Abstract
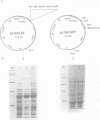
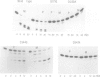
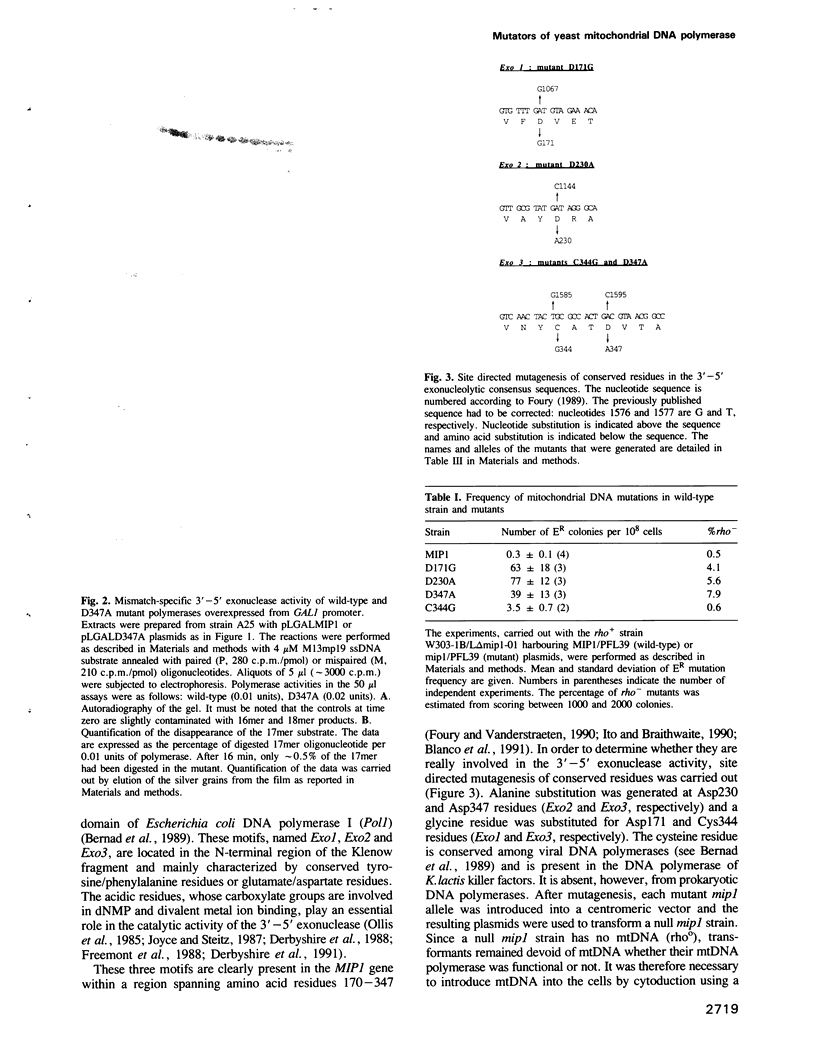
The MIP1 gene which encodes yeast mitochondrial DNA polymerase possesses in its N-terminal region the three motifs (Exo1, Exo2 and Exo3) which characterize the 3'-5' exonucleolytic domain of many DNA polymerases. By site directed mutagenesis we have substituted alanine or glycine residues for conserved aspartate residues in each consensus sequence. Yeast mutants were therefore generated that are capable of replicating mitochondrial DNA (mtDNA) and exhibit a mutator phenotype, as estimated by the several hundred-fold increase in the frequency of spontaneous mitochondrial erythromycin resistant mutants. By overexpressing the mtDNA polymerase from the GAL1 promoter as a major 140 kDa polypeptide, we showed that the wild-type enzyme possesses a mismatch-specific 3'-5' exonuclease activity. This activity was decreased by approximately 500-fold in the mutant D347A; in contrast, the extent of DNA synthesis was only slightly decreased. The wild-type mtDNA polymerase efficiently catalyses elongation of singly-primed M13 DNA to the full-length product. However, the mutant preferentially accumulates low molecular weight products. These data were extended to the two other mutators D171G and D230A. Glycine substitution for the Cys344 residue which is present in the Exo3 site of several polymerases generates a mutant with a slightly higher mtDNA mutation rate and a slightly lower 3'-5' exonucleolytic activity. We conclude that proofreading is an important determinant of accuracy in the replication of yeast mtDNA.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernad A., Blanco L., Lázaro J. M., Martín G., Salas M. A conserved 3'----5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989 Oct 6;59(1):219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L., Bernad A., Salas M. MIP1 DNA polymerase of S. cerevisiae: structural similarity with the E. coli DNA polymerase I-type enzymes. Nucleic Acids Res. 1991 Feb 25;19(4):955–955. doi: 10.1093/nar/19.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Clark-Walker G. D. Contrasting mutation rates in mitochondrial and nuclear genes of yeasts versus mammals. Curr Genet. 1991 Aug;20(3):195–198. doi: 10.1007/BF00326232. [DOI] [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. Genetic and crystallographic studies of the 3',5'-exonucleolytic site of DNA polymerase I. Science. 1988 Apr 8;240(4849):199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Grindley N. D., Joyce C. M. The 3'-5' exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 1991 Jan;10(1):17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin M. J., Patel S. S., Johnson K. A. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry. 1991 Jan 15;30(2):538–546. doi: 10.1021/bi00216a031. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W., Tsui W. C. Kinetic basis of spontaneous mutation. Misinsertion frequencies, proofreading specificities and cost of proofreading by DNA polymerases of Escherichia coli. J Mol Biol. 1982 Mar 25;156(1):37–51. doi: 10.1016/0022-2836(82)90457-0. [DOI] [PubMed] [Google Scholar]
- Foury F. Cloning and sequencing of the nuclear gene MIP1 encoding the catalytic subunit of the yeast mitochondrial DNA polymerase. J Biol Chem. 1989 Dec 5;264(34):20552–20560. [PubMed] [Google Scholar]
- Foury F. The 31-kDa polypeptide is an essential subunit of the vacuolar ATPase in Saccharomyces cerevisiae. J Biol Chem. 1990 Oct 25;265(30):18554–18560. [PubMed] [Google Scholar]
- Freemont P. S., Friedman J. M., Beese L. S., Sanderson M. R., Steitz T. A. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genga A., Bianchi L., Foury F. A nuclear mutant of Saccharomyces cerevisiae deficient in mitochondrial DNA replication and polymerase activity. J Biol Chem. 1986 Jul 15;261(20):9328–9332. [PubMed] [Google Scholar]
- Gibbs J. S., Weisshart K., Digard P., deBruynKops A., Knipe D. M., Coen D. M. Polymerization activity of an alpha-like DNA polymerase requires a conserved 3'-5' exonuclease active site. Mol Cell Biol. 1991 Sep;11(9):4786–4795. doi: 10.1128/mcb.11.9.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insdorf N. F., Bogenhagen D. F. DNA polymerase gamma from Xenopus laevis. I. The identification of a high molecular weight catalytic subunit by a novel DNA polymerase photolabeling procedure. J Biol Chem. 1989 Dec 25;264(36):21491–21497. [PubMed] [Google Scholar]
- Ito J., Braithwaite D. K. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 1991 Aug 11;19(15):4045–4057. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Braithwaite D. K. Yeast mitochondrial DNA polymerase is related to the family A DNA polymerases. Nucleic Acids Res. 1990 Nov 25;18(22):6716–6716. doi: 10.1093/nar/18.22.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni L. S., Olson M. W. Mismatch-specific 3'----5' exonuclease associated with the mitochondrial DNA polymerase from Drosophila embryos. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6469–6473. doi: 10.1073/pnas.86.17.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Mosbaugh D. W. Exonucleolytic proofreading by a mammalian DNA polymerase. Biochemistry. 1989 Feb 7;28(3):988–995. doi: 10.1021/bi00429a011. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Soni A. Exonucleolytic proofreading enhances the fidelity of DNA synthesis by chick embryo DNA polymerase-gamma. J Biol Chem. 1988 Mar 25;263(9):4450–4459. [PubMed] [Google Scholar]
- Lahaye A., Stahl H., Thines-Sempoux D., Foury F. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 1991 Apr;10(4):997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Lodish H. Mitochondrial diseases: gene mapping and gene therapy. Cell. 1990 Jun 15;61(6):925–926. doi: 10.1016/0092-8674(90)90055-j. [DOI] [PubMed] [Google Scholar]
- Longley M. J., Mosbaugh D. W. Properties of the 3' to 5' exonuclease associated with porcine liver DNA polymerase gamma. Substrate specificity, product analysis, inhibition, and kinetics of terminal excision. J Biol Chem. 1991 Dec 25;266(36):24702–24711. [PubMed] [Google Scholar]
- Morrison A., Bell J. B., Kunkel T. A., Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3'----5' exonuclease activity. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netter P., Petrochilo E., Slonimski P. P., Bolotin-Fukuhara M., Coen D., Deutsch J., Dujon B. Mitochondrial genetics. VII. Allelism and mapping studies of ribosomal mutants resistant to chloramphenicol, erythromycin and spiramycin in S. cerevisiae. Genetics. 1974 Dec;78(4):1063–1100. doi: 10.1093/genetics/78.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Patel S. S., Wong I., Johnson K. A. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991 Jan 15;30(2):511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Stocki S., Nonay R. L., Dimayuga E., Goodrich L. D., Konigsberg W. H., Spicer E. K. DNA polymerization in the absence of exonucleolytic proofreading: in vivo and in vitro studies. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2417–2421. doi: 10.1073/pnas.88.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Giot L., Faye G. The 3' to 5' exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991 Aug;10(8):2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Complete DNA sequence coding for the large ribosomal RNA of yeast mitochondria. Nucleic Acids Res. 1983 Jan 25;11(2):339–348. doi: 10.1093/nar/11.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanssens P., Opsomer C., McKeown Y. M., Kramer W., Zabeau M., Fritz H. J. Efficient oligonucleotide-directed construction of mutations in expression vectors by the gapped duplex DNA method using alternating selectable markers. Nucleic Acids Res. 1989 Jun 26;17(12):4441–4454. doi: 10.1093/nar/17.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studwell P. S., O'Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J Biol Chem. 1990 Jan 15;265(2):1171–1178. [PubMed] [Google Scholar]
- Suissa M. Spectrophotometric quantitation of silver grains eluted from autoradiograms. Anal Biochem. 1983 Sep;133(2):511–514. doi: 10.1016/0003-2697(83)90117-3. [DOI] [PubMed] [Google Scholar]
- Tabor S., Huber H. E., Richardson C. C. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987 Nov 25;262(33):16212–16223. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J Biol Chem. 1989 Apr 15;264(11):6447–6458. [PubMed] [Google Scholar]
- Tommasino M., Ricci S., Galeotti C. L. Genome organization of the killer plasmid pGK12 from Kluyveromyces lactis. Nucleic Acids Res. 1988 Jul 11;16(13):5863–5878. doi: 10.1093/nar/16.13.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C. Mitochondrial DNA mutations and neuromuscular disease. Trends Genet. 1989 Jan;5(1):9–13. doi: 10.1016/0168-9525(89)90005-x. [DOI] [PubMed] [Google Scholar]
- Wernette C. M., Conway M. C., Kaguni L. S. Mitochondrial DNA polymerase from Drosophila melanogaster embryos: kinetics, processivity, and fidelity of DNA polymerization. Biochemistry. 1988 Aug 9;27(16):6046–6054. doi: 10.1021/bi00416a033. [DOI] [PubMed] [Google Scholar]
- Zassenhaus H. P., Hofmann T. J., Uthayashanker R., Vincent R. D., Zona M. Construction of a yeast mutant lacking the mitochondrial nuclease. Nucleic Acids Res. 1988 Apr 25;16(8):3283–3296. doi: 10.1093/nar/16.8.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]