Abstract
Methane is the second most important greenhouse gas contributing to about 20% of global warming. Its mitigation is conducted by methane oxidizing bacteria that act as a biofilter using methane as their energy and carbon source. Since their first discovery in 1906, methanotrophs have been studied using a complementary array of methods. One of the most used molecular methods involves PCR amplification of the functional gene marker for the diagnostic of copper and iron containing particulate methane monooxygenase. To investigate the diversity of methanotrophs and to extend their possible molecular detection, we designed a new set of degenerate methane monooxygenase primers to target an 850 nucleotide long sequence stretch from pmoC to pmoA. The primers were based on all available full genomic pmoCAB operons. The newly designed primers were tested on various pure cultures, enrichment cultures and environmental samples using PCR. The results demonstrated that this primer set has the ability to correctly amplify the about 850 nucleotide long pmoCA product from Alphaproteobacteria, Gammaproteobacteria, Verrucomicrobia and the NC10 phyla methanotrophs. The new primer set will thus be a valuable tool to screen ecosystems and can be applied in conjunction with previously used pmoA primers to extend the diversity of currently known methane-oxidizing bacteria.
Electronic supplementary material
The online version of this article (doi:10.1186/s13568-017-0466-2) contains supplementary material, which is available to authorized users.
Keywords: Methane, Particulate methane monooxygenase, Diversity, Methanotroph, Genetic marker
Introduction
Methane is the second most important greenhouse gas contributing to about 20% of global warming (Intergovernmental Panel on Climate Change 2014). The global methane budget is estimated to be around 600 Tg a−1 (Dubey 2001) which is dominated by biogenic sources, where natural wetlands (23%), and rice fields (21%) (Frenzel 2000) account for almost half of the total budget (Chen and Prinn 2005). Methanogenic archaea are assumed to be the sole producers of methane that reside in these environments (Cicerone and Oremland 1988; Conrad et al. 1999; Joulian et al. 1997). These archaea are also present in waste treatment systems, intestines of ruminants and termites and landfills acting as additional CH4 sources. Therefore, microbial methanogenic activity is responsible for nearly 75% of the methane emitted to the atmosphere (Chen and Prinn 2005).
This process, is however, vastly mitigated by methanotrophic microorganisms that oxidize a large part of the produced CH4 (Cappelletti et al. 2016; Crevecoeur et al. 2015; Dumont and Murrell 2005; Reeburgh et al. 1993; Oshkin et al. 2014). It has been estimated that of the primary productivity, roughly 1% ends up in CH4; half of which is emitted into the atmosphere while the other half is consumed by methanotrophs (Reeburgh and Whjalen 2007; Aronson et al. 2013). While anaerobic methane-oxidizing archaea consume more than 75% of the CH4 produced in certain marine sediments (Reeburgh and Whjalen 2007; Beal et al. 2009; Egger et al. 2014), aerobic methane-oxidizing bacteria (MOB) that live at the interface between anoxic and oxic zones in marine environments (Bender and Conrad 1992; Lüke et al. 2016; Padilla et al. 2016), freshwater wetlands and rice fields (Lüke et al. 2014) have been estimated to consume up to 90% of the CH4 produced in these environments (Hanson and Hanson 1996). Alpha- and gammaproteobacterial methanotrophs have further been shown to be dominant methane consumers in acidic peatlands (Esson et al. 2016; Deng et al. 2013; Putkinen et al. 2014). Since their discovery over 100 years ago (Söhngen 1906), many aspects of methanotrophic bacteria have been studied (Whittenbury et al. 1970; Bédard and Knowles 1989; Hanson and Hanson 1996; Lidstrom 2006; Trotsenko and Murrell 2008). At the moment, several groups of aerobic bacteria are known that convert methane by means of a copper- and/or iron-containing enzyme called methane monooxygenase (MMO) (Murrell et al. 2000). Methanotrophic archaea play a prominent role in the anaerobic oxidation of methane and use methyl coenzyme-M reductase (MCR) (Knittel and Boetius 2009; Haroon et al. 2013; Welte et al. 2016).
Two different forms of MMO exist: a soluble MMO (sMMO) encoded by mmoX, mmoY and mmoZ and a particulate MMO encoded by pmoCAB (Lieberman and Rosenzweig 2005). The membrane bound particulate methane monooxygenase (pMMO) catalyzes the hydroxylation of methane. It exists in virtually all methanotrophs while sMMO has only been shown in certain genera such as Methylococcus, Methylosinus, Methylocystis, Methylomonas and Methylocella (Murrell et al. 2000). The more recent discovery of Methylocella silvestris (Crombie and Murrell, 2014), Methyloferula stellata (Dedysh et al. 2015), and Methylocella palustris (Dedysh et al. 2000) has illustrated that some MOB do indeed possess only sMMO and would not be targeted in pMMO-focused molecular studies (Dunfield et al. 2003; Dedysh et al. 2000; Vorobev et al. 2011; Vekeman et al. 2016a). pMMO belongs to the ammonia monooxygenase superfamily and has been shown to be of high biogeochemical and chemical relevance (Bédard and Knowles 1989; Hakemian and Rosenzweig 2007). This is due to the tight correlation that exists between this family and the globally important methane and nitrous oxide fluxes (Conrad 1996). This makes copper containing (Cu) MMO genes extremely useful markers in biological feedback studies looking at global climate change (Singh et al. 2010). Moreover, PCR-based environmental surveys have identified the ecological distribution and relevance of CuMMO-containing organisms correlated to gas flux, land use and climatic conditions (Coleman and the references within 2012). It has also been postulated that this group of enzymes could be correlated to processes other than methanotrophy and ammonia oxidation such as butane-oxidation (Coleman et al. 2012; Crombie and Murrell 2014). Therefore molecular approaches, such as PCR with specific primer sets are a fast and convenient method to screen for the diversity of such enzymes in various environments (Murrell et al. 1998; Mitsumori et al. 2002; Siljanen et al. 2012).
The crystal structure of pMMO has been determined to a resolution of 2.8 Å from Methylococcus capsulatus (Bath) and the enzyme has been found to be a trimer with an α3β3ɣ3 polypeptide arrangement (Lieberman and Rosenzweig 2005). The PmoA subunit contains non-heme iron in its center and for long was proposed to be the site of substrate hydroxylation. The soluble PmoB subunit hosts two metal centers, modelled as mononuclear copper and dinuclear copper, while a third metal center occupied by zinc is located within the membrane (Lieberman and Rosenzweig 2005).
Molecular surveys showed that MOB are present, amongst others, in natural wetlands (Costello et al. 2002; Samad and Bertilsson 2017), marine ecosystems (Vekeman et al. 2016b), permafrost thaw ponds (Crevecoeur et al. 2015), peatlands (Lau et al. 2015) and flooded rice-fields (Krüger et al. 2001; Lüke et al. 2009; Balasubramanian and Rosenzweig 2007; Zheng et al. 2008). Since pMMO was initially assumed to be present in all methane oxidizing bacteria, it has been used in molecular approaches to investigate methanotrophic diversity (Semrau et al. 1995; Holmes et al. 1999; Chi et al. 2012; Saidi-Mehrabad et al. 2013). More specifically pmoA, coding for the beta subunit of pMMO, was found to be highly conserved and as a result used as a functional gene marker (Holmes et al. 1995a, b; Bourne et al. 2001; Costello et al. 2002; Kolb et al. 2003; Luesken et al. 2011b; Wang et al. 2017).
In addition, pmoA amplicon pyrosequencing has been used to look at methanotrophic diversity in depth (Kip et al. 2011; Lüke and Frenzel 2011; Han and Gu 2013; Knief 2015). For all the PCR based methods, the used primers unfortunately do not encompass all different phyla of MOB to the same extent (Bergmann et al. 2011) nor do they cover new phyla such as Verrucomicrobia (Sharp et al. 2014; Erikstad and Birkeland 2015) and NC10. In the latter cases, more phylum specific primers had to be designed to investigate the presence of ‘Candidatus Methylomirabilis oxyfera’ in various ecosystems (Luesken et al. 2011b). Recently several genomes of different MOB have been sequenced by the Omega consortium (Khmelenina et al. 2013; Kits et al. 2013; Khadem et al. 2012; Stephenson et al. 2017) and thus a much larger gene dataset is now available to design new primers to potentially cover a larger methanotroph diversity. Here we introduce a new set of degenerate primers that can be used to examine the diversity of MOB in various environments with the potential ability to target all presently known methanotrophic phyla. The new primers have the capability to target pmoC and pmoA and the intergenic region in between those genes. Application of the primers to various ecosystem resulted in the detection of pmoCA of Alphaproteobacteria, Gammaproteobacteria, Verrucomicrobia and NC10 within their respective habitats. Neither ammonia oxidizers, nor the recently discovered comammox (van Kessel et al. 2015; Pjevac et al. 2016) were detected with these primers. Furthermore, since the binding sites of the primers immediately flank the intergenic region between the genes pmoC and pmoA, they generate MOB lineage specific fragments. This unique property could be used in high throughput sequence analysis experiments for recovering diverse lineages in further environmental studies.
Materials and methods
Construction of pmoCAB operon database and primer design
A total of 83 different full genomic methane monooxygenase along with the isoenzyme PXM and ammonia monooxygenase gene sequences available on MaGe were downloaded (Vallenet et al. 2006; Sievers and Higgins 2014). This included Alpha-, Gamma-, and Betaproteobacteria (AOB), Verrucomicrobia, NC10, Mycobacterium, Nocardia, SAR cluster, divergent PXM operon and second operons from Methylocystis parvus OBBP, Methylocystis sp. BN69, Methylosinus sp. LW3, and Methylosinus sp. LW4 (Table 1). The genes were aligned in pmoCAB operon configuration. In cases where an organism’s genome contained more than one copy of the operon, all copies were included in the pipeline. Sequences were aligned using MUSCLE (Edgar 2004) and the alignment was imported into ARB (Ludwig et al. 2004). Nucleotide sequences were translated into protein sequences and phylogenetic trees were constructed based on the amino acid sequences. Furthermore, using the ‘Probe’ tool, primers that were capable of covering all (or as much as possible) phyla were designed within ARB. The parameters for the primer design were: 18 nucleotides in length, GC content of 50–70%, and minimum group coverage of at least 50%. Furthermore, the primers were made specific to MOB so that they had more than five mismatches with ammonium monooxygenase amo gene sequences of ammonia oxidizing bacteria (AOB).
Table 1.
Aligment of the new pmoCA primers on all the available genomic sequences from different phyla. Wobble positions are shown in yellow
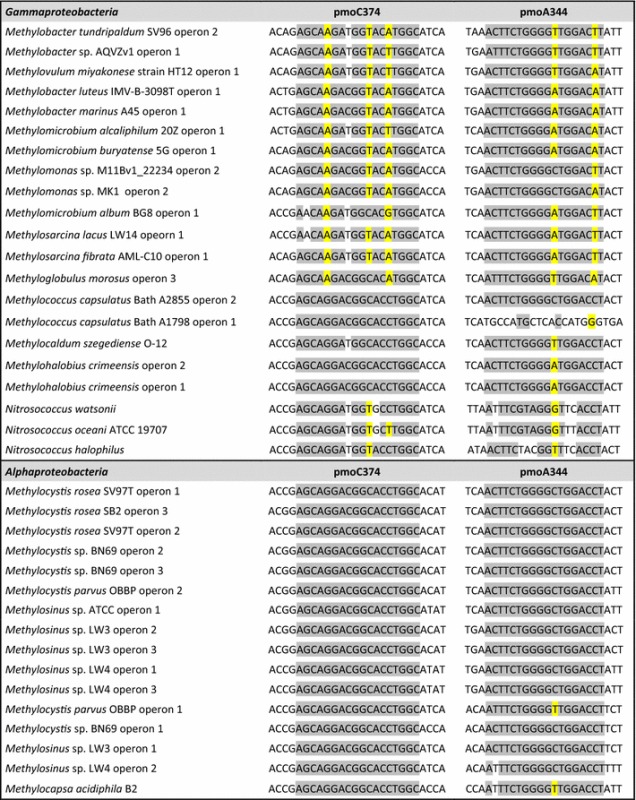
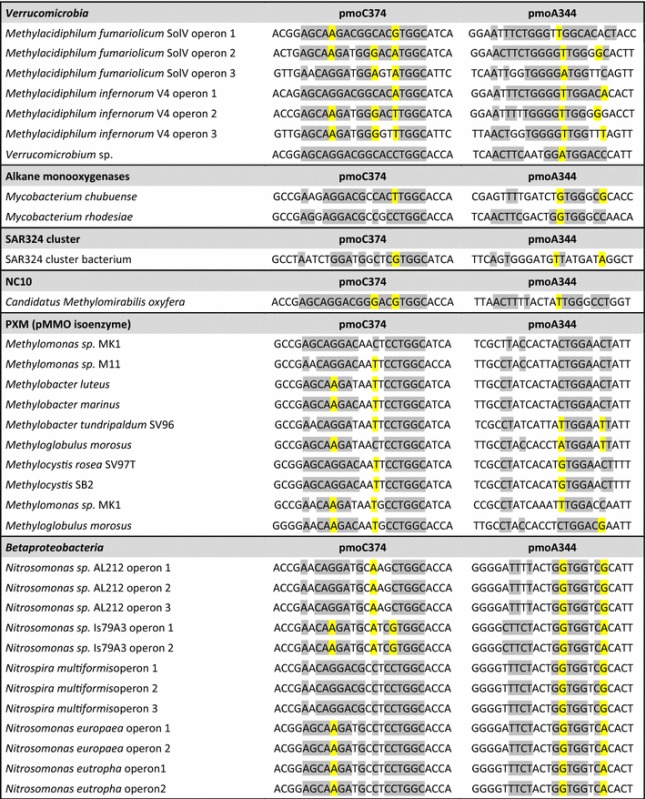
A set of primers covering pmoC, the intergenic region, and pmoA were ultimately designed (Table 2) and ordered from Biolegio (Nijmegen, the Netherlands). The forward primer, called pmoC374, with the reverse primer, called pmoA344 resulted in product length of roughly 850 base pairs (bp) (Table 3). There are slight variations between different lineages. This is caused by variation in on average, 120 bp long intergenic region between pmoC and pmoA.
Table 2.
Comparison of targeting ability between two newly designed degenerate primers and pmoA189
| Phylum | pmoC374 | pmoA344 | pmoA189 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mismatches | Mismatches | Mismatches | ||||||||||
| 0 | 1 | 2 | 3 | 0 | 1 | 2 | 3 | 0 | 1 | 2 | 3 | |
| Gammaproteobacteria | 10/18 | 18/18 | 18/18 | 18/18 | 16/18 | 18/18 | 18/18 | 18/18 | 7/18 | 18/18 | 18/18 | 18/18 |
| Alphaproteobacteria | 16/16 | 16/16 | 16/16 | 16/16 | 14/16 | 16/16 | 16/16 | 16/16 | 16/16 | 16/16 | 16/16 | 16/16 |
| Verrucomicrobia | 3/7 | 5/7 | 5/7 | 6/7 | 0/7 | 0/7 | 1/7 | 3/7 | 0/7 | 0/7 | 0/7 | 3/7 |
| NC10 | 1/1 | 1/1 | 1/1 | 1/1 | 0/1 | 0/1 | 0/1 | 0/1 | 1/1 | 1/1 | 1/1 | 1/1 |
Percent sequence coverage of all pmoCAB available sequences within each phylum were calculated by looking at how many sequences each primer could target. Targeting ability is also shown for zero, one, two and three mismatches within each primer
Table 3.
The new pMMO primers designed based on aligned pmoC, A, and B compared to pmoA189
| Primers | Sequence | MT | %GC |
|---|---|---|---|
| PmoC374 | 5′-AGCARGACGGYACNTGGC-3′ | 42,9 | 56 |
| PmoA189 | 5′-GGNGACTGGGACTTCTGG-3′ | 40,3 | 56 |
| PmoA344 | 5′-ANGTCCAHCCCCAGAAGT-3′ | 42,9 | 50 |
MT melting temperature, %GC GC content in percentage
DNA extraction and PCR conditions
Total DNA was extracted from methanotrophic pure and enrichment cultures and from various environmental samples. Table 4 provides an overview on the cultures and samples used in this study. DNA was extracted using the PowerSoil® DNA Isolation Kit from MO BIO Laboratories (Carlsbad CA, USA) following the protocol of the manufacturer. The primers were tested using polymerase chain reaction (PCR), gradient PCR, touchdown PCR and nested PCR on all of the samples. The optimized protocol consisted of initial denaturation step at 96 °C for 5 min, followed by 35 cycles at 96 °C for 1 min, annealing at 55 °C for 1 min and elongation at 72 °C for 2 min. The final elongation step was done for 10 min at 72 °C.
Table 4.
Over view of the strains, enrichment culture and environmental samples tested in this study to detect pmoCA gene sequences
| Name/sample | Description | Origin/location reference |
|---|---|---|
| Methylocystis rosea | Pure culture Alphaproteobacteria |
DSMZ 17621 |
| Methylosinus sporium | Pure culture Alphaproteobacteria |
DSMZ 17706 |
| Methylomonas lenta | Pure culture Gammaproteobacteria |
Hoefman et al. 2014 |
| Methyloacidimicrobium fagopyrum 3C | Pure culture Verrucomicrobia |
van Teeseling et al. 2014 |
| Methyloacidiphilum fumariolicum SolV | Pure culture Verrucomicrobia |
Pol et al. 2007 |
| Methylomirabilis oxyfera (DAMO) | Enrichment culture NC10 phylum |
Ooijpolder, NL Ettwig et al. 2008 |
| Sludge from waste water treatment plant (WW) | Environmental sample | Lieshout, NL Luesken et al. (2011a, b) |
| Bulk soil form paddy field (BS) | Environmental sample | Vercelli, Italy Vaksmaa et al. (2016) |
| Rhizosphere of rice plants (ROOT) | Environmental sample | Vercelli, Italy Vaksmaa et al. (2016) |
| Enrichment culture with paddy field soil (RV) | Enrichment culture | Vaksmaa et al. (2016) |
| Volcanic mud (VM) | Environmental sample | Campi Flegrei caldera, Italy Pol et al. (2014) |
Excision from gel after gel electrophoresis, purification, ligation and transformation of the amplified PCR products were done following the protocol described by Luesken et al. 2011a. At least 20 random clones were picked for each environmental sample in a blue-white screening and the plasmids were isolated for each PCR product with the GeneJet Miniprep Kit (Fermentas, Vilnus, Lithuania). The samples were sent to BaseClear (Leiden, the Netherlands) for sequencing of the cloned product using M13 forward primer (Luesken et al. 2011a).
Sequence analysis
The resulting sequences were checked for quality using Chromas Lite 2.1.1.0 (Technelysium Pty Ltd). Once erroneous sequences were removed, the results were blasted (BLASTx) using the publically available tools on National Center for Biotechnology Information (NCBI). Sequences matching with AMO superfamily were imported into ARB, translated into protein sequences and aligned to the previously mentioned pmoCAB operon dataset using ARB built-in aligner tools. Phylogenetic tree construction was performed on the amino acid alignment using maximum parsimony and maximum likelihood methods with bootstrapping of 100 times. Consensus sequences based on the fraction and frequency of residues at a specific alignment position within pmoC from all sequences were used to generate the tree.
Sequences are deposited in Genbank with Accession Numbers KY883458–KY883555 (Additional file 1: Table S1).
Results
The design of new primers was obtained by using all available pmoCAB operon sequences from MaGe. Interestingly, pmoB contained no conserved sequence stretch as a potential primer target site. Looking at the full operons, the only conserved regions resided within pmoC and pmoA. A new region at the nucleotide position 374 within the PmoC subunit of Methylococcus capsulatus (Bath), as a reference, was found to be highly conserved amongst all the phyla tested in this experiment. The forward primer binding site encodes for a glutamine residue at 126th base within the crystal structure of pmoC anchored to the membrane in Methylococcus capsulatus (Bath) whereas the reverse primer binding site encodes a phenylalanine residue at 107th base within pmoA. Our newly designed forward primer was compared to Holmes’ forward primer and the results are shown in Tables 1 and 2. As the tables illustrate, with zero mismatches, pmoC374 is able to target three out of the seven available sequences from Verrucomicrobia. If a single mismatch is allowed, five out of the seven sequences from Verrucomicrobia are targeted by pmoC374 whereas pmoA189 (Holmes et al. 1995a, b) with one mismatch still does not target any verrucomicrobial pMMO gene. The details of the novel primer set with regards to number of mismatches are listed in Table 2.
Initially, pmoA189 target region was thought to be a good matching reverse primer, however, a secondary conserved region at the 334th position within the pmoA gene was found. The pmoC374 with pmoA344 combination yielded a PCR product of the correct size in the samples tested, while the same could not always be observed when it was used in combination with pmoA189. In Table 1 and 2, it can be observed that pmoA344 has the ability to target 17 out of the 19 sequences belonging to Gammaproteobacteria with zero mismatches. Based on sequence information, pmoA334 does not have the ability to target NC10 phylum and it needs two or more mismatches to target species belonging to Verrucomicrobia. However, this primer improved the ability to target both Verrucomicrobia and the NC10 phyla in our study when pure isolates were used as positive control in the PCR reaction. The resulting sequences from the various enrichment cultures and environmental samples are depicted in Fig. 1.
Fig. 1.
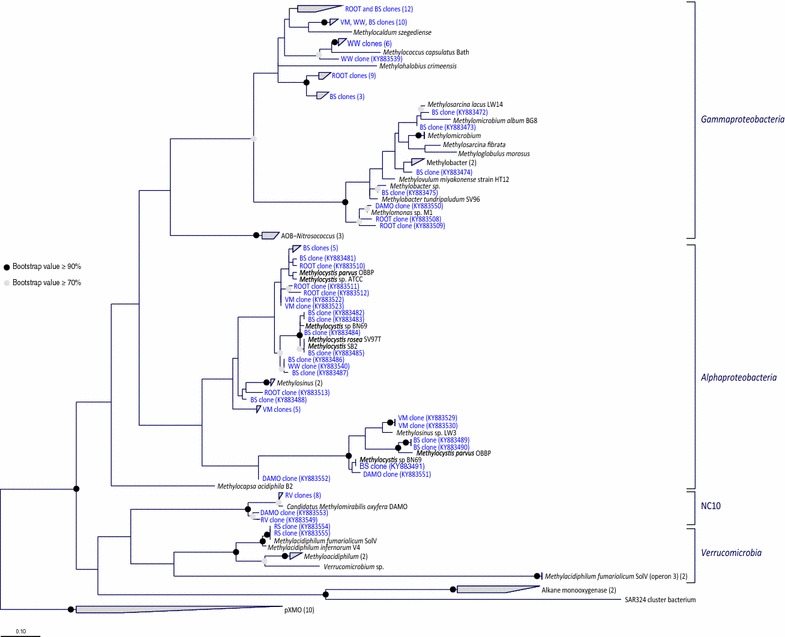
Representing available pMMO sequences including the sequence obtained in this study. The tree was constructed using consensus sequence, based on the fraction and frequency of residues at an alignment position chosen within pmoC using both ARB’s PHYML (amino Acids) tool within the maximum likelihood method and Phylip PROTPARS within the maximum parsimony method. Since the two trees were highly similar, only maximum likelihood is shown here. Due to size limitation, the tree is partially collapsed for an easier illustration and pXMO is used as the outgroup instead of AOB sequences that are omitted from this figure. The tree was built with 100 bootstraps and the ranges of values are shown with the respective colored circles at each node. Clone sequences with their respective accession numbers are highlighted in blue and the numbers in the brackets correspond to the number of sequences within a group. Gammaproteobacteria, Alphaproteobacteria, NC10 and Verrucomicrobia are clearly distinguished in the figure. Origin of clones: BS bulk soil, ROOT rhizosphere, VM volcanic mud, WW waste water sludge, RV bioreactor enrichment from vercelli, RS Methylacidiphilum fumariolicum SolV, DAMO Methylomirabilis oxyfera enrichment culture
The pmoCA sequences obtained from the paddy field sample were closely related to well-known genera including Methylosinus, Methylocystis, Methylococcus, Methylocaldum, Methylohalobius, Methylomicrobium, Methylobacter and Methylomonas. Furthermore, the pmoCA of pure cultures of Methylocystis rosea and Methylosinus sporium belonging to Alphaproteobacteria and Methylomonas lenta (Hoefman et al. 2014) belonging to Gammaproteobacteria could all be amplified with the new primer set. From previous studies, two isozymes of pMMO with various methane oxidation kinetics were found to be present in Methylocystis sp. strain SC2 (Baani and Liesack 2008), the new primers also amplified the second pmoCA in DNA extracted from the paddy soil. Methylocaldum- and Methylococcus-like species were also found in Waste Water samples. Furthermore, both alpha- and gammaproteobacterial pmoCA were found in the volcanic mud sample. Lastly, the pmoCA of the verrucomicrobial methanotroph Methylacidiphilum fumarolicum SolV could be amplified as well from a pure culture (Fig. 1).
In our experiment, only the Verrucomicrobia pMMO sequence most closely related to the ones in Alphaproteobacteria and Gammaproteobacteria could be detected. The new primer set was also used on a pure mesophilic Verrucomicrobia strain Methyloacidimicrobium fagopyrum 3C resulting in gene product of the correct size and gene sequence. The primers do not amplify sequences related to the pmoC3 group. In both anoxic enrichment cultures (DAMO and RV) tested, the pmoCA of NC10 phylum bacterium Methylomirabilis oxyfera could be amplified (Fig. 1). In the case of Methylomonas lenta that does contain the genes for pXMO, only pmoCA gene sequences were detected, while the pXMO was not amplified. Lastly, no AMO (ammonia monooxygenase), PXM (alternative methane monooxygenase) or the recently discovered comammox amo were targeted nor amplified with this primer set in any of the environmental samples or the negative controls used in this study.
Discussion
In the era of ‘omics’, molecular approaches using either specific or degenerate primers are still of high importance, especially in ecological studies where many samples need to be investigated or screened. They allow for a quick and robust detection of uncultivated microbes and aid in hypothesizing the community structure and the key processes that occur in certain environments, at the molecular level. As our knowledge and understanding of these environments expands, the tools that are used to investigate also need to be updated. More specifically, identification of the diverse organisms responsible for the oxidation of methane within various environments will help to better understand the key players involved in the methane cycle and evaluate their potential effectiveness as a biological methane filter. The currently available pmoA based primers are over 10 years old and since known MOB diversity has since been extended, a novel primer set with broader amplification ability would be highly beneficial in molecular studies. It is also important to distinguish between copper monooxygenases belonging to the AMO superfamily to ensure the detection of MOB and not AOB or the more recently discovered comammox (van Kessel et al. 2015; Pjevac et al. 2016; Pinto et al. 2015).
The use of all available pmoCAB operon sequences from MaGe allowed for the design of new primers (Table 1). Interestingly pmoB, which in previous studies has been suggested as the active site of the methane monooxygenase enzyme (Culpepper and Rosenzweig 2012; Lieberman and Rosenzweig 2005) contained no conserved sequence stretch as a potential primer target site. The only conserved regions that could be observed resided within pmoC and pmoA, both of which encode for primarily membrane bound subunits (Lieberman and Rosenzweig 2005). Overall, PmoA is by far the most conserved subunit of this enzyme. Since for long it was thought to be the catalytic subunit as well, primers were designed based on this gene and have since become the academic standard in this line of research and used to date in many studies (Lüke and Frenzel 2011; Rastogi et al. 2009; Kip et al. 2011). However, due to the two mismatches that occur at the 10th position within pmoA target region, previously unknown phyla (i.e. Verrucomicrobia or NC10) remain undetected and demand the design of phylum specific primers (Luesken et al. 2011b). This variation in sequence identity is also one of the reasons why this study focused on the whole pmoCAB operon instead of the PmoA subunit alone (Table 2).
Previous studies have looked into analysis of MOB community in rice fields by targeting 16S rRNA, pMMO and methanol dehydrogenase (Henckel et al. 1999) and observed a large variety of MOB. The new primer set used in this study was also able to detect a wide array of pmoCA sequences from both the bulk soil as well as the rhizosphere of an Italian rice paddy field, a waste water treatment sample, and volcanic mud samples. Further in anoxic Methylomirabilis oxyfera enrichment cultures started with paddy field or Ooijpolder sediment, many different pmoCA sequences could be retrieved (Fig. 1).
Furthermore, the pmoCA of the verrucomicrobial methanotroph Methylacidiphilum fumarolicum SolV could be amplified. This strain contains three complete pmoCAB operon structures that resemble the one observed in proteobacterial methanotrophs, plus a fourth pmoC copy. As expected, the primers do not amplify sequences related to the pmoC3 group as it is further downstream in the genome and the primers do not bind there.
Most sequences from the Waste Water Treatment Plant biomass used in this study were closely related to Methylococcus genus as was previously observed (Luesken et al. 2011a). Lastly, no AMO (ammonia monooxygenase), PXM (alternative methane monooxygenase) or the recently discovered commamox amo were targeted nor amplified with this primer set in any of the environmental samples which is an indication of the specificity. However, with some modification of the primer sequence, the same or similar sites can be used to only target AOB instead of MOB (Pjevac et al. 2016; Wang et al. 2017).
This study illustrates that when primer pmoC374 was used in combination with pmoA344, PCR amplification yielded the correct gene product from various environmental samples and MOB strains. Such observation could not be made when pmoA189 was used as the reverse primer. At times, there were multiple bands that occurred at the expected size within the gel. When each band was excised from the gel, all corresponded to the correct product. Since the pmoCA sequence covers the intergenic region, the slightly different nucleotide length observed in the PCR product is possibly due to the variation that exists in this region. This was more apparent when environmental samples were used as opposed to pure isolates, which further supports our hypothesis.
The obtained results expand our knowledge with regard to primer target ability based solely on in silico coverage as supposed to experimental results, since the new targeting sites would not be desirable due to occurring mismatches. Furthermore, the new pMMO primer set was able to amplify the correct product and sequence from all currently known methanotrophic phyla. If used in conjunction with Holmes’ forward primer, the resulting product could be used in future next generation sequencing studies for a more extensive look at the bacterial community structure. The concurrent use of this primer set along with ones based solely on pmoA would allow for a much lesser bias when it comes to studies that look at the general diversity of the methanotrophic community within various environments. It also permits for the simultaneous detection of Alphaproteobacteria, Gammaproteobacteria, Verrucomicrobia and NC10 phyla with broader sequence variation.
Authors’ contributions
CL conceived of the study. MG performed all experiments, computational analysis, and wrote the paper with input from CL and MSMJ. All authors read and approved the final manuscript.
Acknowledgements
We thank our colleagues Simon Guerrero, Sepehr Mohammady, Annika Vaksmaa, Huub Op den Camp, Sebastian Luecker and Harry Harhangi for generous sharing of their samples and discussions.
Competing interests
All authors declare that they have no competing interests.
Availability of data and materials
Sequences are deposited in Genbank with Accession Numbers KY883458–KY883555 (Additional file 1: Table S1). The data will be publically available as of 1 April 2018.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This study was funded by ERC Advanced Grant EcoMOM 339880.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- MOB
methane oxidizing bacteria
- MMO
methane monooxygenase
- MCR
methyl coenzyme-M reductase
- sMMO
soluble methane monooxygenase
- pMMO
particulate methane monooxygenase
- CuMMO
copper containing methane monooxygenase
- PCR
polymerase chain reaction
- rRNA
ribosomal ribonucleic acid
Additional file
Additional file 1: Table S1. Sequences belonging to each environmental samples and their respective accession numbers from Genbank.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13568-017-0466-2) contains supplementary material, which is available to authorized users.
Contributor Information
Mohammad Ghashghavi, Email: mghash@science.ru.nl.
Mike S. M. Jetten, Email: m.jetten@science.ru.nl
Claudia Lüke, Email: c.lueke@science.ru.nl.
References
- Aronson EL, Allison SD, Helliker BR. Environmental impacts on the diversity of methane-cycling microbes and their resultant function. Front Microbiol. 2013;4:225. doi: 10.3389/fmicb.2013.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baani M, Liesack W. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc Natl Acad Sci. 2008;105(29):10203–10208. doi: 10.1073/pnas.0702643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R, Rosenzweig AC. Structural and mechanistic insights into methane oxidation by particulate methane monooxygenase. Acc Chem Res. 2007;40(7):573–580. doi: 10.1021/ar700004s. [DOI] [PubMed] [Google Scholar]
- Beal EJ, House CH, Orphan VJ. Managense- and iron- dependent marine methane oxidation. Science. 2009;325(5937):184–187. doi: 10.1126/science.1169984. [DOI] [PubMed] [Google Scholar]
- Bédard CH, Knowles RO. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Rev. 1989;53(1):68–84. doi: 10.1128/mr.53.1.68-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender M, Conrad R. Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol Ecol. 1992;101:261–270. doi: 10.1111/j.1574-6941.1992.tb01663.x. [DOI] [Google Scholar]
- Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, Fierer N. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem. 2011;43(7):1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne DG, McDonald IR, Murrell JC. Comparison of pmoA PCR primer sets as tools for investigating methanotroph diversity in three danish soils. Appl Environ Microbiol. 2001;67(9):3802–3809. doi: 10.1128/AEM.67.9.3802-3809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelletti M, Ghezzi D, Zannoni D, Capaccioni B, Fedi S. Diversity of methane oxidizing bacteria in soils from “Hot Lands of Medolla” (Italy) featured by anomalous high-temperatures and biogenic CO2 emissions. Microbes Environ. 2016;4:369–377. doi: 10.1264/jsme2.ME16087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Prinn RG. Atmospheric modeling of high- and low-frequency methane observations: importance of interannually varying transport. J Geophys Res. 2005;110:D10303. doi: 10.1029/2004JD005542. [DOI] [Google Scholar]
- Chi Z, Lu W, Wang H, Zhao Y. Diversity of methanotrophs in a simulated modified biocover reactor. J Environ Sci (China) 2012;6:1076–1082. doi: 10.1016/S1001-0742(11)60889-9. [DOI] [PubMed] [Google Scholar]
- Cicerone RJ, Oremland RS. Biogeochemical aspects of atmospheric methane. Global Biogeochem Cycl. 1988;2:299–327. doi: 10.1029/GB002i004p00299. [DOI] [Google Scholar]
- Coleman NV, Le NB, Ly MA, Ogawa HE, McCarl V, Wilson NL, Holmes AJ. Hydrocarbon monooxygenase in Mycobacterium: recombinant expression of a member of the ammonia monooxygenase superfamily. ISME J. 2012;6:171–182. doi: 10.1038/ismej.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO. Microbiol Rev. 1996;60(4):609–640. doi: 10.1128/mr.60.4.609-640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad ME, Templeton AS, Daley PF, Alvarez-Cohen L. Seasonally-induced fluctuations in microbial production and consumption of methane during bioremediation of aged subsurface refinery contamination. Environ Sci Technol. 1999;33:4061–4068. doi: 10.1021/es990582o. [DOI] [Google Scholar]
- Costello AM, Auman AJ, Macalady JL, Scow KM, Lidstrom ME. Estimation of methanotroph abundance in a freshwater lake sediment. Environ Micro. 2002;8:443–450. doi: 10.1046/j.1462-2920.2002.00318.x. [DOI] [PubMed] [Google Scholar]
- Crevecoeur S, Vincent WF, Comte J, Lovejoy C. Bacterial community structure across environmental gradients in permafrost thaw ponds: methanotroph-rich ecosystems. Front Microbiol. 2015;6:192. doi: 10.3389/fmicb.2015.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombie AT, Murrell CJ. Trace-gas metabolic versatility of the facultative methanotroph Methylocella Silvestris. Nature. 2014;510:148–151. doi: 10.1038/nature13192. [DOI] [PubMed] [Google Scholar]
- Culpepper MA, Rosenzweig AC. Architecture and active site of particulate methane monooxygenase. Crit: Rev Biochem Mol Biol; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedysh SN, Liesack W, Khmelenina VN, Suzina NE, Trotsenko YA, Semrau JD, Bares AM, Panikov NS, Tiedje JM. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int J Syst Evol Microbiol. 2000;50:955–969. doi: 10.1099/00207713-50-3-955. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Naumoff DG, Vorobev AV, Kyrpides N, Woyke T, Shapiro N, Crombie AT, Murrell C, Kalyuzhnaya MG, Smirnova AV, Dunfield PF. Draft genome sequence of Methyloferula stellata AR4, an obligate methanotroph possessing only a soluble methane monooxygenase. Genom Announc. 2015 doi: 10.1128/genomeA.01555-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Cui X, Lüke C, Dumont MG. Aerobic methanotroph diversity in riganqiao peatlands on the Qinghai-Tibetan Plateau. Environ Microbiol Rep. 2013;4:566–574. doi: 10.1111/1758-2229.12046. [DOI] [PubMed] [Google Scholar]
- Dubey SK. Methane emission and rice agriculture. Curr Sci. 2001;81(4):345–346. [Google Scholar]
- Dumont MG, Murrell JC. Community-level analysis: key genes of aerobic methane oxidation. Methods Enzymol. 2005;397:413–427. doi: 10.1016/S0076-6879(05)97025-0. [DOI] [PubMed] [Google Scholar]
- Dunfield PF, Khmelenina VN, Suzina NE, Trotsenko YA, Dedysh SN. Methylocella silvestris sp. nov., a novel methanotroph isolated from an acidic forest cambisol. Int J Syst Evol Microbiol. 2003;53:1231–1239. doi: 10.1099/ijs.0.02481-0. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;5:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M, Rasigraf O, Sapart CJ, Jilbert T, Jetten MSM, Rockmann T, van der Veen C, Banda N, Kartal B, Ettwig KF, Slomp C. Iron-mediated anaerobic oxidation of methane in brackish coastal sediments. Environ Sci Technol. 2014;49:277–283. doi: 10.1021/es503663z. [DOI] [PubMed] [Google Scholar]
- Erikstad HA, Birkeland NK. Draft Genome Sequence of “Candidatus Methylacidiphilum kamchatkense” strain Kam1, a thermoacidophilic methanotrophic verrucomicrobium. Genome Announc. 2015;3(2):pii: e00065–15. doi: 10.1128/genomeA.00065-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esson KC, Lin X, Kumaresan D, Chanton JP, Murrell JC, Kostka JE. Alpha- and gammaproteobacterial methanotrophs codominate the active methane-oxidizing communities in an acidic boreal peat bog. Appl Environ Microbiol. 2016;82(8):2363–2371. doi: 10.1128/AEM.03640-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, Shima S, de Pas-Schoonen V, Katinka T, Kahnt J, Medema MH, Op Den Camp HJ, Jetten MS, Strous M. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ Microbiol. 2008;10(11):3164–3173. doi: 10.1111/j.1462-2920.2008.01724.x. [DOI] [PubMed] [Google Scholar]
- Frenzel P. Plant-associated methane oxidation in rice fields and wetlands. Adv Microb Ecol. 2000;16:85–114. [Google Scholar]
- Hakemian AS, Rosenzweig AC. The biochemistry of methane oxidation. Annu Rev Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- Han P, Gu JD. A newly designed degenerate PCR primer based on pmoA gene for detection of nitrite-dependent anaerobic methane-oxidizing bacteria from different ecological niches. Appl Microbiol Biotechnol. 2013;97(23):10155–10162. doi: 10.1007/s00253-013-5260-8. [DOI] [PubMed] [Google Scholar]
- Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60(2):439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature. 2013;500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65(5):1980–1990. doi: 10.1128/aem.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefman S, Heylen K, De Vos P. Methylomonas lenta sp. nov., a methanotroph isolated from manure and a denitrification tank. Int J Syst Evol. 2014 doi: 10.1099/ijs.0.057794-0. [DOI] [PubMed] [Google Scholar]
- Holmes AJ, Owens NJP, Murrell JC. Detection of novel marine methanotrophs using phylogenetic and functional gene probes after methane enrichment. Microbiol. 1995;141:1947–1955. doi: 10.1099/13500872-141-8-1947. [DOI] [PubMed] [Google Scholar]
- Holmes AJ, Costello A, Lidstrom ME, Murrell JC. Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS microbiol Lett. 1995;132(3):203–208. doi: 10.1111/j.1574-6968.1995.tb07834.x. [DOI] [PubMed] [Google Scholar]
- Holmes AJ, Roslev P, McDonald IR, Iversen N, Henriksen KA, Murrell JC. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl Environ Microbiol. 1999;65(8):3312–3318. doi: 10.1128/aem.65.8.3312-3318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intergovernmental Panel on Climate Change (2014) Climate Change 2014: the physical science basis. Summary for policymakers. Contribution of working group I to the fourth assessment report of the intergovernmental panel on climate change. Paris: summary for policymakers formally approved at the 10th session of working group I of the IPCC
- Joulian C, Escoffier S, Le Mer J, Neue HU, Roger PA. Populations and potential activities of methanogens and methanotrophs in rice fields: relations with soil properties. Eur J Soil Biol. 1997;33(2):105–116. [Google Scholar]
- Khadem AF, Wieczorek AS, Pol A, Vuilleumier S, Harhangi HR, Dunfield PF, Kalyuzhnaya MG, Murrell C, Francoijs KJ, Stunnenberg HG, Stein LY, DiSpirito AA, Semrau JD, Lajus AL, Medigue C, Klotz MG, Jetten MSM, Op den Camp HJM. Draft genome sequence of the volcano-inhabiting thermoacidophilic methanotroph Methylacidiphilum fumariolicum strain SolV. J Bacteriol. 2012;194(14):3729–3730. doi: 10.1128/JB.00501-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelenina VN, Beck DA, Munk C, Davenport K, Daligault H, Erkkila T, Goodwin L, Gu W, Lo CC, Scholz M, Teshima H, Xu Y, Chain P, Bringel F, Vuilleumier S, Dispirito A, Dunfield P, Jetten MS, Klotz MG, Knief C, Murrell JC, Op den Camp HJ, Sakai Y, Semrau J, Svenning M, Stein LY, Trotsenko YA, Kalyuzhnaya MG. Draft Genome sequence of Methylomicrobium buryatense strain 5G, a haloalkaline-tolerant methanotrophic bacterium. Genome Announc. 2013;1(4):pii: e00053–13. doi: 10.1128/genomeA.00053-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kip N, Dutilh BE, Pan Y, Bodrossy L, Neveling K, Kwint MP, Jetten MSM, Op den Camp HJM. Ultra-deep pyrosequencing of pmoA amplicons confirms the prevalence of Methylomonas and Methylocystis in Sphagnum mosses from a Dutch peat bog. Environ Microbiol Rep. 2011;6:667–673. doi: 10.1111/j.1758-2229.2011.00260.x. [DOI] [PubMed] [Google Scholar]
- Kits KD, Kalyuzhnaya MG, Klotz MG, Jetten MS, Op den Camp HJ, Vuilleumier S, Bringel F, Dispirito AA, Murrell JC, Bruce D, Cheng JF, Copeland A, Goodwin L, Hauser L, Lajus A, Land ML, Lapidus A, Lucas S, Médigue C, Pitluck S, Woyke T, Zeytun A, Stein LY. Genome sequence of the obligate Gammaproteobacterial methanotroph Methylomicrobium album strain BG8. Genome Announc. 2013;1(2):e0017013. doi: 10.1128/genomeA.00170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol. 2015;6:1346. doi: 10.3389/fmicb.2015.01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittel K, Boetius A. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol. 2009;63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- Kolb S, Knief C, Stubner S, Conrad R. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl Environ Microbiol. 2003;69(5):2423–2429. doi: 10.1128/AEM.69.5.2423-2429.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger M, Frenzel P, Conrad R. Microbial processes influencing methane emission from rice fields. Glob Change Biol. 2001;7:49–63. doi: 10.1046/j.1365-2486.2001.00395.x. [DOI] [Google Scholar]
- Lau E, Iv EJ, Dillard ZW, Dague RD, Semple AL, Wentzell WL. High throughput sequencing to detect differences in methanotrophic Methylococcaceae and Methylocystaceae in surface peat, forest soil, and Sphagnum Moss in Cranesville Swamp preserve, west Virginia. USA. Microorganisms. 2015;3(2):113–136. doi: 10.3390/microorganisms3020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidstrom ME. Aerobic methylotrophic prokaryotes. Prokaryotes. 2006;2:618–634. doi: 10.1007/0-387-30742-7_20. [DOI] [Google Scholar]
- Lieberman RL, Rosenzweig AC. The quest for the particulate methane monooxygenase active site. Dalton Trans. 2005;21:3390–3396. doi: 10.1039/b506651d. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesken FA, van Alen TA, van der Biezen E, Frijters C, Toonen G, Kampman C, Hendrickx TL, Zeeman G, Temmink H, Strous M, Op den Camp HJ, Jetten MS. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl Microbiol Biotechnol. 2011;92:845–854. doi: 10.1007/s00253-011-3361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesken FA, Zhu BL, van Alen TA, Butler MK, Diaz MR, Song B, den Camp HJMO, Jetten MSM, Ettwig KF. pmoA primers for detection of anaerobic methanotrophs. Appl Environ Microbiol. 2011;77(11):3877–3880. doi: 10.1128/AEM.02960-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüke C, Frenzel P. Potential of pmoA amplicon pyrosequencing for methanotroph diversity studies. Appl Environ Microbiol. 2011;77(17):6305–6309. doi: 10.1128/AEM.05355-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüke C, Krause S, Cavigiolo S, Greppi D, Lupotto E, Frenzel P. Biogeography of wetland rice methanotrophs. Environ Microbiol. 2009 doi: 10.1111/j.1462-2920.2009.02131.x. [DOI] [PubMed] [Google Scholar]
- Lüke C, Frenzel P, Ho A, Fiantis D, Schad P, Schneider B, Schwark L, Utami SR. Macroecology of methane-oxidizing bacteria: the β-diversity of pmoA genotypes in tropical and subtropical rice paddies. Environ Microbiol. 2014;16(1):72–83. doi: 10.1111/1462-2920.12190. [DOI] [PubMed] [Google Scholar]
- Lüke C, Speth DR, Kox MA, Villanueva L, Jetten MS. Metagenomic analysis of nitrogen and methane cycling in the Arabian Sea oxygen minimum zone. Peer J. 2016;7(4):e1924. doi: 10.7717/peerj.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsumori M, Ajisaka N, Tajima K, Kajikawa H, Kurihara M. Detection of Proteobacteria from the rumen by PCR using methanotroph-specific primers. Lett Appl Microbiol. 2002;35(3):251–255. doi: 10.1046/j.1472-765X.2002.01172.x. [DOI] [PubMed] [Google Scholar]
- Murrell JC, McDonald IR, Bourne DG. Molecular methods for the study of methanotroph ecology. FEMS Microbiol: Ecol; 1998. [Google Scholar]
- Murrell JC, Gilbert B, McDonald IR. Molecular biology and regulation of methane monooxygenase. Arch: Microbiol; 2000. [DOI] [PubMed] [Google Scholar]
- Oshkin IY, Wegner CE, Lüke C, Glagolev MV, Filippov IV, Pimenov NV, Liesack W, Dedysh SN. Gammaproteobacterial methanotrophs dominate cold methane seeps in floodplains of west Siberian rivers. Appl Environ Microbiol. 2014;80(19):5944–5954. doi: 10.1128/AEM.01539-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla CC, Bristow LA, Sarode N, Garcia-Robledo E, Gómez Ramírez E, Benson CR, Bourbonnais A, Altabet MA, Girguis PR, Thamdrup B, Stewart FJ. NC10 bacteria in marine oxygen minimum zones. ISME J. 2016;10(8):2067–2071. doi: 10.1038/ismej.2015.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AJ, Marcus DN, Ijaz UZ, Bautista-de Lose Santos QM, Dick GJ, Raskin L. metagenomic evidence for the presence of comammox Nitrospira-like bacteria in a drinking water. Syst mSphere. 2015 doi: 10.1128/mSphere.00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pjevac P, Schauberger C, Poghosyan L, Herbold CW, van Kessel MAHJ, Daebeler A, Steinberger M, Jetten MSM, Lücker S, Wagner M, Daims H. AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. BioRxiv. 2016 doi: 10.3389/fmicb.2017.01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MSM, Op den Camp HJM. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature. 2007;450(7171):874–878. doi: 10.1038/nature06222. [DOI] [PubMed] [Google Scholar]
- Pol A, Barends TR, Dietl A, Khadem AF, Eygensteyn J, Jetten MSM, Op den Camp HJM. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol. 2014;16(1):255–264. doi: 10.1111/1462-2920.12249. [DOI] [PubMed] [Google Scholar]
- Putkinen A, Larmola T, Tuomivirta T, Siljanen HM, Bodrossy L, Tuittila ES, Fritze H. Peatland succession induces a shift in the community composition of Sphagnum-associated active methanotrophs. FEMS Microbiol Ecol. 2014;88(3):596–611. doi: 10.1111/1574-6941.12327. [DOI] [PubMed] [Google Scholar]
- Rastogi G, Ranade DR, Yeole TY, Gupta AK, Patole MS, Shouche YS. Novel methanotroph diversity evidenced by molecular characterization of particulate methane monooxygenase A (pmoA) genes in a biogas reactor. Microbiol Res. 2009;164(5):536–544. doi: 10.1016/j.micres.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Reeburgh WS, Whjalen SC. Oceanic methane biogeochemistry. Chem Rev. 2007;107:486–513. doi: 10.1021/cr050362v. [DOI] [PubMed] [Google Scholar]
- Reeburgh WS, Whjalen SC, Alperin MJ (1993) The role of methylotrophy in the global methane budget in Microbial growth on C1 compounds. In: Murrell JC, Kelly DP (eds ) (Intercept, Andover, England). pp 1–14
- Saidi-Mehrabad A, He Z, Tamas I, Sharp CE, Brady AL, Rochman FF, Bodrossy L, Abell GC, Penner T, Dong X, Sensen CW, Dunfield PF. Methanotrophic bacteria in oilsands tailings ponds of northern Alberta. ISME J. 2013;5:908–921. doi: 10.1038/ismej.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad MS, Bertilsson S. Seasonal variation in abundance and diversity of bacterial methanotrophs in five temperate lakes. Front Microbiol. 2017;8:142. doi: 10.3389/fmicb.2017.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrau JD, Chistoserdov A, Lebron J, Costello A, Davagnino J, Kenna E, Holmes AJ, Finch R, Murrell JC, Lidstrom ME. Particulate methane monooxygenase genes in methanotrophs. J Bacteriol. 1995;177(11):3071–3079. doi: 10.1128/jb.177.11.3071-3079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp CE, Smirnova AV, Graham JM, Stott MB, Khadka R, Moore TR, Grasby SE, Strack M, Dunfield PF. Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ Microbiol. 2014;16(6):1867–1878. doi: 10.1111/1462-2920.12454. [DOI] [PubMed] [Google Scholar]
- Sievers F, Higgins DG (2014) Clustal Omega. Published online: 12 DEC 2014. doi: 10.1002/0471250953.bi0313s48
- Siljanen HM, Saari A, Bodrossy L, Martikainen PJ. Seasonal variation in the function and diversity of methanotrophs in the littoral wetland of a boreal eutrophic lake. FEMS Microbiol Ecol. 2012;80(3):548–555. doi: 10.1111/j.1574-6941.2012.01321.x. [DOI] [PubMed] [Google Scholar]
- Singh BP, Hatton BJ, Singh B, Cowie AL, Kathuria A. Influence of biochars on nitrous oxide emission and nitrogen leaching from two contrasting soils. J Environ Qual. 2010;39:1224–1235. doi: 10.2134/jeq2009.0138. [DOI] [PubMed] [Google Scholar]
- Söhngen NL. Über bakterien, welche methan als kohlenstoffnahrung und energiequelle gebrauchen. Z Bakteriol Parazitenk (Infektionster) 1906;15:513–517. [Google Scholar]
- Stephenson J, Kumaresan D, Hillebrand-Voiculescu AM, Brooks E, Whiteley AS, Murrell JC. Draft genome sequence of the methane-oxidizing bacterium “Candidatus Methylomonas sp. LWB” isolated from movile cave. Genome Announc. 2017;5(3):pii: e01491–16. doi: 10.1128/genomeA.01491-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotsenko YA, Murrell JC. Metabolic aspect of aerobic obligate methanotrophy. Adv Appl Microbiol. 2008 doi: 10.1016/S0065-2164(07)00005-6. [DOI] [PubMed] [Google Scholar]
- Vaksmaa A, Lüke C, van Alen T, Valè G, Lupotto E, Jetten MSM, Ettwig KF. Distribution and activity of the anaerobic methanotrophic community in a nitrogen-fertilized Italian paddy soil. FEMS Microbiol Ecol. 2016;92(12):pii: fiw181. doi: 10.1093/femsec/fiw181. [DOI] [PubMed] [Google Scholar]
- Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, Cruveiller S, Lajus A, Pascal G, Scarpelli C, Médigue C. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006;34(1):53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel MA, Speth DR, Albertsen M, Nielsen PH, den Camp HJ, Kartal B, Jetten MS, Lücker S. Complete nitrification by a single microorganism. Nature. 2015;528(7583):555. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeseling MC, Pol A, Harhangi HR, van der Zwart S, Jetten MS, Op den Camp HJ, van Niftrik L. Expanding the verrucomicrobial methanotrophic world: description of three novel species of Methylacidimicrobium gen. nov. Appl Environ Microbiol. 2014;80(21):6782–6791. doi: 10.1128/AEM.01838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekeman B, Kerckhof FM, Cremers G, de Vos P, Vandamme P, Boon N, Op den Camp HJ, Heylen K. new methyloceanibacter diversity from North Sea sediments includes methanotroph containing solely the soluble methane monooxygenase. Environ Microbiol. 2016;18(12):4523–4536. doi: 10.1111/1462-2920.13485. [DOI] [PubMed] [Google Scholar]
- Vekeman B, Speth D, Wille J, Cremers G, De Vos P, Op den Camp HJ, Heylen K. Genome characteristics of two novel type I methanotrophs enriched from North Sea sediments containing exclusively a lanthanide-dependent XoxF5-type methanol dehydrogenase. Microb Ecol. 2016;72(3):503–509. doi: 10.1007/s00248-016-0808-7. [DOI] [PubMed] [Google Scholar]
- Vorobev AV, Baani M, Doronina NV, Brady AL, Liesack W, Dunfield PF, Dedysh SN. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium possessing only a soluble methane monooxygenase. Int J Syst Evol Microbiol. 2011;61(10):2456–2463. doi: 10.1099/ijs.0.028118-0. [DOI] [PubMed] [Google Scholar]
- Wang JG, Xia F, Zeleke J, Zou B, Rhee SK, Quan ZX. An improved protocol with a highly degenerate primer targeting copper-containing membrane-bound monooxygenase genes for community analysis of methane- and ammonia-oxidizing bacteria. FEMS Microbiol Ecol. 2017;93(3):pii: fiw244. doi: 10.1093/femsec/fiw244. [DOI] [PubMed] [Google Scholar]
- Welte CU, Rasigraf O, Vaksmaa A, Versantvoort W, Arshad A, Op den Camp HJ, Jetten MS, Lücke C, Reimann J. Nitrate- and nitrite-dependent anaerobic oxidation of methane. Environ Microbiol Rep. 2016;8(6):941–955. doi: 10.1111/1758-2229.12487. [DOI] [PubMed] [Google Scholar]
- Whittenbury R, Phillips KC, Wilkinson JG. Enrichment, isolation and some properties of methane utilizing bacteria. J Gen Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Zhang L-M, Zheng Y-M, Di H, He J-Z. Abundance and community composition of methanotrophs in a Chinese paddy soil under long-term fertilization practices. J Soils Sediments. 2008;8:406–414. doi: 10.1007/s11368-008-0047-8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences are deposited in Genbank with Accession Numbers KY883458–KY883555 (Additional file 1: Table S1). The data will be publically available as of 1 April 2018.


