Abstract
The SoxC transcription factors (Sox4, Sox11, and Sox12) play important roles in the development of the vertebrate eye and retina. However, their expression and function during retinal and optic nerve regeneration remain elusive. In this study, we investigated the expression and possible functions of the SoxC genes after retinal and optic nerve injury in adult zebrafish. We found that among the five SoxC members, Sox11b was strongly induced in BrdU-positive cells in the inner nuclear layer (INL) after retinal injury, and morpholino-mediated Sox11b-knockdown significantly reduced the number of proliferating cells in the INL at 4 days post-injury. After optic nerve lesion, both Sox11a and Sox11b were strongly expressed in retinal ganglion cells (RGCs), and knockdown of both Sox11a and Sox11b inhibited RGC axon regrowth in retinal explants. Our study thus uncovered a novel expression pattern of SoxC family genes after retinal and optic nerve injury, and suggests that they have important functions during retinal and optic nerve regeneration.
Keywords: SoxC, Sox11, Expression, Zebrafish, Retina, Optic nerve, Regeneration
Introduction
In the adult mammalian central nervous system (CNS), most neurons are not replaced following traumatic injury or disease. In contrast, teleost fish such as zebrafish exhibit a remarkable capacity for CNS regeneration [1, 2]. Why lower vertebrates like zebrafish possess such a strong regenerative potential while mammals do not is a very important and fundamental question in neuroscience. The mechanisms underlying CNS repair in zebrafish could therefore be used to design novel therapeutic strategies for improving CNS regeneration in mammals.
One good model to study CNS repair is the zebrafish visual system. As part of the CNS, the visual system is more accessible than other regions, making it an ideal system to study mechanisms of CNS regeneration. Following retinal injury in adult zebrafish, the resident Müller glia rapidly dedifferentiate and proliferate into a cycling population of neurogenic precursors that eventually regenerate new neurons and restore vision [3, 4]. Retinal regeneration in zebrafish is regulated by many extracellular cues such as the Hbegf/Wnt/Fgf/TNFα/TGFβ signaling pathways [5–9], as well as cell-intrinsic transcription factors [10–12]. Zebrafish also mount a robust regenerative response after optic nerve injury that regrows severed axons and restores lost vision [13, 14].
The Sox (Sry-related box) transcription factors are named after their shared motif called the SRY box, which is a high-mobility-group (HMG) DNA binding domain [15]. Based on the sequence similarity and genomic alignment, Sox proteins are subdivided into groups A to J [15]. The mammalian SoxC family has three intronless genes: Sox4, Sox11, and Sox12 [15]. Known as transcription activators, the SoxC genes are widely expressed in neuronal progenitors in the brain as well as in other organs during embryogenesis [16, 17]. It has been reported that SoxC members are regulators of cell survival and proliferation, as well as cell fate and differentiation [16, 18, 19]. In the developing vertebrate eye, SoxC genes exhibit a partially-overlapping expression pattern in the neural retina and lens placode [16, 20, 21]. As retinal neurons mature, SoxC expression is down-regulated and very little expression is detectable in the adult mammalian retina [22]. SoxC genes play multiple roles during vertebrate eye development, including ocular morphogenesis, lens development, and retinal neurogenesis [23]. In mice, Sox4/Sox11 conditional knockout leads to a significant loss of retinal ganglion cells (RGCs) and other retinal neurons [24]. Interestingly, Sox4- and Sox11-deficient zebrafish exhibit no significant defects in RGC development. Instead, they have reduced number of mature rod photoreceptors in the developing retina [21], suggesting a role of SoxC genes in photoreceptor differentiation.
Although the role of SoxC transcription factors in eye development has been reported previously, their expression and function during retinal and optic nerve regeneration remain largely unknown. Here we report the temporal and spatial expression patterns of SoxC genes during retinal and optic nerve regeneration in zebrafish. We also used morpholino (MO)-based gene knockdown to explore their functions during these regenerative events.
Materials and Methods
Animals and Eye Injury
The zebrafish used in this study were treated in accordance with the guidelines for animal use and care at Nantong University. The Tg(1016tuba1a:GFP) transgenic line was generated using the 1016-bp goldfish tuba1a promoter as described previously [25]. The retina was injured as described previously [25]. Briefly, fish were anesthetized by immersion in 0.02% Tricaine (Sigma-Aldrich Corp., St. Louis, MO) and the right retina was poked four times, once in each quadrant, with a 30G needle. The needle was inserted through the sclera to the length of the bevel (~0.5 mm). The optic nerve injury was performed as described previously [13]. Briefly, the right eye was gently pulled out of the socket and the dorsal connective tissue was cut. The optic nerve was cut with iridectomy scissors, taking care not to damage the ophthalmic artery. The eye was then gently replaced in the socket and the fish allowed to recover in system water. The intact left optic nerve served as the control.
RT-PCR and Quantitative PCR (qPCR)
Retinas were isolated and the total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA). One microgram of total RNA was reverse-transcribed into cDNA using a Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science, Penzberg, Upper Bavaria, Germany). The primers for PCR and quantitative PCR (qPCR) are listed in Table 1. qPCR was carried out in triplicate using FastStart Universal SYBR Green Master Mix (Roche Applied Science) on a real-time PCR detection system (CFX96TM Real-Time System, Bio-Rad, Hercules, CA). The relative mRNA expression in intact and injured retinas was determined using the ΔΔCt method and normalized to gapdh mRNA expression.
Table 1.
Primers used in this study.
| Gene name | Primer sequence (5′-3′) |
|---|---|
| gapdh-F | ATGACCCCTCCAGCATGA |
| gapdh-R | GGCGGTGTAGGCATGAAC |
| ascl1a-F | GGTGGCCAGACGGAACGAG |
| ascl1a-R | TTCCCGCTCTTCCCTCTTAGTCC |
| sox4a-F | TCGCGAGCGGAAAGGAAGA |
| sox4a-R | CCGCGCAATGCCTTCCTC |
| sox4b-F | CCAGCGGCCACATCAAGAGA |
| sox4b-R | GTATCCGCGTCCTCCTTGCC |
| sox11a-F | GCCACCGGACACATAAAGCG |
| sox11a-R | GCGCGGAGGAGGCGAAGT |
| sox11b-F | ACGACGATGATGATGATGACGAC |
| sox11b-R | CACAGCACAGTCAATGTTTGGACC |
| sox12-F | ATGCGGGACAGAGGAAGAAAACA |
| sox12-R | GGGGCTTGCTGGATTTCATAGG |
| RT-sox4a-F | GAGCAGTCGCCGGACATGC |
| RT-sox4a-R | AGTTTTCGAGCTCTTTTTGGATGC |
| RT-sox4b-F | ACACCGGCAGCATGCGC |
| RT-sox4b-R | ACCTCGGGCGTGCAGTAATCG |
| RT-sox11a-F | TCCCAAACTGAAAGCGAACAAGAC |
| RT-sox11b-F | AGCAGCCGCAGGGAAGAAGT |
| RT-sox12-R | TGATGCAGCGCTGAGATCTA |
Tissue Preparation and Immunofluorescence
Fish were euthanized with an overdose of Tricaine. The eyes were dissected out and fixed in 4% paraformaldehyde at 4 °C overnight. Fixed samples were cryosectioned and prepared for immunofluorescence as previously described [10]. The primary antibodies for immunofluorescence were rabbit anti-GFP (1:1000, Life Technologies, Carlsbad, CA), and rat anti-BrdU (1:500, Abcam, Cambridge, MA). For BrdU staining, sections were first treated with 2 N HCl at 37 °C for 25 min, rinsed in 0.1 mol/L sodium borate (pH 8.5) for 10 min, and then processed using standard procedures.
BrdU Incorporation and In Situ Hybridization (ISH)
For BrdU incorporation, 20 μL of 20 mmol/L BrdU was injected i.p. into the anesthetized fish 3 h before sacrifice. ISH was performed using DIG-labeled cRNA probes on retinal cryosections (DIG-RNA labeling kit, Roche Applied Science) as described previously [10].
Quantification of BrdU+ Cells at the Injury Site
GFP and BrdU immunofluorescence on retinal cryosections showed that each injury-responsive zone (4 injuries per retina) was clearly distinguishable from other areas. The counting area for each injury site was ~400–600 μm in width, based on the actual size. The number of BrdU+ cells per injury in the inner nuclear layer (INL) was counted using the Cell Counter plugin of ImageJ software (Plugin/Analyze/Cell Counter, https://imagej.nih.gov/ij/).
Morpholino Treatments
For retinal injury, 1 μL of 1 mmol/L lissamine-tagged antisense MO (Gene Tools, Philomath, OR) was injected with a Hamilton syringe into the vitreous at the time of injury. Electroporation of MOs was performed as previously described [26]. MO-mediated target gene knockdown in RGCs was performed as previously described [27]. Briefly, gelfoam was cut into small pieces and applied with 1 μL of 1 mmol/L lissamine-tagged MO. The small piece of gelfoam was placed at the cut site of the lesioned optic nerve for 24 h and then removed. A standard control MO was used as the negative control. Translation-blocking MOs targeting zebrafish sox11a and sox11b are referred to as sox11a MO (5′-CCGTTGCCGTGCGTTGTCAGTCCAA-3′) and sox11b MO (5′-CATGTTCAAACACACTTTTCCCTCT-3′). Both have been previously described and validated [27].
Retinal Explant Assay
Retinal explant assays were performed as previously described [13, 28]. Briefly, 4 days after optic nerve transection, fish were anesthetized and retinas were isolated. Retinas were cut into 0.5-mm squares, digested with 1 mg/mL hyaluronidase in L15 medium (Life Technology) for 15 min, and then washed 3 times in culture medium (8% fetal calf serum, 3% zebrafish embryo extract, and 1× antibiotic/antimycotic in L15). The 35-mm plates were pre-coated with 100 μg/mL poly-L-lysine and 10 μg/mL laminin overnight. Retinal squares were then plated in the culture medium (one retina per plate) and maintained at 28 °C for 4 days in a humidified air chamber. Axon length and density were measured from images of adherent explants with Leica Qwin software (Leica Camera AG, Wetzlar, Germany). Axon density was defined as the number of neurites per explant. At least three retinas were used for each group and images of 10–15 explants from each retina were captured and quantified.
Microscopy and Statistical Analysis
Bright-field and fluorescence images of retinal cryosections were captured with a Leica DM4000B or a Zeiss Imager M2 upright microscope (Carl Zeiss AG, Oberkochen, Germany). Images of retinal explants were captured with an Olympus IX51 inverted microscope (Olympus Corporation, Tokyo, Japan). All experiments were repeated at least three times. For single comparisons, a two-tailed unpaired Student’s t-test was used. For multiple comparisons, one-way analysis of variance (ANOVA) followed by the Tukey test was used.
Results
Expression of SoxC Genes in the Retina After Needle-Poke Injury
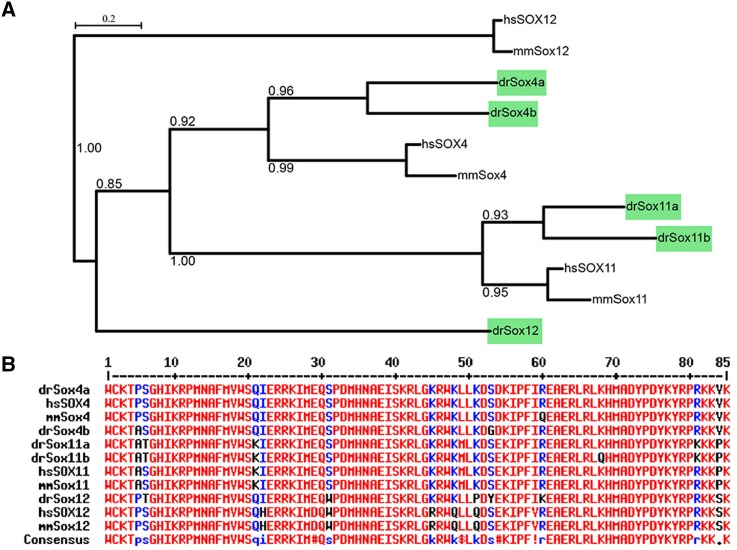
By searching the zebrafish genome (http://www.ensembl.org, Zv9), five SoxC members were found: Sox4a, Sox4b, Sox11a, Sox11b, and Sox12. Like mammalian SoxC genes, the zebrafish SoxC members are intronless except for Sox12, which has two exons (data not shown). Zebrafish SoxC members are homologs of their mammalian counterparts, as shown by phylogenetic analysis (Fig. 1A). The amino-acid sequences of the HMG domain in zebrafish and mammalian SoxC proteins have a high similarity (Fig. 1B). These results indicate that the SoxC proteins were highly conserved during vertebrate evolution, suggesting the importance of their functions.
Fig. 1.
Zebrafish SoxC genes are homologs of their mammalian counterparts. A Phylogenetic tree depicting the evolutionary relationships of human, mouse, and zebrafish SoxC genes generated by PhyML software (http://www.atgc-montpellier.fr/phyml/). Bootstrap values are indicated at each branch point. B Multiple alignment of the amino-acid sequences of the HMG domain of SoxC genes. High consensus sequences (>90%) are shown in red, and low consensus sequences (>50%) are in blue; hs, Homo sapiens; mm, Mus musculus; dr, Danio rerio.
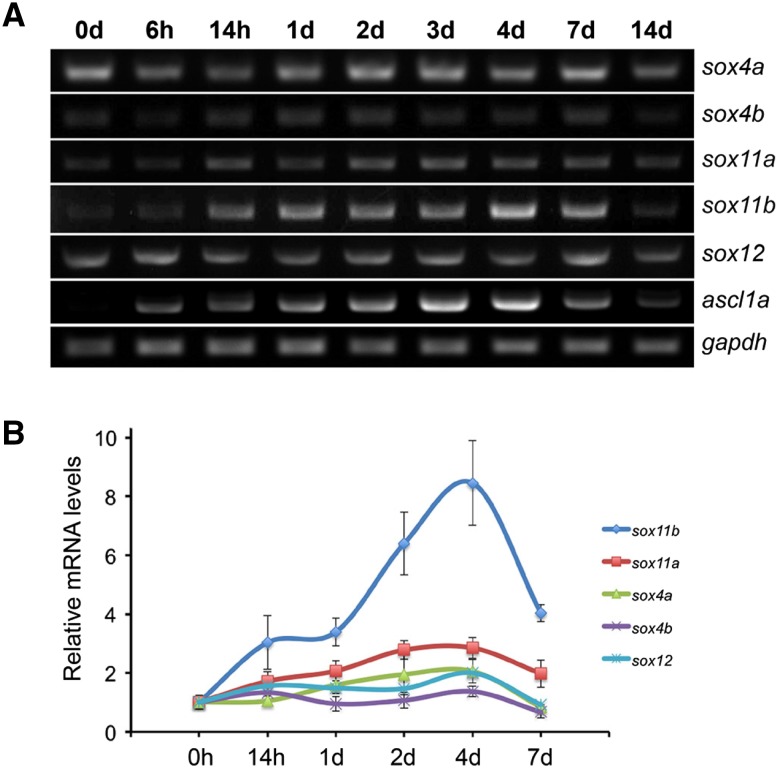
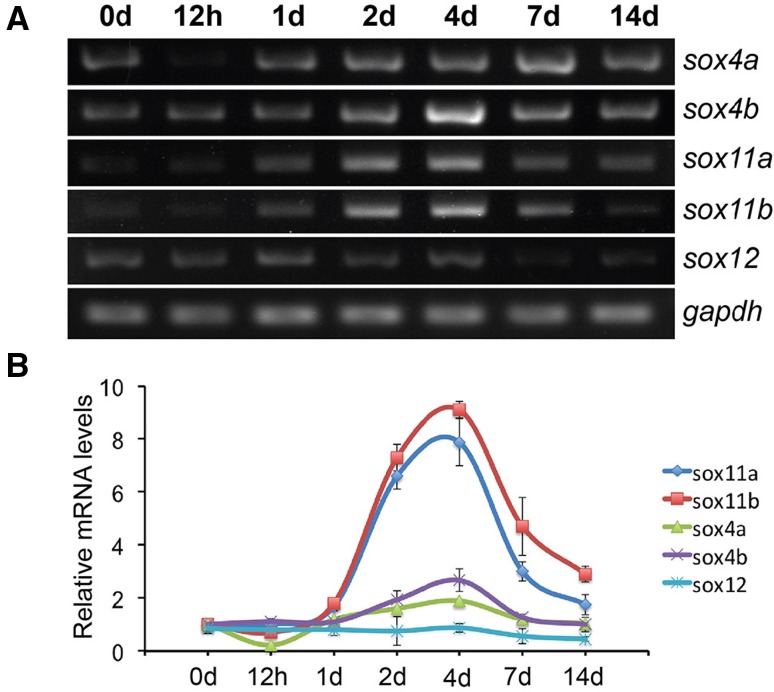
To investigate the temporal expression of zebrafish SoxC genes after retinal injury, retinas were collected at different time points after injury and the total RNA was extracted. RT-PCR results showed that among the five SoxC genes, only sox11a and sox11b were induced after the injury (Fig. 2A). Specifically, sox11a was weakly induced from 14 h post-injury (hpi) to 7 days post-injury (dpi) and declined at 14 dpi (Fig. 2A). The expression of sox11b was up-regulated at 14 hpi, reached a peak at 4 dpi, then declined and returned to near the basal level from 7 to 14 dpi (Fig. 2A). Although the transcripts of other SoxC genes were also detected, they showed no evident changes after injury (Fig. 2A). Interestingly, the expression pattern of sox11b following injury was similar to that of ascl1a (Fig. 2A), which is a key transcription factor regulating retinal regeneration [8, 10]. We next quantified the expression levels of the SoxC genes by qPCR analysis. At 4 dpi, qPCR detected a 2.8-fold increase of sox11a and an 8.4-fold increase of sox11b mRNA in the retina (Fig. 2B). The fold changes of other SoxC genes were no more than 2 (Fig. 2B). These results indicate that Sox11b is the major SoxC member induced in the retina after needle-poke injury.
Fig. 2.
Temporal expression of zebrafish SoxC genes during retinal regeneration. A PCR results of the expression of zebrafish SoxC genes (sox4a, sox4b, sox11a, sox11b, and sox12) in the whole retina at the indicated time points. The pro-neural gene ascl1a served as a positive control for retinal regeneration, and gapdh as a loading control. B qPCR analysis of the expression of zebrafish SoxC genes after retinal injury at indicated time points (n = 3 biological replicates).
Sox11b is Expressed in BrdU+ Cells in the INL After Needle-Poke Injury
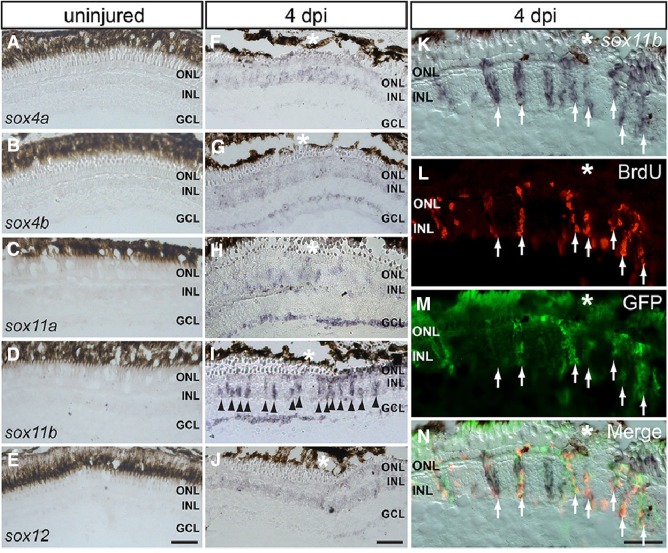
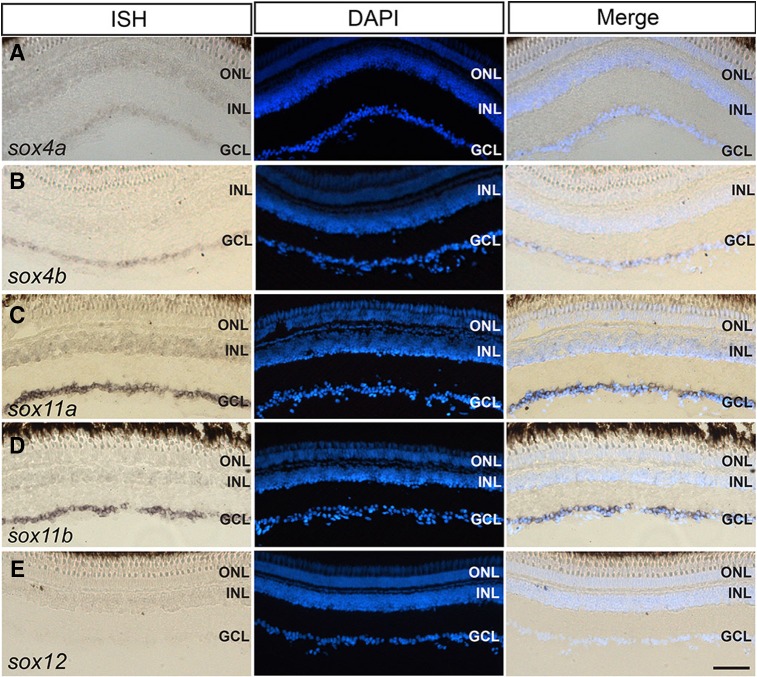
To determine where the SoxC genes are expressed in the intact and injured retina, ISH was carried out on retinal cryosections at 0 and 4 dpi. None of the SoxC genes showed a detectable signal in the uninjured retina (Fig. 3A–E), while very weak signals were found for sox4a, sox4b, and sox12 at the injury site at 4 dpi (Fig. 3F, G, J). Interestingly, sox11a was only weakly induced in the INL but showed a stronger signal in the ganglion cell layer (GCL) around the injury site at 4 dpi (Fig. 3H), suggesting that this gene plays a more important role during optic nerve regeneration than during retinal regeneration. Consistent with the RT-PCR and qPCR results, sox11b displayed a strong in situ signal both in the INL and the GCL at the injury site at 4 dpi (Fig. 3I). These results suggest that Sox11b, rather than Sox11a, plays an important role during retinal regeneration in adult zebrafish.
Fig. 3.
Spatial expression patterns of SoxC genes in the injured and intact retina. A–E In situ hybridization (ISH) of all SoxC genes on cryosections of the uninjured retina. F–J ISH showing the expression of SoxC genes in the injured retina at 4 dpi. K–N ISH of sox11b combined with BrdU/GFP immunofluorescence showing its expression in BrdU+/GFP+ cells in the INL at 4 dpi (arrows, co-localization of in situ, GFP and BrdU signals; *injury site; ONL outer nuclear layer, INL inner nuclear layer, GCL ganglion cell layer; scale bars 50 μm).
It has been shown that almost all BrdU+ cells in the INL at 4 dpi are derived from Müller glia and are multipotent progenitor cells [25, 29]. BrdU+ cells in the INL at 4 dpi thus represent injury-induced Müller glia-derived progenitor cells (MGPCs). By using the transgenic line Tg(1016tuba1a:GFP), in which GFP is specifically induced in de-differentiated Müller glia and MGPCs after injury [25], we found that sox11b mRNA co-localized with BrdU and GFP signals in the INL at 4 dpi (Fig. 3K–N, arrows), suggesting that sox11b is likely expressed in proliferating MGPCs after retinal injury.
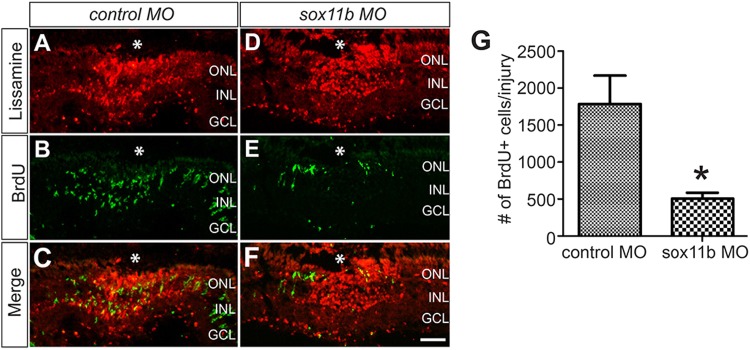
Sox11b Knockdown Decreases the Number of BrdU+ Cells in the INL at 4 dpi
To investigate whether Sox11b is necessary for retinal regeneration, we knocked it down by electroporation of lissamine-tagged control or sox11b antisense MOs into the retina at the time of injury. It is important to note that the efficacy of the sox11a and sox11b MOs used in this study has been described and validated in a previous paper [27]. Cryosections of the electroporated retina showed strong red fluorescence at the injury site at 4 dpi (Fig. 4A, D), indicating that the lissamine-tagged control and sox11b MOs were efficiently transferred into the retina. Strikingly, knockdown of Sox11b resulted in a significantly reduced number of BrdU+ cells in the INL at the injury site compared to the control at 4 dpi (Fig. 4B–G). This suggests that Sox11b plays an important role during retinal regeneration in adult zebrafish.
Fig. 4.
Sox11b knockdown decreased the number of BrdU+ cells in the INL. A–F MO-mediated Sox11b-knockdown reduced the number of BrdU+ cells (green) in the INL at 4 dpi. Successful electroporation of lissamine-tagged MOs into the retina was confirmed by red fluorescence at the injury site (*injury site; scale bar 50 μm). G Quantification of the number of BrdU+ cells in the INL at the injury site (n = 9 biological replicates; *P < 0.05; MO morpholino, ONL outer nuclear layer, INL inner nuclear layer, GCL ganglion cell layer).
Expression of SoxC Genes During Zebrafish Optic Nerve Regeneration
The induction of sox11a and sox11b in the GCL after a needle-poke injury that damaged all the retinal layers in a small region (Fig. 2) suggested that SoxC genes may be induced in the regenerating retinal ganglion cells after optic nerve transection. We therefore performed PCR and qPCR to examine the temporal expression of SoxC genes in the uninjured and optic nerve-lesioned retina. Indeed, PCR and qPCR revealed a strong upregulation of sox11a and sox11b after the injury, whereas other SoxC members showed no induction except for sox4b, which displayed a small induction at 4 dpi (Fig. 5A, B). Specifically, sox11a and sox11b were both upregulated at 1 dpi, showed strong induction during 2–7 dpi, and then returned close to the basal level at 14 dpi (Fig. 5A, B). At 4 dpi, qPCR detected a 7.9-fold increase of sox11a and a 9.1-fold increase of sox11b (Fig. 5B). ISH results showed that at 4 dpi, sox4a and sox4b were weakly expressed in the GCL (Fig. 6A, B), and sox12 was undetectable (Fig. 6E). In contrast, sox11a and sox11b were strongly induced in the GCL (Fig. 6C, D), suggesting that they play roles in optic nerve regeneration.
Fig. 5.
Temporal expression of SoxC genes after optic nerve transection. A PCR results of the expression of zebrafish SoxC genes (sox4a, sox4b, sox11a, sox11b, and sox12) in the whole retina at indicated time points (gapdh was used as a loading control). B qPCR analysis of the expression of zebrafish SoxC genes after optic nerve injury at indicated time points (n = 3 biological replicates).
Fig. 6.
ISH of zebrafish SoxC genes combined with DAPI staining on cryosections of the injured retina at 4 days after optic nerve injury. Weak in situ signals of sox4a (A) and sox4b (B) were detected in the GCL. Strong in situ signals of sox11a (C) and sox11b (D) were detected in the GCL, and no sox12 signal was detectable (E) (scale bar 50 μm).
Sox11a and Sox11b are Required for Optic Axon Regeneration
To investigate whether Sox11a and Sox11b are required for optic nerve regeneration in adult zebrafish, we combined in vivo MO-mediated protein knockdown in RGCs with retina explants to assay RGC axon regeneration [13]. Retinas were isolated 4 days after optic nerve lesion and MO treatment, diced, and cultured as explants for 4 days. RGC axon outgrowth was then quantified as previously described [28]. Our results showed that explants prepared from control MO-treated retinas displayed robust axon outgrowth (Fig. 7A). Separate knockdown of Sox11a or Sox11b had no significant effect on optic axon regrowth compared to control (Fig. 7A–C, E, F). Sox11a+11b knockdown slightly reduced the average length of regenerated axons but the difference was not significant (Fig. 7A, D, E). However, the average number of axons per explant from Sox11a+11b MO-treated retinas was significantly lower than that of control (Fig. 7A, D, F). The results suggest that Sox11a and Sox11b play important roles during optic nerve regeneration in vivo.
Fig. 7.
MO-mediated Sox11a+11b knockdown suppressed optic axon regrowth in explants. A–D Representative images of retinal explants showing the outgrowth of regenerated RGC axons from control (A) and Sox11a/11b MO-treated retinas (B–D). Retinas were isolated 4 days after optic nerve transection, diced, and cultured for another 4 days before analysis of neurite outgrowth. The red fluorescence is from lissamine-tagged control or Sox11a/11b MOs (scale bar, 200 μm). E, F Quantification of axon length and density (mean ± SD, n = 6 individual fish; con-MO, control MO; **P < 0.01 compared to control).
Discussion
SoxC transcription factors play important roles during vertebrate ocular morphogenesis and retinal development. However, their expression and function in retinal and optic nerve regeneration remain elusive. In this report, we showed that among the five SoxC genes, only sox11b was strongly induced in BrdU+ cells in the INL after retinal injury, and was required for retinal regeneration in zebrafish. During optic nerve regeneration, both sox11a and sox11b were upregulated in RGCs and were required for optic axon regrowth. Our study thus uncovered the expression pattern of SoxC genes and suggested their important roles during zebrafish retinal and optic nerve regeneration.
During ocular and retinal development, Sox4 and Sox11 showed partially-overlapping expression patterns and redundant functions [23]. In mice, Sox4 and Sox11 are both widely expressed in the neuroblast layer and GCL of the developing retina and share a similar expression pattern [24]. During zebrafish embryonic development, Sox4 (Sox4a/4b) and Sox11 (Sox11a/11b) are also strongly expressed in the developing retina from 24 to 72 h post-fertilization [21, 30]. Because tissue regeneration following injury or disease is often similar to embryonic development, we hypothesized that both Sox4 and Sox11 would be strongly expressed in the regenerating retina. Surprisingly, our results demonstrated that only Sox11 (Sox11a/11b) was strongly induced during retinal and optic nerve regeneration. In contrast, Sox4 (Sox4a/4b) and Sox12 were either not expressed or expressed very weakly after retinal and optic nerve injury. Why Sox11 is the only SoxC gene induced during regeneration is still unknown. As Sox4 and Sox11 are functionally redundant, it is possible that expression of Sox11 itself is sufficient to regenerate a damaged retina. Our finding is therefore a good example showing that although development and regeneration are similar, they still differ in many ways.
Sox11 plays important roles during ocular morphogenesis and retinal development. In zebrafish, MO-mediated Sox11 knockdown results in delayed and abnormal lens formation, coloboma, and reduced rod photoreceptors due to elevated Hedgehog signaling [21]. In mice, Sox11 conditional knock-out (KO) leads to a moderate reduction of RGC generation in the retina [24]. In another study, Sox11 KO mice showed delayed neurogenesis and differentiation of RGCs and cone photoreceptors [31]. These studies demonstrate that Sox11 is an important transcription factor regulating retinal neurogenesis during embryonic development. Here, we showed that after retinal injury, Sox11b was rapidly induced in the retina and was expressed in BrdU+/GFP+ cells (presumably MGPCs) in the INL (Figs. 2 and 3). In addition, MO-mediated Sox11b knockdown significantly decreased the number of proliferating cells in the INL at the injury site, suggesting that Sox11b is essential for MGPC formation and retinal regeneration. The molecular mechanism underlying the function of Sox11b in retinal regeneration is currently unknown and requires further investigation. Since Sox11 regulates cell proliferation [32], it is possible that Sox11b promotes the proliferation of Müller glia and MGPCs during retinal regeneration. It is also possible that Sox11b is required in early retinal regeneration, such as Müller glia reprogramming and de-differentiation.
Sox11 is widely expressed in the developing CNS and peripheral nervous system (PNS) neurons during periods of axon growth and is then down-regulated in the adult stage [32]. In response to injury, Sox11 is rapidly induced in peripheral neurons and promotes axon regeneration [33–36]. However, Sox11 induction is missing after injury in the mammalian CNS where regeneration does not occur [37, 38]. Recently, it was reported that forced expression of Sox11 in mouse corticospinal tract (CST) neurons promotes CST regeneration after spinal injury [38]. Therefore, Sox11 may function as a key transcription factor that enhances the intrinsic regenerative capacity of CNS neurons. Consistently, we showed that Sox11a and Sox11b were strongly induced in RGCs following optic nerve transection in adult zebrafish, and were required for optic axon regrowth in a retinal explant assay. The phenotype of axon regrowth after Sox11a/11b knockdown was moderate and this may be due to the presence of other SoxC genes in RGCs such as Sox4a and Sox4b. Our result differs from a previous report showing that Sox11a/11b are not required for RGC axon regrowth in adult zebrafish [27]. We speculate that the difference might be due to the different fish lines used, the conditions for retinal explants, and the amounts of MOs retained in the RGCs after gelfoam treatment. Further in vivo experiments, particularly functional studies, are required to clarify the roles of Sox11a/11b in RGCs after optic nerve injury.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (31401234), Natural Science Foundation of Jiangsu Province, China (BK20140428), the Basic Research Program of Education Department of Jiangsu Province, China (14KJB180019), and the Science and Technology Project of Nantong Municipality, Jiangsu Province, China (MS22015002).
Footnotes
Zhaoxia Mu and Shuqiang Zhang have contributed equally to this work.
Contributor Information
Nan Hu, Email: hunaneye@hotmail.com.
Hui Xu, Email: huixu82@126.com.
References
- 1.Kizil C, Kaslin J, Kroehne V, Brand M. Adult neurogenesis and brain regeneration in zebrafish. Dev Neurobiol. 2012;72:429–461. doi: 10.1002/dneu.20918. [DOI] [PubMed] [Google Scholar]
- 2.Becker CG, Becker T. Adult zebrafish as a model for successful central nervous system regeneration. Restor Neurol Neurosci. 2008;26:71–80. [PubMed] [Google Scholar]
- 3.Goldman D. Muller glial cell reprogramming and retina regeneration. Nat Rev Neurosci. 2014;15:431–442. doi: 10.1038/nrn3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenkowski JR, Raymond PA. Muller glia: Stem cells for generation and regeneration of retinal neurons in teleost fish. Prog Retin Eye Res. 2014;40:94–123. doi: 10.1016/j.preteyeres.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lenkowski JR, Qin Z, Sifuentes CJ, Thummel R, Soto CM, Moens CB, et al. Retinal regeneration in adult zebrafish requires regulation of TGFbeta signaling. Glia. 2013;61:1687–1697. doi: 10.1002/glia.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson CM, Ackerman KM, O’Hayer P, Bailey TJ, Gorsuch RA, Hyde DR. Tumor necrosis factor-alpha is produced by dying retinal neurons and is required for Muller glia proliferation during zebrafish retinal regeneration. J Neurosci. 2013;33:6524–6539. doi: 10.1523/JNEUROSCI.3838-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin Z, Kidd AR, 3rd, Thomas JL, Poss KD, Hyde DR, Raymond PA, et al. FGF signaling regulates rod photoreceptor cell maintenance and regeneration in zebrafish. Exp Eye Res. 2011;93:726–734. doi: 10.1016/j.exer.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramachandran R, Zhao XF, Goldman D. Ascl1a/Dkk/beta-catenin signaling pathway is necessary and glycogen synthase kinase-3beta inhibition is sufficient for zebrafish retina regeneration. Proc Natl Acad Sci U S A. 2011;108:15858–15863. doi: 10.1073/pnas.1107220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan J, Ramachandran R, Goldman D. HB-EGF is necessary and sufficient for Muller glia dedifferentiation and retina regeneration. Dev Cell. 2012;22:334–347. doi: 10.1016/j.devcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramachandran R, Zhao XF, Goldman D. Insm1a-mediated gene repression is essential for the formation and differentiation of Muller glia-derived progenitors in the injured retina. Nat Cell Biol. 2012;14:1013–1023. doi: 10.1038/ncb2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thummel R, Enright JM, Kassen SC, Montgomery JE, Bailey TJ, Hyde DR. Pax6a and Pax6b are required at different points in neuronal progenitor cell proliferation during zebrafish photoreceptor regeneration. Exp Eye Res. 2010;90:572–582. doi: 10.1016/j.exer.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsaeidi F, Bemben MA, Zhao XF, Goldman D. Jak/Stat signaling stimulates zebrafish optic nerve regeneration and overcomes the inhibitory actions of Socs3 and Sfpq. J Neurosci. 2014;34:2632–2644. doi: 10.1523/JNEUROSCI.3898-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleisch VC, Fraser B, Allison WT. Investigating regeneration and functional integration of CNS neurons: lessons from zebrafish genetics and other fish species. Biochim Biophys Acta. 2011;1812:364–380. doi: 10.1016/j.bbadis.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/S1534-5807(02)00223-X. [DOI] [PubMed] [Google Scholar]
- 16.Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins–Sox4, Sox11 and Sox12–exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoser M, Potzner MR, Koch JM, Bosl MR, Wegner M, Sock E. Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol Cell Biol. 2008;28:4675–4687. doi: 10.1128/MCB.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergsland M, Ramskold D, Zaouter C, Klum S, Sandberg R, Muhr J. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25:2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattaram P, Penzo-Mendez A, Sock E, Colmenares C, Kaneko KJ, Vassilev A, et al. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat Commun. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cizelsky W, Hempel A, Metzig M, Tao S, Hollemann T, Kuhl M, et al. sox4 and sox11 function during Xenopus laevis eye development. PLoS One. 2013;8:e69372. doi: 10.1371/journal.pone.0069372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pillai-Kastoori L, Wen W, Wilson SG, Strachan E, Lo-Castro A, Fichera M, et al. Sox11 is required to maintain proper levels of Hedgehog signaling during vertebrate ocular morphogenesis. PLoS Genet. 2014;10:e1004491. doi: 10.1371/journal.pgen.1004491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Usui A, Iwagawa T, Mochizuki Y, Iida A, Wegner M, Murakami A, et al. Expression of Sox4 and Sox11 is regulated by multiple mechanisms during retinal development. FEBS Lett. 2013;587:358–363. doi: 10.1016/j.febslet.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 23.Pillai-Kastoori L, Wen W, Morris AC. Keeping an eye on SOXC proteins. Dev Dyn. 2015;244:367–376. doi: 10.1002/dvdy.24235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y, Ding Q, Xie X, Libby RT, Lefebvre V, Gan L. Transcription factors SOX4 and SOX11 function redundantly to regulate the development of mouse retinal ganglion cells. J Biol Chem. 2013;288:18429–18438. doi: 10.1074/jbc.M113.478503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fausett BV, Goldman D. A role for alpha1 tubulin-expressing Muller glia in regeneration of the injured zebrafish retina. J Neurosci. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao XF, Wan J, Powell C, Ramachandran R, Myers MG, Jr, Goldman D. Leptin and IL-6 family cytokines synergize to stimulate Muller glia reprogramming and retina regeneration. Cell Rep. 2014;9:272–284. doi: 10.1016/j.celrep.2014.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312:596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 28.Veldman MB, Bemben MA, Goldman D. Tuba1a gene expression is regulated by KLF6/7 and is necessary for CNS development and regeneration in zebrafish. Mol Cell Neurosci. 2010;43:370–383. doi: 10.1016/j.mcn.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen W, Pillai-Kastoori L, Wilson SG, Morris AC. Sox4 regulates choroid fissure closure by limiting Hedgehog signaling during ocular morphogenesis. Dev Biol. 2015;399:139–153. doi: 10.1016/j.ydbio.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Usui A, Mochizuki Y, Iida A, Miyauchi E, Satoh S, Sock E, et al. The early retinal progenitor-expressed gene Sox11 regulates the timing of the differentiation of retinal cells. Development. 2013;140:740–750. doi: 10.1242/dev.090274. [DOI] [PubMed] [Google Scholar]
- 32.Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol. 2010;42:425–428. doi: 10.1016/j.biocel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jankowski MP, Cornuet PK, McIlwrath S, Koerber HR, Albers KM. SRY-box containing gene 11 (Sox11) transcription factor is required for neuron survival and neurite growth. Neuroscience. 2006;143:501–514. doi: 10.1016/j.neuroscience.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jankowski MP, McIlwrath SL, Jing X, Cornuet PK, Salerno KM, Koerber HR, et al. Sox11 transcription factor modulates peripheral nerve regeneration in adult mice. Brain Res. 2009;1256:43–54. doi: 10.1016/j.brainres.2008.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanabe K, Bonilla I, Winkles JA, Strittmatter SM. Fibroblast growth factor-inducible-14 is induced in axotomized neurons and promotes neurite outgrowth. J Neurosci. 2003;23:9675–9686. doi: 10.1523/JNEUROSCI.23-29-09675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jing X, Wang T, Huang S, Glorioso JC, Albers KM. The transcription factor Sox11 promotes nerve regeneration through activation of the regeneration-associated gene Sprr1a. Exp Neurol. 2012;233:221–232. doi: 10.1016/j.expneurol.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore DL, Goldberg JL. Multiple transcription factor families regulate axon growth and regeneration. Dev Neurobiol. 2011;71:1186–1211. doi: 10.1002/dneu.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Reynolds A, Kirry A, Nienhaus C, Blackmore MG. Overexpression of Sox11 promotes corticospinal tract regeneration after spinal injury while interfering with functional recovery. J Neurosci. 2015;35:3139–3145. doi: 10.1523/JNEUROSCI.2832-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]