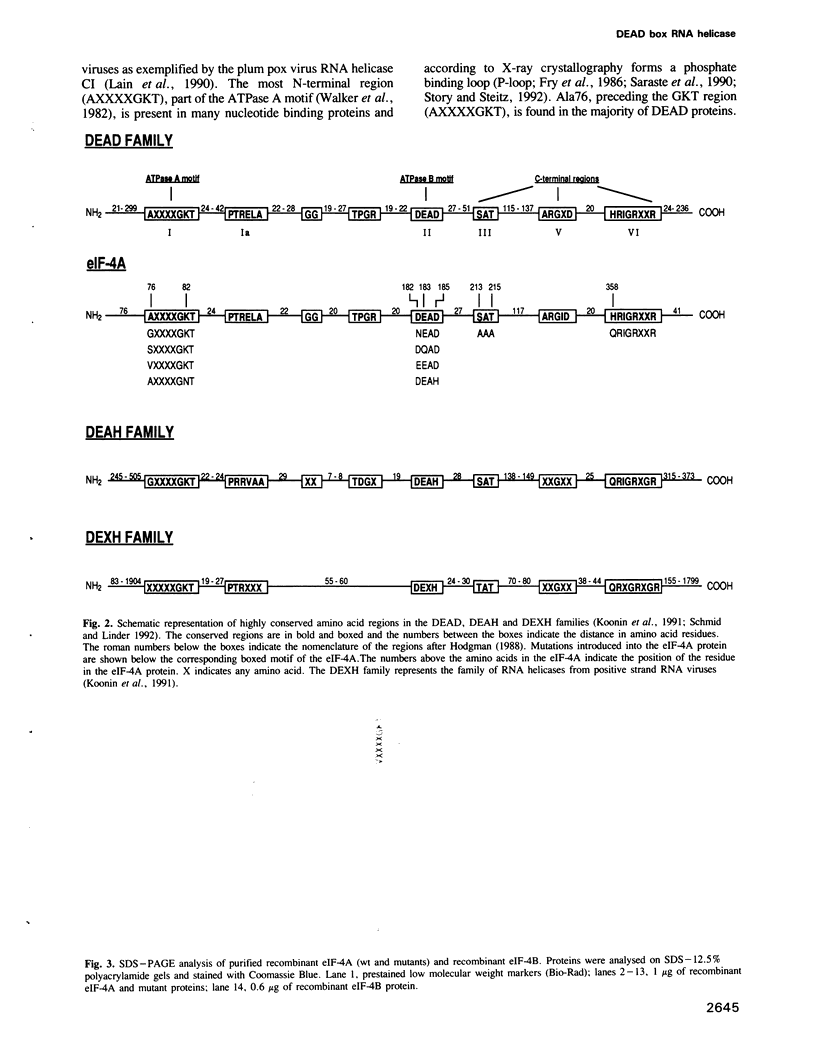
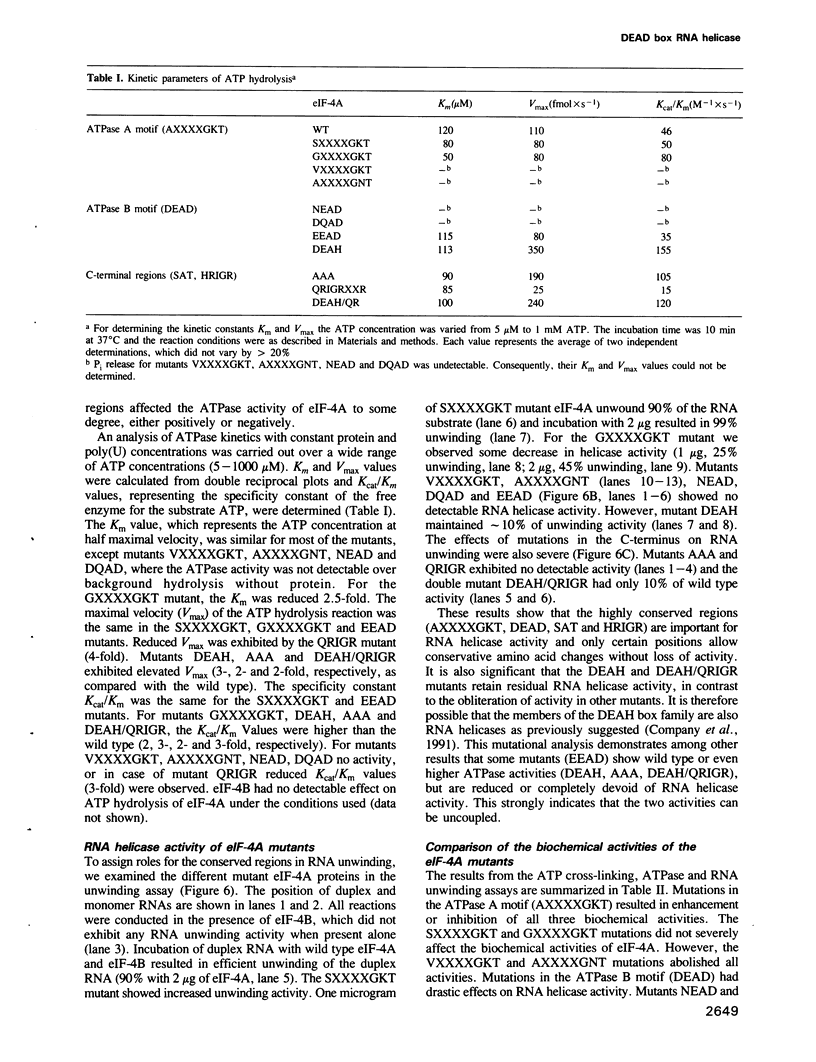
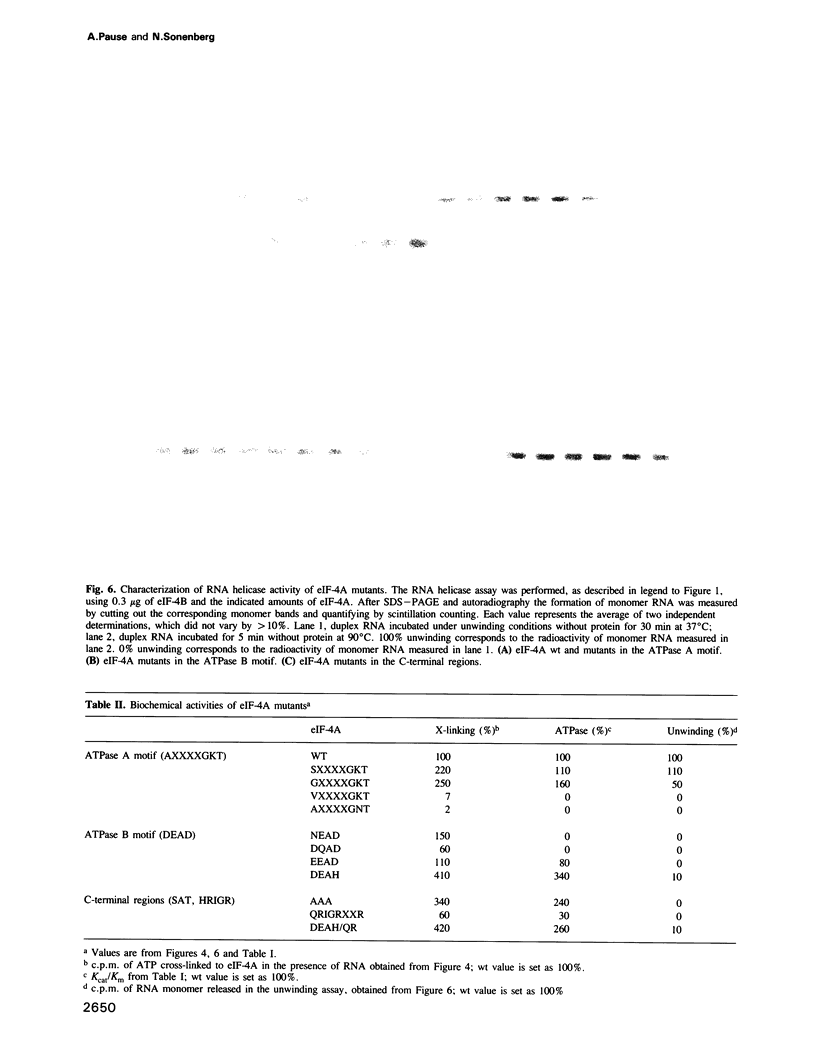
eIF-4A is a translation initiation factor that exhibits bidirectional RNA unwinding activity in vitro in the presence of another translation initiation factor, eIF-4B and ATP. This activity is thought to be responsible for the melting of secondary structure in the 5' untranslated region of eukaryotic mRNAs to facilitate ribosome binding. eIF-4A is a member of a fast growing family of proteins termed the DEAD family. These proteins are believed to be RNA helicases, based on the demonstrated in vitro RNA helicase activity of two members (eIF-4A and p68) and their homology in eight amino acid regions. Several related biochemical activities were attributed to eIF-4A: (i) ATP binding, (ii) RNA-dependent ATPase and (iii) RNA helicase. To determine the contribution of the highly conserved regions to these activities, we performed site-directed mutagenesis. First we show that recombinant eIF-4A, together with recombinant eIF-4B, exhibit RNA helicase activity in vitro. Mutations in the ATPase A motif (AXXXXGKT) affect ATP binding, whereas mutations in the predicted ATPase B motif (DEAD) affect ATP hydrolysis. We report here that the DEAD region couples the ATPase with the RNA helicase activity. Furthermore, two other regions, whose functions were unknown, have also been characterized. We report that the first residue in the HRIGRXXR region is involved in ATP hydrolysis and that the SAT region is essential for RNA unwinding. Our results suggest that the highly conserved regions in the DEAD box family are critical for RNA helicase activity.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abramson R. D., Dever T. E., Lawson T. G., Ray B. K., Thach R. E., Merrick W. C. The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J Biol Chem. 1987 Mar 15;262(8):3826–3832. [PubMed] [Google Scholar]
- Abramson R. D., Dever T. E., Merrick W. C. Biochemical evidence supporting a mechanism for cap-independent and internal initiation of eukaryotic mRNA. J Biol Chem. 1988 May 5;263(13):6016–6019. [PubMed] [Google Scholar]
- Altmann M., Blum S., Pelletier J., Sonenberg N., Wilson T. M., Trachsel H. Translation initiation factor-dependent extracts from Saccharomyces cerevisiae. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):155–159. doi: 10.1016/0167-4781(90)90158-x. [DOI] [PubMed] [Google Scholar]
- Benne R., Hershey J. W. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978 May 10;253(9):3078–3087. [PubMed] [Google Scholar]
- Berkower C., Michaelis S. Mutational analysis of the yeast a-factor transporter STE6, a member of the ATP binding cassette (ABC) protein superfamily. EMBO J. 1991 Dec;10(12):3777–3785. doi: 10.1002/j.1460-2075.1991.tb04947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum S., Mueller M., Schmid S. R., Linder P., Trachsel H. Translation in Saccharomyces cerevisiae: initiation factor 4A-dependent cell-free system. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6043–6046. doi: 10.1073/pnas.86.16.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Company M., Arenas J., Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991 Feb 7;349(6309):487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- Conroy S. C., Dever T. E., Owens C. L., Merrick W. C. Characterization of the 46,000-dalton subunit of eIF-4F. Arch Biochem Biophys. 1990 Nov 1;282(2):363–371. doi: 10.1016/0003-9861(90)90130-q. [DOI] [PubMed] [Google Scholar]
- Dalbadie-McFarland G., Abelson J. PRP5: a helicase-like protein required for mRNA splicing in yeast. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4236–4240. doi: 10.1073/pnas.87.11.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombroski A. J., Brennan C. A., Spear P., Platt T. Site-directed alterations in the ATP-binding domain of rho protein affect its activities as a termination factor. J Biol Chem. 1988 Dec 15;263(35):18802–18809. [PubMed] [Google Scholar]
- Duncan R., Hershey J. W. Regulation of initiation factors during translational repression caused by serum depletion. Covalent modification. J Biol Chem. 1985 May 10;260(9):5493–5497. [PubMed] [Google Scholar]
- Edery I., Hümbelin M., Darveau A., Lee K. A., Milburn S., Hershey J. W., Trachsel H., Sonenberg N. Involvement of eukaryotic initiation factor 4A in the cap recognition process. J Biol Chem. 1983 Sep 25;258(18):11398–11403. [PubMed] [Google Scholar]
- Fry D. C., Kuby S. A., Mildvan A. S. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc Natl Acad Sci U S A. 1986 Feb;83(4):907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifo J. A., Abramson R. D., Satler C. A., Merrick W. C. RNA-stimulated ATPase activity of eukaryotic initiation factors. J Biol Chem. 1984 Jul 10;259(13):8648–8654. [PubMed] [Google Scholar]
- Grifo J. A., Tahara S. M., Morgan M. A., Shatkin A. J., Merrick W. C. New initiation factor activity required for globin mRNA translation. J Biol Chem. 1983 May 10;258(9):5804–5810. [PubMed] [Google Scholar]
- Hirling H., Scheffner M., Restle T., Stahl H. RNA helicase activity associated with the human p68 protein. Nature. 1989 Jun 15;339(6225):562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- Hodgman T. C. A new superfamily of replicative proteins. Nature. 1988 May 5;333(6168):22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- Jackson R. J. The ATP requirement for initiation of eukaryotic translation varies according to the mRNA species. Eur J Biochem. 1991 Sep 1;200(2):285–294. doi: 10.1111/j.1432-1033.1991.tb16184.x. [DOI] [PubMed] [Google Scholar]
- Jagus R., Anderson W. F., Safer B. The regulation of initiation of mammalian protein synthesis. Prog Nucleic Acid Res Mol Biol. 1981;25:127–185. doi: 10.1016/s0079-6603(08)60484-5. [DOI] [PubMed] [Google Scholar]
- Jaramillo M., Browning K., Dever T. E., Blum S., Trachsel H., Merrick W. C., Ravel J. M., Sonenberg N. Translation initiation factors that function as RNA helicases from mammals, plants and yeast. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):134–139. doi: 10.1016/0167-4781(90)90154-t. [DOI] [PubMed] [Google Scholar]
- Jaramillo M., Dever T. E., Merrick W. C., Sonenberg N. RNA unwinding in translation: assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF-4B. Mol Cell Biol. 1991 Dec;11(12):5992–5997. doi: 10.1128/mcb.11.12.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Choi G. H., Nuss D. L., Shapira R., Carrington J. C. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10647–10651. doi: 10.1073/pnas.88.23.10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. Similarities in RNA helicases. Nature. 1991 Jul 25;352(6333):290–290. doi: 10.1038/352290c0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawson T. G., Lee K. A., Maimone M. M., Abramson R. D., Dever T. E., Merrick W. C., Thach R. E. Dissociation of double-stranded polynucleotide helical structures by eukaryotic initiation factors, as revealed by a novel assay. Biochemistry. 1989 May 30;28(11):4729–4734. doi: 10.1021/bi00437a033. [DOI] [PubMed] [Google Scholar]
- Laín S., Riechmann J. L., García J. A. RNA helicase: a novel activity associated with a protein encoded by a positive strand RNA virus. Nucleic Acids Res. 1990 Dec 11;18(23):7003–7006. doi: 10.1093/nar/18.23.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Lasko P. F., Ashburner M., Leroy P., Nielsen P. J., Nishi K., Schnier J., Slonimski P. P. Birth of the D-E-A-D box. Nature. 1989 Jan 12;337(6203):121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- Milburn S. C., Hershey J. W., Davies M. V., Kelleher K., Kaufman R. J. Cloning and expression of eukaryotic initiation factor 4B cDNA: sequence determination identifies a common RNA recognition motif. EMBO J. 1990 Sep;9(9):2783–2790. doi: 10.1002/j.1460-2075.1990.tb07466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn S. C., Pelletier J., Sonenberg N., Hershey J. W. Identification of the 80-kDa protein that crosslinks to the cap structure of eukaryotic mRNAs as initiation factor eIF-4B. Arch Biochem Biophys. 1988 Jul;264(1):348–350. doi: 10.1016/0003-9861(88)90604-2. [DOI] [PubMed] [Google Scholar]
- Morley S. J., Traugh J. A. Differential stimulation of phosphorylation of initiation factors eIF-4F, eIF-4B, eIF-3, and ribosomal protein S6 by insulin and phorbol esters. J Biol Chem. 1990 Jun 25;265(18):10611–10616. [PubMed] [Google Scholar]
- Nielsen P. J., McMaster G. K., Trachsel H. Cloning of eukaryotic protein synthesis initiation factor genes: isolation and characterization of cDNA clones encoding factor eIF-4A. Nucleic Acids Res. 1985 Oct 11;13(19):6867–6880. doi: 10.1093/nar/13.19.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. J., Trachsel H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 1988 Jul;7(7):2097–2105. doi: 10.1002/j.1460-2075.1988.tb03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K., Morel-Deville F., Hershey J. W., Leighton T., Schnier J. An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly. Nature. 1988 Dec 1;336(6198):496–498. doi: 10.1038/336496a0. [DOI] [PubMed] [Google Scholar]
- Pai E. F., Krengel U., Petsko G. A., Goody R. S., Kabsch W., Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990 Aug;9(8):2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B. K., Lawson T. G., Kramer J. C., Cladaras M. H., Grifo J. A., Abramson R. D., Merrick W. C., Thach R. E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985 Jun 25;260(12):7651–7658. [PubMed] [Google Scholar]
- Rozen F., Edery I., Meerovitch K., Dever T. E., Merrick W. C., Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990 Mar;10(3):1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen F., Pelletier J., Trachsel H., Sonenberg N. A lysine substitution in the ATP-binding site of eucaryotic initiation factor 4A abrogates nucleotide-binding activity. Mol Cell Biol. 1989 Sep;9(9):4061–4063. doi: 10.1128/mcb.9.9.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990 Mar 2;247(4946):1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- Safer B., Kemper W., Jagus R. Identification of a 48 S preinitiation complex in reticulocyte lysate. J Biol Chem. 1978 May 25;253(10):3384–3386. [PubMed] [Google Scholar]
- Saraste M., Sibbald P. R., Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990 Nov;15(11):430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Sarkar G., Edery I., Sonenberg N. Photoaffinity labeling of the cap-binding protein complex with ATP/dATP. Differential labeling of free eukaryotic initiation factor 4A and the eukaryotic initiation factor 4A component of the cap-binding protein complex with [alpha-32P]ATP/dATP. J Biol Chem. 1985 Nov 5;260(25):13831–13837. [PubMed] [Google Scholar]
- Scheffner M., Knippers R., Stahl H. RNA unwinding activity of SV40 large T antigen. Cell. 1989 Jun 16;57(6):955–963. doi: 10.1016/0092-8674(89)90334-6. [DOI] [PubMed] [Google Scholar]
- Schmid S. R., Linder P. Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol Cell Biol. 1991 Jul;11(7):3463–3471. doi: 10.1128/mcb.11.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991 Feb 7;349(6309):494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- Sonenberg N. Cap-binding proteins of eukaryotic messenger RNA: functions in initiation and control of translation. Prog Nucleic Acid Res Mol Biol. 1988;35:173–207. doi: 10.1016/s0079-6603(08)60614-5. [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Morgan M. A., Merrick W. C., Shatkin A. J. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5'-terminal cap in mRNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story R. M., Steitz T. A. Structure of the recA protein-ADP complex. Nature. 1992 Jan 23;355(6358):374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Trachsel H., Erni B., Schreier M. H., Staehelin T. Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J Mol Biol. 1977 Nov;116(4):755–767. doi: 10.1016/0022-2836(77)90269-8. [DOI] [PubMed] [Google Scholar]
- Valencia A., Chardin P., Wittinghofer A., Sander C. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry. 1991 May 14;30(19):4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. RNA splicing. Alive with DEAD proteins. Nature. 1991 Feb 7;349(6309):463–464. doi: 10.1038/349463a0. [DOI] [PubMed] [Google Scholar]
- Yue V. T., Schimmel P. R. Direct and specific photochemical cross-linking of adenosine 5'-triphosphate to an aminoacyl-tRNA synthetase. Biochemistry. 1977 Oct 18;16(21):4678–4684. doi: 10.1021/bi00640a023. [DOI] [PubMed] [Google Scholar]







