Abstract
Mushrooms represent a formidable source of bioactive compounds. Some of these may be considered as biological response modifiers; these include compounds with a specific biological function: antibiotics (e.g. plectasin), immune system stimulator (e,g, lentinan), antitumor agents (e.g. krestin, PSK) and hypolipidemic agents (e.g. lovastatin) inter alia. In this study, we focused on the Chinese medicinal mushroom “yun zhi”, Trametes versicolor, traditionally used for (cit.) “replenish essence and qi (vital energy)”. Previous studies indicated the potential activity of extracts from culture filtrate of asexual mycelia of T. versicolor in controlling the growth and secondary metabolism (e.g. mycotoxins) of plant pathogenic fungi. The quest of active principles produced by T. versicolor, allowed us characterising an exo-polysaccharide released in its culture filtrate and naming it Tramesan. Herein we evaluate the biological activity of Tramesan in different organisms: plants, mammals and plant pathogenic fungi. We suggest that the bioactivity of Tramesan relies mostly on its ability to act as pro antioxidant molecule regardless the biological system on which it was applied.
Introduction
Polysaccharides alone or conjugated to proteins are biopolymers exhibiting a variety of biological functions; from a structural point of view, this is due to the huge amount of different chemical and conformational motifs they can possess using different i) sugar composition, ii) glycosidic linkages and iii) branching. Actually, only 8x103 tripeptides arise using 20 amino acids, but 6x106 structurally different trisaccharides can be arranged using 20 different monosaccharides. Cells have evolved receptors with particular domains for carbohydrates [1] and polysaccharides can be recognized as such or as glycoproteins moieties. Indeed, the density and number of glycan epitopes can modulate the ligand/receptor interactions [2] and ultimately drive the fate of host-pathogen challenge [3].
In host-fungal interactions, glucans, cell wall-bound as well as secreted ones, are gaining momentum within scientific community since the incidence of fungal infections has been steadily rising in the past decades, in plant crops as well as in humans [4,5]. Among various polysaccharides, β-glucans of different sizes and branching patterns display significant and variable immune potency [6] and a significant number of studies on fungal β-glucans and their host receptors (such as Toll Like Receptors, TLR, and Dectin-1) have been performed [6].
Most fungal pathogens (e.g. Candida albicans, Aspergillus fumigatus) as well as non-toxic (edible) fungi (e.g. Lentinula edodes) display a species-specific pattern of cell wall glycans [7]. Two groups of glucans are present in fungi, α- and β-glucans. Some fungi have α-1,4-linked glucose units in their cell walls either to interconnect the linear α-1,3-glucan chains or in an alternated way with α-1,3-linked glucose units (e.g. nigeran in Aspergillus niger) [8]. The most abundant β-glucan in the fungal cell wall is a β-1,3-glucan which account for between 65% and 90% of the whole β-glucan content [9] together with the cell wall linked water insoluble β-1,4-linked poly-N-acetyl-D-glucosamine (chitin) [10]. These glucans may act as molecular/pathogen associated molecular patterns (MAMP or PAMP). Hosts have evolved an array of pattern recognition receptors (PRRs) able to sense the different molecular moieties that compose fungal cell walls. PRRs for fungal PAMPs include Toll-like receptors (TLRs), C-type lectin receptors (CLRs) and galectin family proteins [5,10,11].
Although poorly studied, glycans are important in cell-to-cell communication even in fungi. The most studied issue within fungus/fungus interactions is related to self/non-self-recognition between vegetative mycelia. This mechanism relies on het gene complex and ultimately leads fungus to programmed cell death under vegetative incompatibility circumstances [12,13]. Notably, TOL receptor in Neurospora crassa possesses a LRR domain, similar to plant and humans PRR, which allows fungus to distinguish self from non-self [12].
Host cell may react similarly towards fungal glucans. After having recognised them as non-self/MAMP, host cells activate defence strategies such as the cell antioxidant system [5,12]. Examples may be provided for mammals, plants and fungi. In mammals, dectin-1, assisted by TLRs, recognizes fungal ß-glucans and triggers ROS formation and activation of the master regulator of the antioxidant cellular defence Nrf-2. The downstream pathways may lead to cell death or antimicrobial immunity [14,15]. As an example in plants, Cerk1, together with CEBiP, recognizes fungal chitin and activates defence responses [16,17]. Fungi induce formation of ROS with a consequent alteration of cell redox balance. Recently, the close link between ROS perception and defence activation was described for the tomato WRKY transcriptional factor SlDRW1 required for disease resistance against Botrytis cinerea and tolerance to oxidative stress [18]. In fungi, the semi-purified fraction of lentinan, a ß-glucan of the basidiomycete Lentinula edodes, modulates oxidant/antioxidant balance in Aspergillus sect. Flavi by manipulating the expression of the oxidative stress-related transcription factor ApyapA [19,20]. In turn, several features of A. parasiticus are modified by recognition of lentinan: morphology, growth and secondary metabolism [19]. The lectins recognition is required for coiling around the prey mycelium and formation of helix shaped hyphae in mycoparasitism. In Hypocrea virens the sucrose transporter is induced in the early stages of root colonization [21]. Thus, fungal glucans acting as elicitors may trigger several responses in different hosts. Amongst these, fungal polysaccharides may elicit the activation of cell redox balancers such as the oxidative stress-related transcription factors Yap-1 whose mechanism of action was firstly described in Saccharomyces cerevisiae [20].
Despite the increasing biological relevance of fungal polysaccharides, the majority of glucans investigated are derived from fungal extracts [6] obtained applying poor purification procedures which hinder result standardization and render the characterization of their biological activity, confused and, in some cases, contradictory.
In this study, we describe the identification of a biological active saccharidic fraction in the filtrate of the medicinal mushroom T. versicolor and the partial chemical characterization of the polysaccharide therein contained (henceforth referred as Tramesan). Its ability to trigger an antioxidant response in different biological systems (human cell lines, plants, and fungi) is reported.
Materials and methods
Fungal strain and growth conditions
Trametes versicolor strain C used in this study was registered at CABI biosciences (UK) and deposited in the culture collection of Department of Environmental Biology of Sapienza University of Rome as ITEM 117. T. versicolor C was grown for 7 days in potato dextrose broth (PDB, Himedia) and incubated at 25°C under shaken conditions (100 rpm). The liquid culture was homogenized, in sterile condition in Waring blender 8012. After homogenization, an aliquot (5% v/v) of the fungal culture was inoculated in 500 mL of PDB in 1L-Erlenmeyer flasks and incubated for 14 days at 25°C under rotary shaken conditions (100 rpm). The mycelia were then separated from the culture filtrates by subsequent filtrations with different size filters (Whatmann) to eliminate all the mycelia. Mycelia-purified culture filtrate was lyophilized and utilized for subsequent analyses. The isolates were kept in Potato Dextrose Agar at 4°C and the cultures were sub-cultured every 30 days.
Purification of polysaccharide fractions
A suspension of the lyophilized T. versicolor culture filtrate (1g in 30 mL of H2O) was filtered to separate insoluble from soluble part. The remaining solution was cooled and precipitated with 4 volumes of cold ethanol. The precipitate was recovered by centrifugation at 1200 xg (20 min at 4°C), dissolved in 8 mL of 10 mM phosphate buffer pH 7.5 and treated with pronase E (Sigma Aldrich) at 37°C for 16 hr. After dialysis (12000 Da membrane Cut Off), the sample was recovered by lyophilisation (yield = 60 mg/L).
The polysaccharide fraction was separated by low-pressure size exclusion chromatography on a Sephacryl S-300 column (fractionation domain: 1–400 kDa for dextrans; gel bed volume: 1.6 id x 90 cm), using 50 mM NaNO3 as eluent at a flow rate of 6 mL/h. About 30 mg of the sample was dissolved in 1.9 mL of eluent and centrifuged before being loaded onto the column. Fractions of 2 mL were collected at 20 min intervals. Elution was monitored using a refractive index detector (WGE Dr. Bures, LabService Analytica), connected to a paper recorder and interfaced with a computer via PicoLog software. Polysaccharide molecular mass (MM) was evaluated by high performance size exclusion chromatography (HP-SEC) on an Agilent Technologies 1200 series HPLC equipped with three columns in series (Tosoh Bioscience, TSKgel G3000PW, G5000PW and G6000PW, i.d. 7.5 mm, length 30 cm) kept at 40°C with a thermostat (Waters Millipore). Calibration of the chromatographic system was performed using pullulan standards exhibiting molecular masses in the range 1.66x106–5.90x103 (Polymer Laboratories, Germany and Sigma for pullulan with MM = 1.66x106). The calibration curve is reported in Figure A in S1 File. Elution was performed with 0.15 M NaCl, with a flow rate of 0.5 mL/min and monitored using a refractive index detector (Knauer, Labservice Analytica), interfaced with a computer via Agilent software.
Composition analysis of Tramesan
In order to determine the composition in terms of neutral monosaccharides, the polysaccharide fraction was hydrolysed with 2 M trifluoroacetic acid (TFA) at 125°C for 1 h and the monosaccharides obtained were derivatised to alditol acetates [22]. The products were separated by gas–liquid chromatography (GLC) on a Perkin-Elmer Autosystem XL gas chromatograph equipped with a flame ionisation detector, an SP2330 capillary column (Supelco, 30 m), using He as carrier gas. The temperature program applied was 200–245°C at 4°C/min. Identification of the monosaccharides in Tramesan was achieved by comparing the retention times of each peak with those of standard sugars, previously derivatized in the same way. Quantification was obtained using inositol as internal standard.
Linkage analysis of Tramesan
The glycosidic linkage position for each sugar residue was achieved by methylation analysis. The polysaccharide was permethylated according to the protocol by Harris [23], hydrolysed with 2 M TFA at 125°C for 1 h, and the products were then derivatized to alditol acetates [22], thus obtaining a mixture of partially methylated alditol acetates (PMAA). The alditols are substituted with a methyl group on the hydroxyls groups that were originally not engaged in linkages while acetyl esters are formed on the hydroxyl functions that were originally engaged in glycosidic linkages and in ring formation. The elution time in the GLC chromatogram led to the identification of the sugar type, and integration of the peak areas, after correcting by the effective carbon response factors [24], gave quantitative information. GLC-MS mass spectra identified the ring size and the position of glycosidic bonds. The PMAA were analysed by GLC using the same instrument and settings as for the composition analyses but with the temperature program: 150–250°C at 4°C/min. GLC-MS analyses were carried out on an Agilent Technologies 7890A gas chromatograph coupled to an Agilent Technologies 5975C VL MSD.
Partial hydrolysis of Tramesan and characterisation of the products
Since the glycosidic linkages of 6-deoxy-sugars are more labile compared to those engaged by other monosaccharides, the polysaccharide was subjected to mild acid treatment in order to produce oligosaccharides that are suitable to electro-spray ionization mass spectrometry (ESI-MS). The mass spectra gave information on the monosaccharide composition and fragmentation of the parent ions revealed the position of the 6-deoxy sugars thanks to the different molecular mass of 6-deoxy-sugars and hexoses. 2 mg of Tramesan were treated with 0.5 M TFA at 100°C for 2 h. The hydrolysate was dried under a N2 flow, dissolved in 50% aqueous methanol—11 mM NH4OAc and subjected to ESI-MS experiments using a Bruker Esquire 4000 ion trap mass spectrometer connected to a syringe pump for the injection of the samples. The instrument was calibrated using a tune mixture provided by Bruker. In order to increase the detection sensitivity, oligosaccharides were reduced with NaBH4 [25], to label the reducing end and subsequently permethylated [26], dissolved in a 1:1 chloroform: methanol mixture, 11mM NH4OAc and subjected to ESI-MS and MS2 analyses. Samples were injected at 180 μL/h and detection was performed in the positive ion mode. After having verified that the hydrolysis conditions produced the desired oligosaccharides, the partial hydrolysis reaction was repeated on 10 mg of Tramesan, the hydrolysate was rotovaporated to dryness under reduced pressure at 45°C to eliminate residual TFA, taken to pH = 7.2, rotovaporated to dryness again, dissolved in 1.9 mL of 50 mM NaNO3, centrifuged and separated by size exclusion chromatography on a Bio Gel P2 column (fractionation domain: 100–1800 Da; gel bed volume: 1.6 cm i.d. × 90 cm), using 50 mM NaNO3 as eluent at a flow rate of 6 mL/h and the same system set up reported above. Fractions of 1.5 mL were collected at 15 min intervals and those belonging to the same peak were pooled together, and desalted on a Bioline preparative chromatographic system equipped with a Superdex G30 column (fractionation domain: up to 10 kDa; gel bed volume: 90 cm x 1.0 cm i.d., flow rate 1.5 mL/min) previously equilibrated in H2O. Elution of each peak was monitored with a Refractive Index detector (Knauer, LabService Analytica). The purified oligosaccharides were analysed by NMR spectroscopy.
NMR spectroscopy
When necessary, polysaccharide fractions were de-O acetylated with 10 mM NaOH at room temperature for 5 h, under N2 flow. Samples were exchanged two times with 99.9% D2O by lyophilisation and then dissolved in 0.6 mL of 99.96% D2O. Spectra were recorded on a 500 MHz VARIAN spectrometer operating at 50°C for polysaccharides solution and at 25°C for oligosaccharide solution. 2D experiments were performed using standard VARIAN pulse sequences and pulsed field gradients for coherence selection when appropriate. HSQC spectra were recorded using 140 Hz (for directly attached 1H–13C correlations). TOCSY spectra were acquired using 120 ms spin-lock time and 1.2 s relaxation time. NOESY experiments were recorded with 200 ms mixing time and 1.5 s relaxation time. Chemical shifts are expressed in ppm using acetone as internal reference (2.225 ppm for 1H and 31.07 ppm for 13C). NMR spectra were processed using MestreNova software.
Assay of Tramesan in fungi
A. flavus (Speare) NRRL 3357and A. parasiticus (Speare) NRRL 2999, both producers of aflatoxin B1 (AFL B1), were grown on PDA at 30°C for 7 days and from these cultures a suspension of 100 conidia, of each strain, independently, in 10 μL of sterilised distilled water was inoculated in 190 μL of PDB in presence or absence of Tramesan 0.38 μM, using 96-wells microplates. The cultures were incubated at 30°C for 3 days. The assay allowed us to test all the fractions in minimal amount and to generate hundreds of replications in a very short time (aflatoxin microtiter-based bioassay). Different cultures were independently filtrated with Millipore filters (0.22 μm). Then, the mycelia were lyophilized and weighted. The aflatoxin B1 was extracted adding, for each condition (control and treated with 0.19 and 0.38 μM of Tramesan), chloroform/methanol (2:1, v/v). The mixture was vortexed for 1 min, centrifuged and then the lower phases was drawn off. The extraction was repeated twice and the samples were concentrated under a N2 stream, re-dissolved in 50 μL of acetonitrile/water/acetic acid (20:79:1 v/v) and quantified by triple quad LC/MS 6420 (Agilent) with a method reported by Sulyok et al. [27], with minor modifications. Such modifications regarded mainly the use of Mycospin (Romer Labs) for the cleaning up of the samples prior analysis. The amount of aflatoxin B1 was evaluated by using an ISTD-normalised method in MassHunter workstation software, quantitative analysis version B.07.00. Aflatoxin B1-13C-d3 (Clearsynth) at 2 μM final concentration was used as ISTD. Aflatoxin B1 amount was expressed in ppb.
For the gene expression evaluation, total RNA from the mycelia of A. flavus and A. parasiticus strains was extracted, as reported by Scala et al. [28], 7 days after inoculation in PDB cultures amended or not (control) with Tramesan 0.38 μM and used to develop reverse-transcriptase quantitative PCR (RT-qPCR) assays for AfyapA (XM_002382086.1), ApyapA (DQ104418.2), (yapA primers:; for 5’- GGTTGTTTGAGCCGTTGAGT -3’; rev 5’- ACGGCCTCAATAACAACGAC -3’), sod1 (AFLA_099000; for 5’- AGTCGGTAAGGCAAACTGGG -3’; rev 5’- GAATTCGCCAGGACCAGACA -3’). RT-PCR were performed using SensiFAST™ SYBR® No-ROX™ (Bioline, Italy) at 95°C for 2 min; 30 cycles of 95°C for 15 s, 54–58°C (according to primer selected) for 30 s and 72°C for 15 s. The specificity of the reaction was verified by melt curve analysis and the efficiency of each primer was checked using the standard curve method. Primers with slopes between −3.1 and −3.6, and reaction efficiencies between 90 and 110% were selected for the analysis. Gene expression in the fungal strains was calculated by using the 2-ΔΔCt method, i.e. by normalizing transcript levels of the gene of interest (GOI) onto the transcript of a housekeeping gene β-tubulin for A. flavus (HF937107.1; for 5’- GCTGGAGCGTATGAACGTCT-3’; rev 5’- GTACCAGGCAGAACGAGGAC-3’) and A. parasiticus (L49386.1; for 5’- TCACCTGCTCTGCCATCTTG-3’; rev 5’- TGTTGTTGGGGATCCACTCG-3’) as reported in Reverberi et al., 2011 [29] and onto their value in the untreated control (no Tramesan added). The housekeeping gene β-tubulin proved as being the most stable after analysis with the Normfinder algorithm (https://moma.dk/normfinder-software). The software for relative expression quantification provided with the Line GeneK thermocycler (Bioer, PRC) was used.
Assay of Tramesan in planta
Parastagonospora nodorum (Berk.) was isolated from naturally infected durum wheat leaves (cv. Ciccio) cultivated in Italy. The strain ITEM 17131 was registered and conserved in the Culture Collection Agro Food Important Toxigenic Fungi-Item, Institute of Science of Food Production (ISPA), National Research Council (CNR), Via Amendola, 122/O, 70126 Bari, Italy (http://server.ispa.cnr.it/ITEM/Collection/). For in planta analysis, a growth in phytotron was used. Temperature, humidity and light were regulated in the chamber and notably, T = 20°C; humidity: 80%; light: 18 h of light with 150 μmol of photon/m2s, from 6.00 am to 22.00 pm). The plots were positioned in a rotary floor, so every plants were submitted to same conditions. Only a susceptible durum wheat variety (Svevo) was used in these tests. Kernels were disinfected by sodium hypochlorite (1%) during 10 min with permanent agitation of 150 rpm, and then rinsed three times with sterile distilled water during 5 min with permanent agitation of 150 rpm. The kernels germinated in vitro in water medium (0.5%). After, they were incubated at 20°C in dark during 24 h; 4°C in dark during 48 h; 20°C in dark during 24 h. The kernels were transferred in a two time autoclaved (20 min at 121°C) soil mixture (20 L of soil / 5 L of perlite), in pots of 0.5 L. The plants were irrigated three time a week, twice with 1 L of osmotic water. Plant leaves were sprayed with a solution of Tramesan 0.38 μM (100 mL per 64 plots) 48 hours prior pathogen inoculation. This latter occurred by spraying a picnidiospore suspension of P. nodorum on wheat second “real” leaf after flag leaf emergence (BBCH39). Visual identification of the disease and microscopic identification of P. nodorum pathogen were performed. In the phytotron, the SNB infection was visually assessed every 7–10 days post inoculation (dpi) as disease severity on flag leaf and as severity and incidence of the disease on ear using the Liu’ scale [30]. To assess fungal growth, we calculated fungal DNA present into plant tissues. Total DNA extracted from wheat leaves and seeds according to Färber method with minor modifications [19] and used for developing a specific SYBR green qPCR method by designing primers (for_ TGGGTACGCTTTTGAT CTCC; rev_ AACGAGGTGGTTCAGGTCAC) in the β-tubulin of P. nodorum (NCBI Gene Bank Ac. No. AY786332) as reported by Iori et al. [31]. For gene expression analysis, aliquots of 25 mg of lyophilized wheat leaves were powdered in liquid nitrogen and treated for RNA extraction. RNA extraction was performed with the TRI REAGENT method (Sigma-Aldrich, USA) and following manufacturer’ instructions and cDNA obtained using first Strand cDNA synthesis SUPER SCRIPT II for RT-PCR (Invitrogen, USA) kit. Real-time PCR was performed as described by Nobili et al. [32], using SensiFAST™ SYBR® No-ROX ™ (Bioline, Italy) at 95°C for 2 min; 30 cycles of 95°C for 15 s, 54–58°C (according to primer selected) for 30 s and 72°C for 15 s. The specificity of the reaction was verified by melt curve analysis and the efficiency of each primer was checked using the standard curve method. Primers with slopes between −3.1 and −3.6, and reaction efficiencies between 90 and 110% were selected for the analysis. The housekeeping gene β-tubulin of T. turgidum susp. durum (AJ971820.1; for 5’- GCTGCTGTATTGCAGTTGGC-3’; rev 5’- AAGGAATCCCTGCAGACCAG-3’), proved as being the most stable after analysis with the Normfinder algorithm (https://moma.dk/normfinder-software), was used as a reference for data normalization. The relative expression, as 2− ΔΔ Ct values, of PR9 wheat gene PR9 (EU264058.1; for 5’-CAAGGTGAACTCGTGATGGA-3’; rev 5’-TTGAGGATTCAACCGTCGTT-3’), was evaluated by using as calibrator the Ct values of this gene in the infected, but not treated with Tramesan, samples (control).
Assay of Tramesan on mammalian cell lines
The analysis were carried out on murine melanoma B16-F10 stabilized cells, grown in DMEM and incubated at 37°C with 5% CO2 for different times. An amount of cells/well (1x105) were plated in 12-wells plate and incubated o/n. The day after, the cells were treated with Tramesan 0.38 μM and incubated for 48 h. Subsequently, the cells were centrifuged for 7 min at 1000 rpm and the pellet was washed with PBS. The pellet was then resuspended in 50 μL of 1M NaOH and incubated 1 h at 60°C. The quantity of extracted melanin was determined at 405 nm, with a Perkin-Elmer Lambda 25 UV/Vis spectrometer. Melanin content was calculated by interpolating the results with a standard curve, generated by absorbance of known concentrations of synthetic melanin and corrected for the number of cells. Three determinations were performed in duplicate, the results were expressed as μg of total melanin/number of cells, and values were reported as percentage of control. At the same time, experiments were carried out treating the melanoma cells (about 4 x 104 cells for well) with Tramesan 0.38 μM. The melanoma cells were then incubated at 37°C for 24 and 48 h and the cell counts were performed with light microscopy at these time points. For evaluating gene expression, an amount of B16-F10 stabilized cells (3x105) was plated and incubated. After 24 h of incubation, the cells were treated with Tramesan 0.75 μM and after 6 h from the treatment, the cells were harvested. Before RT-PCR analysis, the cells were washed with PBS and total RNA was isolated using RNeasy Minikit (Qiagen). Subsequently, cDNA was synthesized using oligo-dT primers and ImProm-IITM reverse transcriptase (Promega) according to the manufacturer’s instructions. RT-PCR was carried out in 15 μL (total volume) with SYBR green PCR Master Mix (Bio-Rad) and 200 nM of each primer. The sequences of primers were forward and reverse: β-actin 5’-GACAGGATGCAGAAGGAGATTACT-3’ and 5’-TGATCCACATCTGCTGGAAGGT-3’; nrf-2 for 5’ -CGCTGGAAAAAGAAGTGG- 3’ and rev 5’-AGTGACTGACTGATGGCAGC- 3’. The housekeeping gene β-actin proved as being the most stable after analysis with the Normfinder algorithm (https://moma.dk/normfinder-software). RT-PCR reactions were carried out in triplicate using the Real Time Detection System (iQ5 Bio-Rad) equipped with ICYCLER IQ5 optical system software version 2.0 (Bio-Rad). The condition of thermal cycling were: initial denaturation step at 95°C for 3 min, followed by 40 cycles at 95°C for 10 sec and 60°C for 30 sec.
Results
Bio-based purification assays
In a previous study, we assessed that culture filtrates of T. versicolor inhibited aflatoxin synthesis by enhancing the antioxidant capacity of A. flavus [33]. Notably, aflatoxin synthesis, as well as other secondary metabolites in pathogenic fungi, is controlled by the cell redox status via AP-1 like factors [20,33,34]. Thus, we demonstrated that bioactive compounds present in the culture filtrate of T. versicolor enhanced Ap1-like gene expression in A. flavus that, in turn, switched off toxin synthesis [32,35,36].
In order to reduce volumes, and mainly the time needed to verifying aflatoxin inhibition, we set a more handy assay using 96-wells microplates. This is based on the ability of A. flavus to grow and produce toxins in a small volume, as reported in materials and methods section. Firstly, we separated the main components of a fraction of T. versicolor culture filtrate particularly active in inhibiting aflatoxins (fraction A), [36]. Fungal secretome is essentially composed of polysaccharides, proteins (glycosylated and not, with enzymatic or different activities), polyphenols and small metabolites [37–40]. In relation to this, considering the fraction A as essentially free from polyphenols, small metabolites and small peptides [36], we separated fractions enriched with polysaccharides or proteins. A scheme of the procedure is present in Figure B in S1 File. Thus, we originated six fraction (from B to G) and we tested them as dry pellets added to fungal medium (1% w/v) with our microtiter-based bioassay (Table 1). In this screening, we included also a commercially available “extract” of T. versicolor, used as diet supplementation (C-TV).
Table 1. Inhibition of aflatoxin B1 biosynthesis in A. flavus 3357 by T. versicolor culture filtrate fractions.
A. flavus was grown for 3 days into 200-μL multiwells plate, at 30°C in dark conditions and treated with different fractions (B-G) originated from fraction A as indicated in the scheme presented into Figure B in S1 File. Commercially available “extract” of T. versicolor, used as diet supplementation (C-TV) was included too. Results represents the mean of 3 (biological) x 12 (technical) replicates ± SE.
| Fractions | Aflatoxin B1 inhibition (%) |
|---|---|
| Fraction B | 76.3 ± 2.2 |
| Fraction C | 90.3 ± 3.1 |
| Fraction D | 10.2 ± 0.7 |
| Fraction E | 60.2 ± 0.5 |
| Fraction F | 58.3 ± 3.5 |
| Fraction G | 7.5 ± 3.1 |
| C-TV | 5.0 ± 2.2 |
| Fraction A | 75.2 ± 1.2 |
Fraction C proved to be the most efficient in inhibiting aflatoxin synthesis (up to 90%) without a significant fungi static effect (data not shown). Indeed, other fractions had a lower aflatoxin-inhibitory effect whilst preserving a certain degree (up to 50%) of fungal growth inhibition (data not shown). It is possible that for these fractions a certain amount of polysaccharide co-precipitated with proteins that, in turn, could present a slight antimicrobial ability. In relation to this, aflatoxin inhibition may be related also to fungal growth reduction.
The aflatoxin microtiter-based bioassay allowed us to identify fraction C as the one active limiting consistently mycotoxin synthesis by A. flavus.
Purification and characterization of fraction C
Fraction C was analysed by 1H-NMR and the spectrum showed the typical pattern of saccharidic molecules (data not shown) indicating that its major component was a polysaccharide. A scheme of fraction C fractionation and characterization is reported in Figure C in S1 File In addition, fractions obtained from different T. versicolor cultivations resulted to contain the same polysaccharide as confirmed by NMR analysis. The polysaccharide produced by T. versicolor strain C was compared with that present in a T. versicolor commercial powder by means of composition analysis. The data (Table A in S1 File) indicated that the two samples are very different: Fraction C contained Fuc, Man, Gal and Glc, while the commercial powder contained mainly Man and Glc, a small amount of Gal, but no Fuc. Therefore, these preliminary data suggested that T. versicolor strain C produced a novel polysaccharide, not present in the commercial sample.
Fraction C was subjected to size exclusion chromatography and three fractions (referred as CI, CII and CIII see Figure D in S1 File) were obtained. They were tested using aflatoxin inhibition in A. flavus as indicator of biological activity (as described above): fraction CI did not show any effect while fractions CII and CIII presented a significant bioactivity (Figure F in S1 File). These fractions were analysed by 1H NMR spectroscopy (Figure G in S1 File) which, a part the 1.3 and 4.2 ppm peaks, due to free lactate often present as contaminant from hands, showed that fractions CII and CIII were structurally identical. Therefore, further 2D NMR experiments were carried out only on fraction CIII, taking advantage of its lower molecular mass. In fact, high-pressure size exclusion chromatography using pullulan standards, (Figure A in S1 File) showed that fraction CII had a molecular mass of about 23000 Da and fraction CIII of about 6000 Da. Compositional analysis of fraction CIII showed Fuc:Man:Gal:Glc in the molar ratios 1.00:2.25:2.22:0.26. Methylation analysis for glycosidic linkages determination showed a rather complex pattern reported in Table 2. The presence of 2,6-Gal and 2,6-Hex indicated the occurrence of two branch sites as also confirmed by the presence of terminal non-reducing sugars, t-Fuc and t-Man. The sum of all terminal monosaccharides (t-Fuc + t-Man) is 1.01, in very good agreement with the sum of all branched monosaccharides (2,6-Gal + 2,6-Hex), equal to 0.94. The most important finding is the presence of fucose, an uncommon component of fungal polysaccharides found only in Mucorales and in some basidiomycetes [41].
Table 2. Glycosidic linkage positions in Tramesan fraction III and in the polysaccharide sample eluted at the void volume (V0) of the Bio Gel P2 column after partial hydrolysis.
| Monosaccharides | Fraction CIII | Fraction V0 |
|---|---|---|
| t-Fuc | 0.12 | - |
| 3-Fuc | 0.47 | 0.20 |
| t-Man | 0.89 | 0.91 |
| 2-Man | 1.00 | 1.00 |
| 6-Gal | 0.80 | 1.76 |
| 2,6-Gal | 0.66 | 0.16 |
| 2,6-Hex | 0.28 | 0.71 |
Numbers indicate the position of glycosidic linkages, t-Fuc = terminal non-reducing Fuc, t-Man = terminal non-reducing Man.
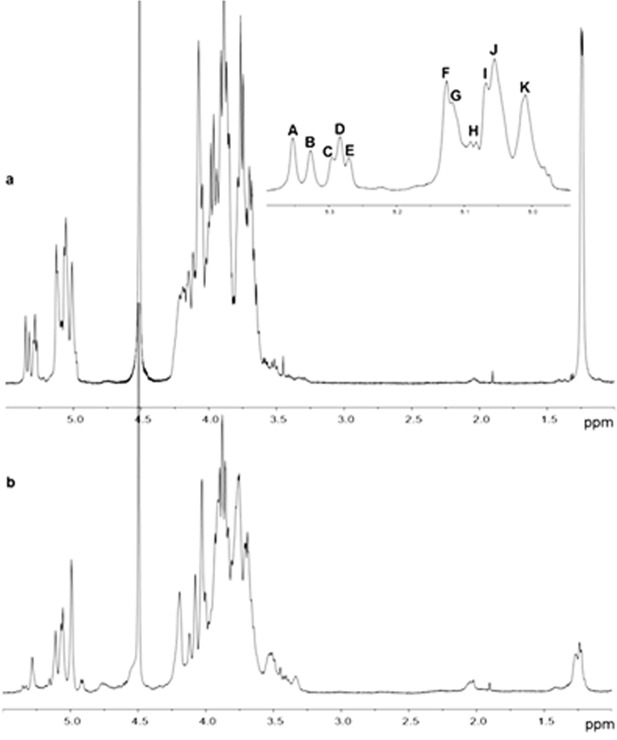
Since the 1H NMR spectrum indicated the presence of O-acetyl groups (Figure F in S1 File) which increase the complexity of the spectrum [42], fraction CIII was de-O-acetylated for further NMR analysis. The 1H NMR spectrum (Fig 1A) is typical of a polysaccharide: the anomeric proton region, between 5.5–4.8 ppm, (inset of Fig 1A) exhibited a complex pattern, reflecting the glycosidic linkage data and suggesting the absence of regular repeating units. The resonance at 1.25 ppm was attributed to the methyl groups of 6-deoxy residues, confirming the presence of Fuc.
Fig 1. 1H NMR spectrum (500 MHz, 50°C, D2O) of Tramesan.
A) Tramesan purified from T. versicolor culture broth (fraction CIII). The enlargement of the anomeric protons resonance ppm range is shown as insert where the most intense anomeric protons are named from A to K. B) 1H NMR spectrum (500 MHz, 50°C, D2O) of the total excluded high molecular mass fraction (V0) obtained by partial hydrolysis of Tramesan.
After inspection of the 1H NMR spectrum and of the COSY plot, the most intense anomeric protons were named from A to K (inset of Fig 1A) in order of decreasing chemical shift, and TOCSY, NOESY and HSQC experiments were recorded. Heavy overlapping of most peaks prevented complete chemical shifts assignments; nevertheless, some useful information could be gained.
Resonances belonging to H1 and H2 of each spin system were easily identified from the COSY plot. Taking advantage of the small JH1-H2 coupling constant of Man, which results in the absence of magnetisation transfer from H2 to H3 in the TOCSY plot, this monosaccharide was easily identified with signals A-E, F, and J. Resonances of carbon nuclei linked to the respective protons were then assigned from the HSQC plot (Figure G in S1 File) and the data collected can be found in Table B in S1 File. Resonances from A to E had H2s at relatively low field and C2s chemical shifts were shielded with respect to values for unbound Man [43–45], indicating engagement in glycosidic linkage; therefore, they were attributed to 2-Man residues. The very small difference of chemical shifts of these Man residues suggested that they may be part of oligosaccharide side chains of different sizes [46], thus experiencing slightly different electronic environments. The signals F and J were tentatively assigned to t-Man. Spin system G was completely assigned and attributed to 3-Fuc, following the magnetisation transfer in the TOCSY plot starting from H1 as well as from H6, which is well separated from the other signals. Furthermore, C6 nuclei engaged in glycosidic linkages were clearly detected in the HSQC spectrum as negative peaks (Figure G in S1 File, blue traces), shifted to higher ppm values with respect to the corresponding unbound residue (from about 62 to about 67.5 ppm), confirming the methylation analysis findings.
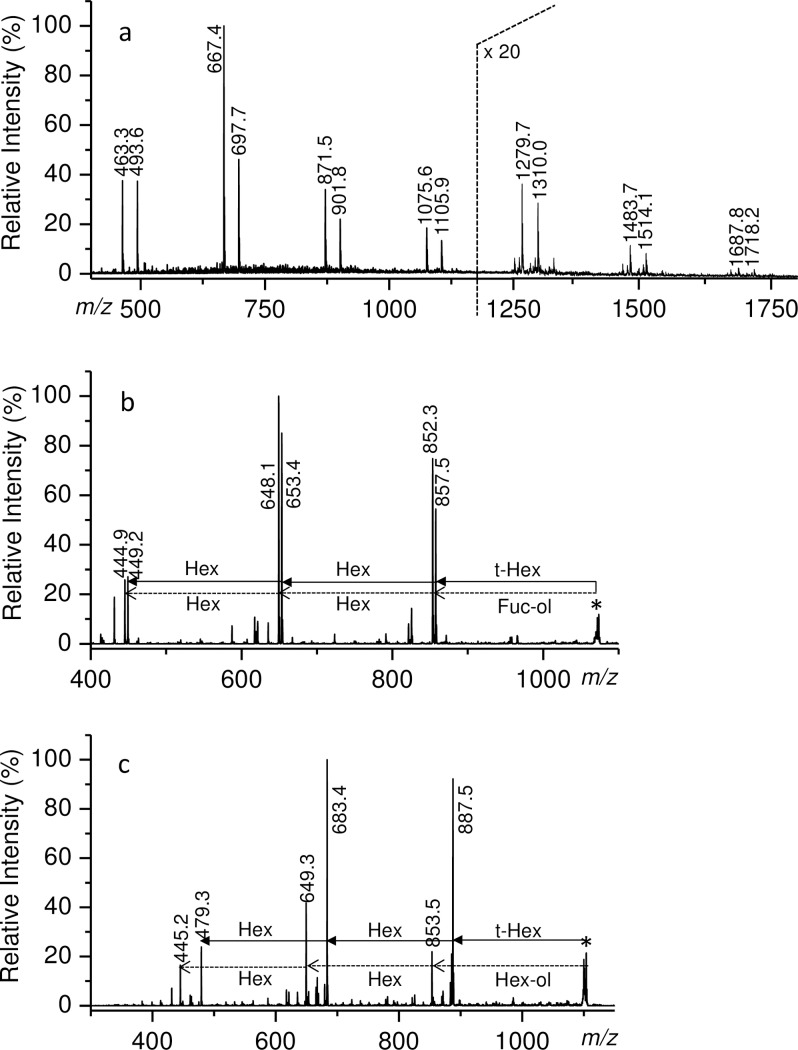
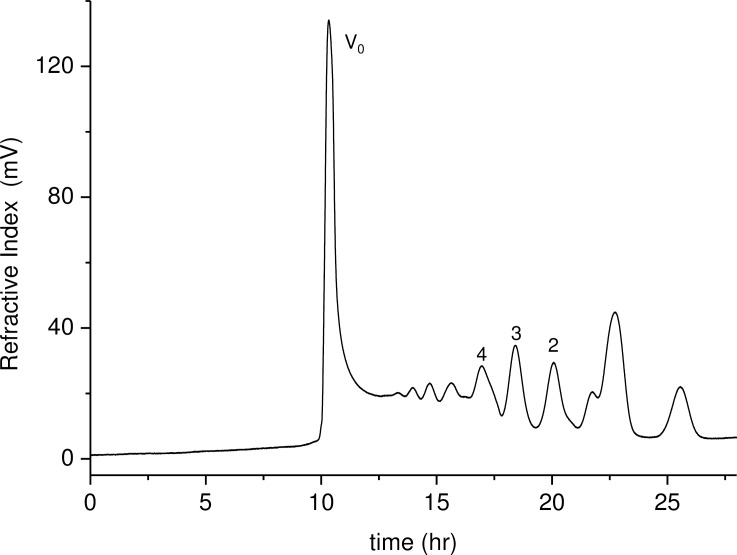
In order to gain more insights on the primary structure of Tramesan, a sample was subjected to partial hydrolysis, taking advantage of the more labile glycosidic linkage of Fuc residues. A small amount of hydrolysate was subjected to ESI-MS and the spectrum (data not shown) indicated a series of oligosaccharides from disaccharides to octasaccharides. The sample mixture was then reduced with sodium borohydride and permethylated. ESI-MS (Fig 2A) showed pairs of (M + Na)+ parent ions corresponding to oligosaccharides, differing for the presence of one Fuc residue in place of one Hex residue. MS2 (Fig 2B and 2C) of the pentasaccharides gave a good fragmentation pattern establishing the exclusive reducing terminal position of the Fuc residues. The hydrolysate was separated by size exclusion chromatography on a Bio Gel P2 column (Fig 3); fractions belonging to same peak were pooled together and subjected to NMR spectroscopy.
Fig 2.
A) ESI-MS spectrum of the reduced and permethylated oligosaccharide mixture obtained by partial hydrolysis, ESI-MS2 spectra of the parent ions at 1075.6 (B) and 1105.9 (C) m/z. The fragmentation schemes (B and C) starting from the non-reducing end are reported with full lines, those starting from the reducing end with dotted lines. Fuc-ol and Hex-ol stands for fucitol and hexitol, respectively.
Fig 3. Size exclusion chromatographic pattern of the mixture obtained from Tramesan by partial hydrolysis.
V0 indicates the total excluded high molecular mass fraction. Numbers 2–4 refer to sugar residues in oligosaccharides separated by the Bio Gel P2 resin.
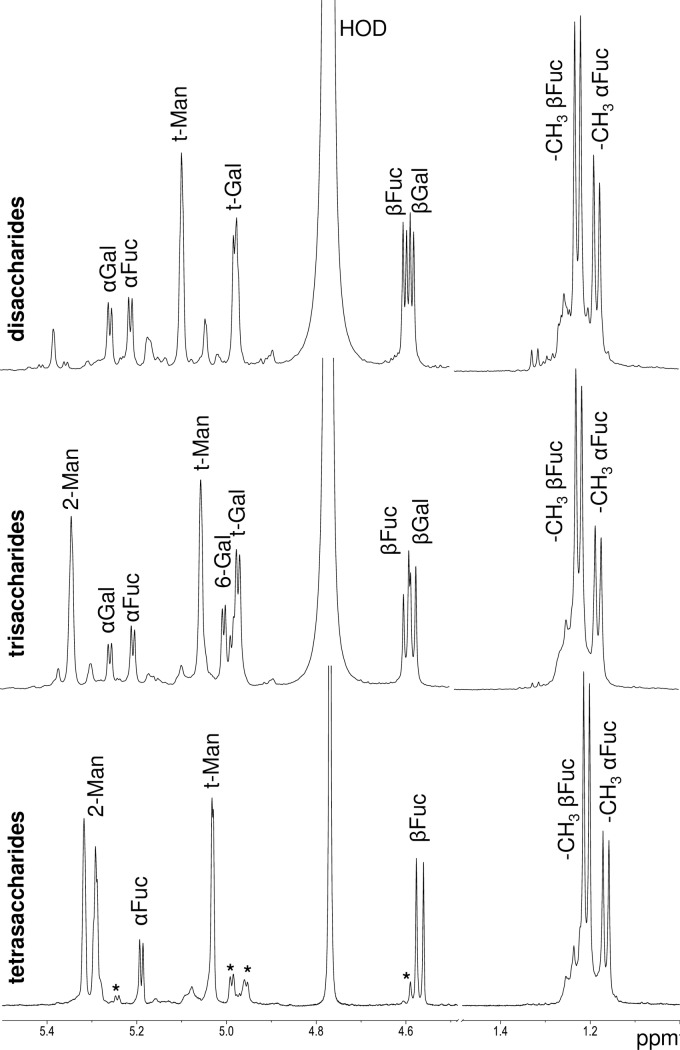
The expansions of the anomeric and methyl regions of the 1H NMR spectrum of the disaccharides mixture is reported in Fig 4A; only the major signals were considered and they were assigned after examining the COSY, TOCSY, NOESY and HSQC spectra as well. The methyl doublets at 1.23 and 1.18 ppm belong to the H6 of Fuc residue and reflect the anomeric equilibrium between α- and β-forms, thus confirming its reducing end position.
Fig 4. 1H NMR spectra (500 MHz, 50°C, D2O) of oligosaccharides obtained after partial hydrolysis and SEC-chromatography.
From top to bottom: A) disaccharides, B) trisaccharides and C) tetrasaccharides.
The resonances at 5.21 and 4.60 ppm were assigned to the α- and β- anomeric forms of Fuc, while the resonances at 5.26 and 4.59 ppm were attributed to the α- and β- anomeric forms of Gal, thus showing two different reducing ends, and therefore, two different disaccharides. Based on the JH1-H2 coupling constants, the signals at 5.09 and 4.97 ppm were assigned to Man and Gal, respectively. The coupling constant JH1-H2 = 3.5 Hz for the Gal signal at 4.97 ppm established its α anomeric configuration, while the determination of the Man configuration could not be achieved, because of the very small and similar JH1-H2 of α- and β- Man. Integration values of the peak areas are reported in Table 3 and they are consistent with two different disaccharides in equimolar amounts.
Table 3. Chemical shifts assignment and area integration (Int) values for the anomeric protons and methyl protons of disaccharides, trisaccharides and tetrasaccharides obtained from partial hydrolysis of Tramesan.
| DISACCHARIDES | TRISACCHARIDES | TETRASACCHARIDES | |||
|---|---|---|---|---|---|
| Assignment (ppm) | Int values | Assignment (ppm) | Int values | Assignment (ppm) | Int values |
| 2-Man (5.34) | 1.00 | 2-Man (5.32) | 1.00 | ||
| 2-Man (5.29) | 1.11 | ||||
| α-Gal (5.26) | 0.38 | α-Gal (5.26) | 0.31 | ||
| α-Fuc (5.21) | 0.43 | α-Fuc (5.20) | 0.45 | α-Fuc (5.19) | 0.41 |
| t-Man (5.10) | 1.00 | t-Man (5.05) | 1.23 | t-Man (5.03) | 1.14 |
| t-Gal (4.98) | 1.03 | 6-Gal (5.00) | 1.89 | ||
| 6-Gal (4.99) | |||||
| t-Gal (4.97) | |||||
| β-Fuc (4.60) | 0.68 | β-Fuc (4.60) | 1.38 | β-Fuc (4.57) | 0.76 |
| β-Gal (4.59) | 0.72 | β-Gal (4.58) | |||
| -CH3 of Fuc (1.23, 1.18) |
4.00 | -CH3 of Fuc (1.22, 1.18) |
3.77 | -CH3 of Fuc (1.21, 1.16) |
4.23 |
Inspection of the COSY, TOCSY and HSQC spectra led to the assignment of most of the peaks (see Table C in S1 File); in the HSQC plot (data not shown) C6 signals engaged in glycosidic linkages were visible at about 67 ppm, in agreement with 6-linked Gal. NOESY spectrum (data not shown) showed a strong correlation between H1 of Man and H3 of Fuc, thus establishing the structure Man-(1→3)-Fuc. Therefore, the second disaccharide is α-Gal-(1→6)-Gal. These experimental data were corroborated by the output of the CASPER computer program for the two disaccharides sequences [47,48].
The expansions of the anomeric and methyl regions of the 1H NMR spectrum of the trisaccharides mixture is reported in Fig 4B. By comparison with the 1H NMR spectrum of the disaccharides and resorting to chemical shifts values from the literature, anomeric protons were easily assigned, as shown in Fig 4C, and these assignments were also corroborated by COSY, TOCSY, HSQC and NOESY spectra (data not shown). Integration values of the main peaks are reported in Table 3 and the data collected suggested the presence of two main trisaccharides in roughly equimolar amounts: Man-(1→2)-Man-(1→3)-Fuc and Gal-(1→6)-Gal-(1→6)-Gal. Therefore, the obtained trisaccharides are consistent with the structure of the disaccharides with the addition of one further unit: Man and Gal, respectively.
The enlargement of the anomeric and methyl regions of the 1H NMR spectrum of the tetrasaccharides fraction showed two sets of signals having very different intensities (Fig 4C); the most intense resonances were assigned by comparison with both literature values and with the di- and trisaccharides spectra. Integration data are reported in Table 3. The data collected indicated that in the tetrasaccharide peak the main oligosaccharide present has the structure Man-(1→2)-Man-(1→2)-Man-(1→3)-Fuc, while signals attributable to Gal residues (marked with a star in Fig 3C), and, by analogy with di- and tri-saccharides structures, likely belonging to a galactotetraose, were much less intense.
The fraction eluted at the void volume (V0) of the Bio Gel P2 column obtained from the partial hydrolysis reaction, containing high molecular mass oligomers, was subjected to linkage analysis and 1H NMR spectroscopy. The results of linkage analysis are reported in Table 2: t-Fuc was not detected, while the intensity of 3-Fuc dramatically decreased, in agreement with its presence as reducing end residue in the oligosaccharides. The total amount of branched residues (2,6-Gal + 2,6-Hex = 0.87) is in very good agreement with the amount of t-Man (0.89). The residue 2,6-Gal also decreased with the concomitant increase of 6-Gal, thus indicating that partial hydrolysis removed side chains attached on C2 of Gal, producing the oligosaccharides described above. Therefore, the main chain of Tramesan is a 1–6 linked galactan with (Man)n-Fuc branches in position 2, and according with glycosidic linkage analysis (Table 2, fraction CIII), some side chains can consist only of t-Fuc. Linkage analysis showed that t-Man and 2-Man were present in the V0 fraction, indicating that they are not confined in the 2-linked Gal branches; together with the relative increase of 2,6-Hex (Table 4, fraction V0), data might indicate the presence of (Hex)n-(Man)m branched sequences, either composing a second polysaccharide, or bound to the above described galactomannan. Furthermore, the gas-chromatographic retention time of 2,6-Hex peak was identical to that of a 2,6-Man standard from our laboratory (Figure H in S1 File), and the absence in fraction V0 1H-NMR spectrum (Fig 1B) of resonances typical of Glc units [43] strongly suggested to identify 2,6-Hex with 2,6-Man.
Table 4. Biological activity of Tramesan on A. flavus and A. parasiticus.
In vitro culture of A. flavus and A. parasitucus under aflatoxin permissive conditions (PDB, 30°C) were treated or not (control) with 0.38 μM Tramesan and incubated for 7 days. Aflatoxin B1 production, evaluated by LC-MS/MS, and fungal growth, evaluated by weighting dried mycelia, in Tramesan-treated cultures, were normalised for non-treated ones and the percentage of inhibition calculated consequently. Mycelia was used to evaluate the expression, calculated by 2-ΔΔCt method in RT-PCR, of the oxidative stress related transcription factors AfyapA and ApyapA and the superoxide dismutase encoding gene sod1 in A. flavus and A. parasiticus, respectively.
| Fungal species | treatment | Aflatoxin B1 (ppb) | % of Aflatoxin B1 inhibition compared to untreated control | Fungal growth (mg/mL d.w.) | Ap-1 like | sod1 |
|---|---|---|---|---|---|---|
| A. flavus | control | 125.2 ± 2.5 | 95.2 | 5.1 ± 0.6 | 2.1 ± 0.3 | 25.2 ± 3.2 |
| Tramesan 0.38 µM | 6.02 ± 0.2 | 5.2 ± 0.5 | ||||
| A. parasiticus | control | 185.5 ± 7.2 | 98.7 | 4.5 ± 0.2 | 2.5 ± 0.2 | 22.3 ± 4.1 |
| Tramesan 0.38 µM | 2.4 ± 0.5 | 4.4 ± 0.8 |
The 1H NMR spectrum of fraction V0 in Fig 1B showed interesting changes. The methyl groups of Fuc greatly decreased in intensity, in agreement with methylation analysis. In the anomeric region, H1 resonances of A, B, C, and E of Man residues substantially decreased, in agreement with the removal of Man-containing oligosaccharides, linked on position 2 of Gal residues. Signals F (t-Man) and G (3-Fuc) greatly decreased in intensity, while resonance H disappeared, in agreement with its tentative assignment to t-Fuc. Other changes occurred in the region 5.2–4.8 ppm, but no precise information could be obtained on the type of monosaccharides involved.
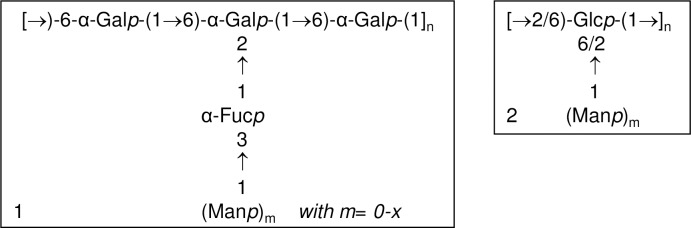
Although for a detailed structural definition Tramesan needs to be deeper investigated, the data obtained in this study on the biologically active polysaccharidic fractions suggested that it consists of two sequences that may or may be not part of a single polymer, similar to other fungal polysaccharides [49]. The proposed structures are shown in Fig 5.
Fig 5. Proposed structure for Tramesan.
Scheme 1 and 2 can be part of the same polysaccharide or forming repeating units of different polymers.
Biological effects
Purified Tramesan was firstly tested on A. flavus and A. parasiticus for its inhibiting effect on fungal growth and aflatoxin B1 production. Tramesan was used at a concentration 0.38 μM as suggested by Scarpari et al. [36]; the amount utilised, amended from fungal culture, did not significantly affected the fungal growth of both strains whereas, the aflatoxin B1 production was markedly inhibited (p<0.001, Table 4). Considering that aflatoxin biosynthesis is under the control of oxidative stress [34,50], the expression of AfyapA and ApyapA, oxidative stress related transcription factors that control the synthesis of aflatoxin upstream to the aflatoxin regulator AflR [20,34] and of the superoxide dismutase encoding sod1 in A. flavus and A. parasiticus, respectively, was monitored. It was observed that Tramesan significantly triggered the expression of AfyapA and of sod1 (Table 4).
To check if Tramesan could elicit antioxidant response in other biological systems, thus acting as a pro antioxidant molecule, we tested its effect in hampering the necrotrophic progression of a foliar pathogen of durum wheat, Parastagonospora nodorum. This pathogen produces a necrotrophic effector that also act by increasing ROS production at the interface for facilitating tissue degeneration and cell death [51]. Thus, we pre-treated durum wheat leaves in greenhouse with a suspension of Tramesan 48 h before inoculating the pathogen. Concomitantly, we tested the antifungal activity of Tramesan on pure P. nodorum culture. Wheat plants treated with Tramesan resulted more protected from P. nodorum infection compared to untreated ones; interestingly, Tramesan had no appreciable effect on fungal growth under in vitro conditions (data not shown), whereas in planta limited its growth. We here suggest that Tramesan enhanced some ROS-scavenging ability (e.g. PR9) of wheat plant leaves (Table 5) disabling necrotrophic weapons for triggering PCD and causing disease.
Table 5. Effect of Tramesan on wheat leaves infected with P. nodorum.
Durum wheat leaves of an Italian commercial variety were sprayed with a solution of 0.38μM Tramesan and after 48 h, inoculated with 105 conidia of P. nodorum; other plants were inoculated with the pathogen but not pre-treated with Tramesan (Infected control). Disease severity was quantified by using Liu’ scale (necrotic spot extension) whereas relative expression of the peroxidase-encoding, PR-9 gene, was calculated using the 2-ΔΔCt method. Fungal growth was assessed by qPCR using P. nodorum specie-specific primers.
| Durum wheat leaves | Necrotic spot | Fungal growth | PR-9 (peroxidase) |
|---|---|---|---|
| rated by Liu’ scale1 | qPCR (ng fungal DNA/μg total DNA) | mRNA relative expression (2-ΔΔCt) | |
| Infected control | 4 | 80.1 ± 2.2 | 1 |
| Tramesan | 0 | 5.0 ± 0.5 | 64.2 ± 2.1 |
1Liu et al., 2004 [30]
To validate the widespread ability of Tramesan to behave as a pro antioxidant molecule, we also tested its effect on a murine cell line of melanoma (B16). In this case, the expected effect is a limitation in cell growth since cancer cells express high level of intrinsic oxidative stress that normally boosts their division [52]. Notably, we aimed at checking if Tramesan was able to enhance the ROS scavenging ability of these cutaneous murine cells. In relation to this, we quantified the amount of an antioxidant molecule (melanin) and tested the expression of Nrf-2 mRNA, since Nrf-2 protects melanocytes against the harmful ROS effects [53]. Cells treatment with 0.38 μM Tramesan increased melanin content of about two-folds whilst enhancing Nrf-2 expression and consistently reducing cell growth (Table 6).
Table 6. Effect of Tramesan on murine cell lines of melanoma (B16).
Murine cell lines of melanoma (B16) were treated with 0.38 μM Tramesan or untreated (control). The amount of melanin was calculated by spectrophotometer (Abs405), normalised for untreated samples and the percentage of its increase was evaluated and compared to untreated samples. Cell growth was calculated by counting the number of viable cells. The results are the mean ± SD of 5 separate experiments Relative expression of the Nrf-2 gene normalised on untreated cells was calculated using the 2-ΔΔCt method.
| Melanin | Cell growth | Nrf-2 | |
|---|---|---|---|
| Percentage of increase compared to untreated samples (%) | Percentage of reduction of cell number (%) | mRNA relative expression (2-ΔΔCt) | |
| Tramesan | 97.5 ± 4.3 | 83.7 ± 2.2 | 9.5 ± 0.2 |
Discussion
Fungal glycans may act as eliciting molecules in host cells. In addition, some fungi produce and secrete glycans to be recognised [54]. Recently, some oligomers of chitin were found to be actively released in the interface between fungal symbionts and their host to “facilitate” recognition processes and repress host defence [21,55]. Apart lignin degrading enzymology, few reports actually concern the biology of T. versicolor. In a previous study, the semi-purified culture filtrate of T. versicolor containing molecules with a supposed molecular mass (MM) >3.0 kDa showed a particular ability in enhancing antioxidant activity in A. flavus, dramatically inhibiting the biosynthesis of aflatoxins [36]. In relation to this, we exploited the ability of the semi-purified culture filtrate of T. versicolor to inhibit aflatoxin biosynthesis as a fast and reliable method to detect active principles present into the MM>3.0 kDa fraction (fraction A in this study). We found that this mushroom produces an exo-polysaccharide that is not simply “leaked” by the complex fungal cell wall, but is secreted into the environment with a still unknown biosynthetic and transportation pathways and, overall, with an unknown biological function. In the present study, we provide some hints for elucidating the latter. Tramesan is a branched fucose-enriched fungal polysaccharide of about 23 kDa with a probable “repetitive” scheme of monosaccharide sequence in the linear (α-1,6-Gal)n backbone as well as in the lateral chain Man-(1→2)-Man-(1→3)-Fuc. It could be suggested that host cells may recognize fungi from their saccharidic “barcode” composed by a scheme of repetitive units in which the single unit acts–de facto–as signalling molecule. This was recently confirmed for signal factors in fungal mycorrhizae that are composing lipochitooligosaccharides [56] and chitooligosaccharides [54]. These fungal complex glucans may elicit calcium waves in different systems and Tramesan, when administered to murine melanoma cell culture, displays this effect too (data from our laboratories). Thus, what we suggest here is that Tramesan may act for T. versicolor as an “informative” molecule able to modify the relation with other organisms into its trophic niche.
In our study, we demonstrated that Tramesan might elicit similar responses in different organisms. Notably, in previous reports non-purified Tramesan proved able to induce the activation of antioxidant defences into mycotoxin producing fungi [33,36]. Several mycotoxigenic fungi are sensitive to variation in the cell redox balance and many exploit augmentation in circulating ROS by vehiculating this oxidative power into toxin synthesis [50,57]. Thus, the hypothesis ongoing after these results proposes Tramesan as a pro antioxidant molecule, able to trigger the expression of oxidative stress related transcription factors (e.g. Nrf-2 like as well as yAP-1 like factors) and, in turn, re-establish the proper cell redox balance and “subtracting” oxidative power that boosts toxin synthesis. Since ROS are often the molecular trigger for opening different responses such as defence and differentiation in plant and animals [58–60], we tested two different “models”: the defence reaction of a plant to a necrotrophic pathogen (P. nodorum x T. turgidum subsp. durum) and the melanin differentiation into murine melanoma. As stated previously, these mechanisms are triggered, or at least accompanied, by a cell redox unbalance due to oxidative burst [53,60]. Tramesan enhances the defence ability of durum wheat as well as melanin biosynthesis in melanoma. Notably, this polysaccharide induces the expression of oxidative stress defence-related genes such as peroxidases and Nrf-2. Even in these organisms as well as in mycotoxin producing fungi, Tramesan can elicit an antioxidant response, probably, by manipulating gene expression. We here suggest that, Tramesan could be recognized by specific receptors that, in turn, activate pathways leading to an antioxidant response. As it is known, several receptors in different organisms act as ligands of fungal glycans and, in turn, trigger various responses ranging from innate immunity to cell death or symbiosis [5,12,61]. Tramesan could act as ligand for a still unknown inter-kingdom conserved receptor able to control antioxidant responses.
In conclusion, we present for the first time the partial elucidation of the structure of Tramesan, a branched fungal glycan secreted into the environment by the lignin degrading fungus T. versicolor. This compound is able to act as a pro antioxidant in different organisms. By enhancing the “natural” antioxidant defences of the “hosts”, Tramesan could represent a useful tool for different challenges in different “contexts” in which limiting ROS production represents a solution for defusing harmful situations such as toxins release into foodstuff, necrotic lesions into plant crops or growth of cancer cells.
Supporting information
Figure A. HP-SEC calibration plot obtained with nine pullulan standards with molecular masses in the range 1.7x106–5.9x103, used to evaluate Tramesan fraction molecular masses.
Figure B. Scheme summarizing the purification steps applied for the obtainment of the different fractions (B-G) used in the aflatoxin inhibition bioassays.
Figure C. Scheme of the characterization of the bioactive polysaccharide fraction C.
Figure D. Size exclusion chromatography on a Sephacryl S-300 column of the filtrate C from T. versicolor TV117 cultures. The three obtained fractions (CI, CII, CIII) are indicated.
Figure E. Aflatoxin inhibition by the different SEC-separated fractions (I-III).
Figure F. 1H-NMR spectra of the fractions II (red) e III (blue) obtained after size exclusion chromatography on a Sephacryl S-300 column.
Figure G. HSQC plots of Trametan fraction III solution recorded at 50°C. A) Expansion of the anomeric region and B) expansion of the ring region. Assignments as reported in S2 Table, relative to acetone (2.225 ppm for 1H and 31.07 ppm for 13C). H6, C6 of hexoses are in blue: those belonging to 6-linked hexoses resonate at about 67 ppm, those that are not linked at about 62 ppm.
Figure H. Comparison of GLC elution profiles between PMAA derivatives of Tramesan and a sample containing 2,6-Man. The peaks of interest are marked with asterisks.
Table A. Composition analysis of the fraction C containing the polysaccharide produced by T. versicolor Tv117 (PLS 117) and of a Trametes versicolor commercial powder sample (CP). Results are expressed in weight %.
Table B. 1H and 13C chemical shift assignments of Tramesan, referred to acetone (2.225 ppm for 1H and 31.07 ppm for 13C).
Table C. 1H and 13C chemical shift assignments of the disaccharides obtained from partial hydrolysis of Tramesan. Signals are referred to acetone (2.225 ppm for 1H and 31.07 ppm for 13C).
(DOCX)
Acknowledgments
Tramesan was patented (N° 0001418693) in Italy with the commercial name of Trametano. We are grateful to Dr. Anna Adele Fabbri for her invaluable and constant advices. We are grateful to Ms V. Farina and Dr. A. Iori for aiding in the execution of experiments on plants.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was partly funded by external contributions provided by Fondazione Benefica Kathleen Foreman Casali, Ministero dell’Istruzione, dell’Università e della Ricerca, award number 9837 05/05/2009. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Meng X, Liang H, Luo L. Antitumor polysaccharides from mushrooms: a review on the structural characteristics, antitumor mechanisms and immunomodulating activities. Carbohydr Res. 2016; 424: 30–41 doi: 10.1016/j.carres.2016.02.008 [DOI] [PubMed] [Google Scholar]
- 2.Dam TK, Brewer CF. Effects of clustered epitopes in multivalent ligand–receptor interactions. Biochem. 2008;47: 8470–8476. [DOI] [PubMed] [Google Scholar]
- 3.Nürnberger T, Brunner F, Kemmerling B, Piater, L. Innate immunity in plants and animals: striking similarities and obvious differences. Immun Rev. 2004;198: 249–266. [DOI] [PubMed] [Google Scholar]
- 4.Savary S, Ficke A, Aubertot J.-N, Hollier C. Crop losses due to diseases and their implications for global food production losses and food security. Food Security (Online) 2012;19 [Google Scholar]
- 5.Cottier F, Pavelka N. Complexity and dynamics of host-fungal interactions. Immunol Res. 2012;53(1–3):127–35 doi: 10.1007/s12026-012-8265-y [DOI] [PubMed] [Google Scholar]
- 6.Chan G.CF, Chan WK, Sze DMY. The effects of β-glucan on human immune and cancer cells. J Hematol Oncol. 2009;2: 25–35. doi: 10.1186/1756-8722-2-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nobuyuki S, Akifumi S, Hidemitsu K, Yoshio O. Chemical structure of the cell-wall mannan of Candida albicans serotype A and its difference in yeast and hyphal forms. Biochem J. 2007;404: 365–372. doi: 10.1042/BJ20070081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruiz-Herrera J. Fungal Cell Wall: Structure, Synthesis, and Assembly, Second Edition—CRC Press Book; 1991 [Google Scholar]
- 9.Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. BioEssays 2006;28 (8): 799–808. doi: 10.1002/bies.20441 [DOI] [PubMed] [Google Scholar]
- 10.Fesel PH, Zuccaro A. β-glucan: Crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genet Biol. 2016;90: 53–60. doi: 10.1016/j.fgb.2015.12.004 [DOI] [PubMed] [Google Scholar]
- 11.Newman MA, Sundelin T, Nielsen JT and Erbs G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci. 2013; 4:139 doi: 10.3389/fpls.2013.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass NL, Dementhon K, Non-self recognition and programmed cell death in filamentous fungi. Curr Opin Microbiol. 2006; 9(6): 553–558. doi: 10.1016/j.mib.2006.09.001 [DOI] [PubMed] [Google Scholar]
- 13.Hall C, Welch J, Kowbel DJ, Glass NL. Evolution and diversity of a fungal self/nonself recognition locus. PLoS ONE 2010;5(11) e14055 doi: 10.1371/journal.pone.0014055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid DM, Gow NA, Brown GD. Pattern recognition: recent insights from Dectin-1. Curr Opin Immunol. 2009;21(1): 30–37. doi: 10.1016/j.coi.2009.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vijayan V, Baumgart-Vogt E, Naidu S, Qian G, Immenschuh S. Bruton's tyrosine kinase is required for TLR-dependent heme oxygenase-1 gene activation via Nrf2 in macrophages. J Immunol. 2011; 187(2):817–27. doi: 10.4049/jimmunol.1003631 [DOI] [PubMed] [Google Scholar]
- 16.Lai Z, Vinod K, Zheng Z, Fan B, Chen Z. Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 2008;8: 68 doi: 10.1186/1471-2229-8-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010; 64: 204–214. doi: 10.1111/j.1365-313X.2010.04324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B, Hong YB, Zhang YF, Li XH, Huang L, Zhang HJ, Li DY, et al. Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress, Plant Sci. 2014, 227: 145–156. doi: 10.1016/j.plantsci.2014.08.001 [DOI] [PubMed] [Google Scholar]
- 19.Reverberi M, Fabbri AA, Zjalic S, Ricelli A, Fanelli C. Antioxidant enzymes stimulation in Aspergillus parasiticus by Lentinula edodes inhibits aflatoxin production. Appl Microbiol Biotechnol. 2005; 69: 207–215. doi: 10.1007/s00253-005-1979-1 [DOI] [PubMed] [Google Scholar]
- 20.Reverberi M, Zjalic S, Ricelli A, Punelli F, Camera E, Fabbri AA, et al. Modulation of antioxidant defence in Aspergillus parasiticus is involved in aflatoxin biosynthesis: a role of the ApyapA gene. Eukaryot Cell 2008; 7: 988–1000. doi: 10.1128/EC.00228-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Druzhinina I, Seidl-Seiboth V, Herrera-Estrella A, Horwitz BA, Kenerley CM, Monte E, et al. Trichoderma: the genomics of opportunistic success. Nat Rev Microbiol. 2011;9(10):749–59. doi: 10.1038/nrmicro2637 [DOI] [PubMed] [Google Scholar]
- 22.Albersheim P, Nevins DJ, English PD, Karr A. A method for the analysis of sugars in plant cell wall polysaccharides by gas-liquid chromatography. Carbohydr. Res. 1967; 5: 340–345. [Google Scholar]
- 23.Harris PJ, Henry RJ, Blakeney AB, Stone BA. An improved procedure for the methylation analysis of oligosaccharides and polysaccharides. Carbohydr. Res. 1984;127: 59–73. [DOI] [PubMed] [Google Scholar]
- 24.Sweet DP, Shapiro RH, Albersheim P. Quantitative analysis by various g.l.c. response-factor theories for partially methylated and partially ethylated alditol acetates. Carbohydr. Res. 1975; 40:217–25. [Google Scholar]
- 25.Dolfi S, Sveronis A, Silipo A, Rizzo R, Cescutti P. A novel rhamno-mannan exopolysaccharide isolated from biofilms of Burkholderia multivorans C1576. Carbohydr. Res. 2015;411: 42–48. doi: 10.1016/j.carres.2015.04.012 [DOI] [PubMed] [Google Scholar]
- 26.Dell A. Preparation and desorption mass spectrometry of permethyl and peracetyl derivatives of oligosaccharides. Method. Enzymol. 1990;193: 647–660. [DOI] [PubMed] [Google Scholar]
- 27.Sulyok M, Berthiller F, Krska R, Schuhmacher R. Development and validation of a liquid chromatography/tandem mass spectrometric method for the determination of 39 mycotoxins in wheat and maize. Rapid Commun. Mass Sp. 2006; 20:2649–2659. [DOI] [PubMed] [Google Scholar]
- 28.Scala V, Camera E, Ludovici M. Dall’Asta C, Cirlini M, Giorni P, et al. Fusarium verticillioides and maize interaction in vitro: relation between oxylipin cross-talk and fumonisin synthesis. World Mycotox J. 2013;6: 343–3514. [Google Scholar]
- 29.Reverberi M, Zjalic S, Ricelli A, Di Meo C. Scarpari M, Fanelli C, et al. Mushrooms versus fungi: natural compounds from Lentinula edodes inhibit aflatoxin biosynthesis by Aspergillus parasiticus. World Mycotox J. 2011;4(3): 217–224. [Google Scholar]
- 30.Liu ZH, Faris JD, Meinhardt SW, Ali S, Rasmussen JB, Friesen TL. Genetic and Physical Mapping of a Gene Conditioning Sensitivity in Wheat to a Partially Purified Host-Selective Toxin Produced by Stagonospora nodorum. Phytopathol. 2004; 94(10): 1056–1060. [DOI] [PubMed] [Google Scholar]
- 31.Iori A, Scala V, Cesare D, Pinzari F, D’Egidio MG, Fanelli C, et al. Hyperspectral and molecular analysis of Stagonospora nodorum blotch disease in durum wheat Eur J Plant Pathol. 2015; 141(4): 689–702. [Google Scholar]
- 32.Nobili C, D’angeli S, Altamura MM, Scala V, Fabbri AA, Reverberi M, et al. ROS and 9-oxylipins are correlated with deoxynivalenol accumulation in the germinating caryopses of Triticum aestivum after Fusarium graminearum infection. Eur J Plant Pathol. 2014;139(2): 423–438. [Google Scholar]
- 33.Zjalic S, Reverberi M, Ricelli A, Granito VM, Fanelli C, Fabbri AA. Trametes versicolor: a possible tool for aflatoxin control. Int J Food Microbiol. 2006;107:243–249 doi: 10.1016/j.ijfoodmicro.2005.10.003 [DOI] [PubMed] [Google Scholar]
- 34.Roze LV, Hong S-Y, Linz JE. Aflatoxin biosynthesis: current frontiers. Annu Rev Food Sci Tech. 2013;4: 293–311. [DOI] [PubMed] [Google Scholar]
- 35.Zaccaria M, Ludovici M, Sanzani SM, Ippolito A, Cigliano RA, Sanseverino W, et al. Menadione-induced oxidative stress re-shapes the oxylipin profile of Aspergillus flavus and its Lifestyle. Toxins 2015;7: 4315–4329. doi: 10.3390/toxins7104315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scarpari M, Parroni A, Zaccaria M, Fattorini L, Bello C, Fabbri AA, et al. Trametes versicolor bioactive compounds stimulate Aspergillus flavus antioxidant system and inhibit aflatoxin synthesis, Plant Biosyst. 2016;150(4): 653–659. [Google Scholar]
- 37.Krijger J-J, Thon MR, Deising HB and Wirsel SGR. Compositions of fungal secretomes indicate a greater impact of phylogenetic history than lifestyle adaptation. BMC Genomics 2014;15:722 doi: 10.1186/1471-2164-15-722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gander JE, Beachy J, Unkefer CJ, Tonn SJ. Toward understanding the structure, biosynthesis, and Function of a membrane-bound fungal glycopeptide. In Fungal Polysaccharides, Paul A. Sanfdfor and Kazuo Matsuda Eds., ACS Symposium Series Vol. 126. 1980; Cpt4: 49–79.
- 39.Rizzo J, Oliveira DL, Joffe LS, Hu G, Gazos-Lopes F, Fonseca FL, et al. Role of the Apt1 protein in polysaccharide secretion by Cryptococcus neoformans. Eukaryot. Cell. 2014;13(6):715–26. doi: 10.1128/EC.00273-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernando MDM, Wijesundera RLC, Soysa SSBDP, de Silva ED, Nanayakkara CM. Antioxidant potential and content of the polyphenolic secondary metabolites of white rot macrofungi; Flavodon flavus (Klotzsch.) and Xylaria feejeensis (Berk.). Plant Sci. J. 2016;1 (1): 1–6. [Google Scholar]
- 41.Liao SF, Liang CH, Ho MY, Hsu TL, Tsai TI, Hsieh YS, et al. Immunization of fucose-containing polysaccharides from Reishi mushroom induces antibodies to tumor-associated Globo H-series epitopes. Proc Natl Acad Sci USA 2013;110(34):13809–13814. doi: 10.1073/pnas.1312457110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cescutti P, Cuzzi B, Liut G, Segonds C, Di Bonaventura G, Rizzo R. A novel highly charged exopolysaccharide produced by two strains of Stenotrophomonas maltophilia recovered from patients with cystic fibrosis. Carbohydr Res. 2011;346: 1916–1923. doi: 10.1016/j.carres.2011.05.007 [DOI] [PubMed] [Google Scholar]
- 43.Bock K, Thøgersen H. Nuclear Magnetic resonance Spectroscopy in the study of mono- and oligosaccharides. Ann Rep NMR Spectr. 1982;13: 1–57. [Google Scholar]
- 44.Bock K, Pedersen C. Carbon-13 Nuclear Magnetic Resonance Spectroscopy of Monosaccharides. Adv Carbohydr Chem Biochem. 1983;41: 27–66. [Google Scholar]
- 45.Bock K, Pedersen C, Pedersen H. Carbon-13 nuclear magnetic resonance data for oligosaccharides. Adv Carbohydr Chem Biochem. 1984;42: 193–225. [Google Scholar]
- 46.Shibata N, Suzuki A, Kobayashi H and Okawa Y. Chemical structure of the cell-wall mannan of Candida albicans serotype A and its difference in yeast and hyphal forms. Biochem J. 2007;404: 365–372. doi: 10.1042/BJ20070081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lundborg M, Widmalm G. Structure Analysis of Glycans by NMR Chemical Shift Prediction. Anal Chem. 2011; 83:1514–1517. doi: 10.1021/ac1032534 [DOI] [PubMed] [Google Scholar]
- 48.Jansson P-E, Stenutz R, Widmalm G. Sequence determination of oligosaccharides and regular polysaccharides using NMR spectroscopy and a novel Web-based version of the computer program CASPER. Carbohydr Res. 2006; 341:1003–1010. doi: 10.1016/j.carres.2006.02.034 [DOI] [PubMed] [Google Scholar]
- 49.Leal JA, Prieto A, Bernabé M., Hawksworth DL. An assessment of fungal wall heteromannans as a phylogenetically informative character in ascomycetes. FEMS Microbiol Rev. 2010;34:986–1014. doi: 10.1111/j.1574-6976.2010.00225.x [DOI] [PubMed] [Google Scholar]
- 50.Roze LV, Laivenieks M, Hong S-Y, Wee J, Wong SS, Vanos B, et al. Aflatoxin biosynthesis is a novel source of reactive oxygen species—a potential redox signal to initiate resistance to oxidative stress? Toxins 2015;7: 1411–1430. doi: 10.3390/toxins7051411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vincent D, Du Fall LA, Livk A, Mathesius U, Lipscombe RJ, et al. A functional genomics approach to dissect the mode of action of the Stagonospora nodorum effector protein SnToxA in wheat. Mol Plant Pathol. 2012;13(5):467–82. doi: 10.1111/j.1364-3703.2011.00763.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010; 44(5):479–96. doi: 10.3109/10715761003667554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gęgotek A, Skrzydlewska E. The role of transcription factor Nrf2 in skin cells metabolism. Arch Dermatol Res. 2015; 307(5):385–396. doi: 10.1007/s00403-015-1554-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Genre A, Chabaud M, Balzergue C, Puech-Pagès V, Novero M, Rey T, et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncagula roots and their production is enhanced by strigolactone. New Phytol. 2013;198: 190–202. doi: 10.1111/nph.12146 [DOI] [PubMed] [Google Scholar]
- 55.Sun J, Miller JB, Granqvist E, Wiley-Kalil A, Gobbato E, Maillet F, et al. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell 2015; 27(3): 823–838. doi: 10.1105/tpc.114.131326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dénarié J, Debellé F, Promé JC. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Ann Rev Biochem. 1996;65: 503–535. doi: 10.1146/annurev.bi.65.070196.002443 [DOI] [PubMed] [Google Scholar]
- 57.Sanzani SM, De Girolamo A, Schena L, Solfrizzo M, Ippolito A, Visconti A. Control of Penicillium expansum and patulin accumulation on apples by quercetin and umbelliferone. Eur Food Res Technol. 2009;228: 381 doi: 10.1007/s00217-008-0944-5 [Google Scholar]
- 58.Lushchak VI. Adaptive response to oxidative stress: Bacteria, fungi, plants and animals. Comp Biochem Physiol C Toxicol Pharmacol. 2011;153(2):175–190. doi: 10.1016/j.cbpc.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 59.Scandalios JG. The rise of ROS. Trends Biochem Sci. 2002;27:483–486 [DOI] [PubMed] [Google Scholar]
- 60.Torres MA. ROS in biotic interactions. Physiol Plant. 2010;138:414–429. doi: 10.1111/j.1399-3054.2009.01326.x [DOI] [PubMed] [Google Scholar]
- 61.Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 2003; 302: 630–633. doi: 10.1126/science.1090074 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure A. HP-SEC calibration plot obtained with nine pullulan standards with molecular masses in the range 1.7x106–5.9x103, used to evaluate Tramesan fraction molecular masses.
Figure B. Scheme summarizing the purification steps applied for the obtainment of the different fractions (B-G) used in the aflatoxin inhibition bioassays.
Figure C. Scheme of the characterization of the bioactive polysaccharide fraction C.
Figure D. Size exclusion chromatography on a Sephacryl S-300 column of the filtrate C from T. versicolor TV117 cultures. The three obtained fractions (CI, CII, CIII) are indicated.
Figure E. Aflatoxin inhibition by the different SEC-separated fractions (I-III).
Figure F. 1H-NMR spectra of the fractions II (red) e III (blue) obtained after size exclusion chromatography on a Sephacryl S-300 column.
Figure G. HSQC plots of Trametan fraction III solution recorded at 50°C. A) Expansion of the anomeric region and B) expansion of the ring region. Assignments as reported in S2 Table, relative to acetone (2.225 ppm for 1H and 31.07 ppm for 13C). H6, C6 of hexoses are in blue: those belonging to 6-linked hexoses resonate at about 67 ppm, those that are not linked at about 62 ppm.
Figure H. Comparison of GLC elution profiles between PMAA derivatives of Tramesan and a sample containing 2,6-Man. The peaks of interest are marked with asterisks.
Table A. Composition analysis of the fraction C containing the polysaccharide produced by T. versicolor Tv117 (PLS 117) and of a Trametes versicolor commercial powder sample (CP). Results are expressed in weight %.
Table B. 1H and 13C chemical shift assignments of Tramesan, referred to acetone (2.225 ppm for 1H and 31.07 ppm for 13C).
Table C. 1H and 13C chemical shift assignments of the disaccharides obtained from partial hydrolysis of Tramesan. Signals are referred to acetone (2.225 ppm for 1H and 31.07 ppm for 13C).
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.