Abstract
Integrin-mediated signaling is crucial for cell-substrate adhesion and can be triggered from both intra- and extracellular interactions. Although talin binding is sufficient for inside-out activation of integrin, other cytoplasmic proteins such as α-actinin and filamin can directly interfere with talin-mediated integrin activation. Specifically, α-actinin plays distinct roles in regulating αIIbβ3 versus α5β1 integrin. It has been shown that α-actinin competes with talin for binding to the cytoplasmic tail of β3-integrin, whereas it cooperates with talin for activating integrin α5β1. In this study, molecular dynamics simulations were employed to compare and contrast molecular mechanisms of αIIbβ3 and α5β1 activation in the presence and absence of α-actinin. Our results suggest that α-actinin impairs integrin signaling by both undermining talin binding to the β3-integrin cytoplasmic tail and inducing a kink in the transmembrane domain of β3-integrin. Furthermore, we showed that α-actinin promote talin association with β1-integrin by restricting the motion of the cytoplasmic tail and reducing the entropic barrier for talin binding. Taken together, our results showed that the interplay between talin and α-actinin regulates signal transmission via controlling the conformation of the transmembrane domain and altering natural response modes of integrins in a type-specific manner.
Introduction
Cell-substrate signaling is critical for various mechano-biological functions of the cell, including rigidity sensing, migration, differentiation, and growth (1, 2, 3). More than 150 signaling and scaffolding molecules are involved in orchestrating mechanochemical communication of cells with their environment through focal adhesions (4, 5). Early adhesions are formed by a few molecules but are transformed to mature focal adhesions as local forces increase (5, 6, 7). Mature focal adhesions are firmly attached to the cytoskeleton, converting the actin retrograde flow to traction forces exerted on the substrate (7).
Talin is a chief focal adhesion binding protein and initiates inside-out signaling by disrupting a key interaction between integrin subunits known as the inner membrane clasp (IMC) (8). However, other molecules such as kindlin, filamin, and α-actinin also associate with the integrin tail and directly couple it with the actin cytoskeleton (9, 10, 11, 12, 13). The effect of these molecules on the process of integrin activation depends strongly upon their interplay with talin (14). More specifically, these molecules can interfere with integrin activation through either competing or cooperating with talin (14). α-Actinin, one of the most prominent players in focal adhesions (5, 15, 16), binds to various molecules including integrin and vinculin and is directly involved in transmitting forces between the cytoskeleton and adhesion sites (17, 18, 19, 20, 21, 22, 23). β3-integrin is necessary for nascent adhesion formation, whereas β1-integrins localize to more mature adhesions. Roca-Cusachs et al. (15), in 2013, reported that α-actinin competes with talin for binding to β3-integrin and suppresses activation, whereas it cooperates with talin for β1-integrin activation.
Proline-induced kinks in the transmembrane domain (TMD) of integrins are shown to be functionally important and associated with diseases in some cases (24). Recent efforts have tried to recognize and reverse the effects of proline mutations. An important example is the A711P mutation, which can compensate for the integrin activation induced by the K716E mutation. The A711P mutation introduces a kink in the β3-TMD that breaks the continuity of the helix and replaces the IMC interaction (25). It should be noted that while they both play important roles in regulating the TMD topography (25), neither K716E nor A711P has been recognized as a disease-causing mutation. However, such proline-induced kinks are evolutionarily significant, and can be used toward mutagenesis-based treatments. Other potential sources of TMD kinks and their impacts on the signal transmission across membrane receptors are still not as well understood.
The composition of proteins at the integrin tail during adhesion formation plays an important role in integrin-mediated singling (14). The molecular mechanism of α-actinin and talin crosstalk is not yet understood, largely due to the limitations of experimental studies in capturing the molecular events at sufficient spatial and temporal resolution. In this study, we employed molecular dynamics (MD) models and simulations to investigate the order of binding events and mechanisms of regulating integrin activation through cytoplasmic interactions with talin and α-actinin.
Materials and Methods
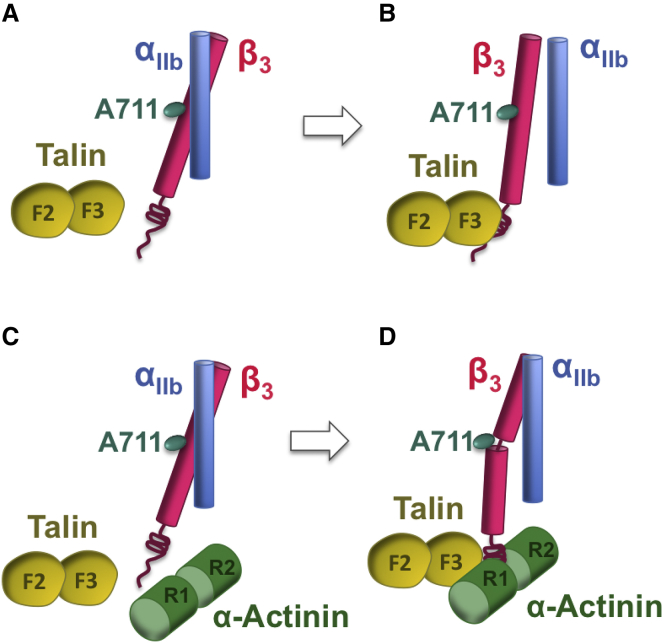
All molecular models were prepared using the Visual Molecular Dynamics software package (26) and simulations were performed in NAMD (27) using the CHARMM27 force field (28). The configuration of the system in the binding simulations included the full-length α-actinin (PDB: 1SJJ), the head domain of talin (F0 to F3 subdomains from PDB: 3IVF), and only the transmembrane and cytoplasmic parts of either αIIbβ3 (PDB: 2KNC) or α5β1 integrins; α-actinin was excluded from the control simulations (Fig. 1). The α5β1 integrin crystal structure is not available, thus the αIIbβ3 was submitted to the PHYRE2 server as the template for homology modeling. The output model was quite reliable, with the confidence level of 99.9% reported by PHYRE2. In the initial configuration of the binding simulations, the integrin binding sites on talin and α-actinin were positioned at equal distances from the integrin cytoplasmic tail, allowing a few water layers to form in the space between the molecules, whereas the TMD domain of the integrin heterodimer was embedded in the lipid membrane.
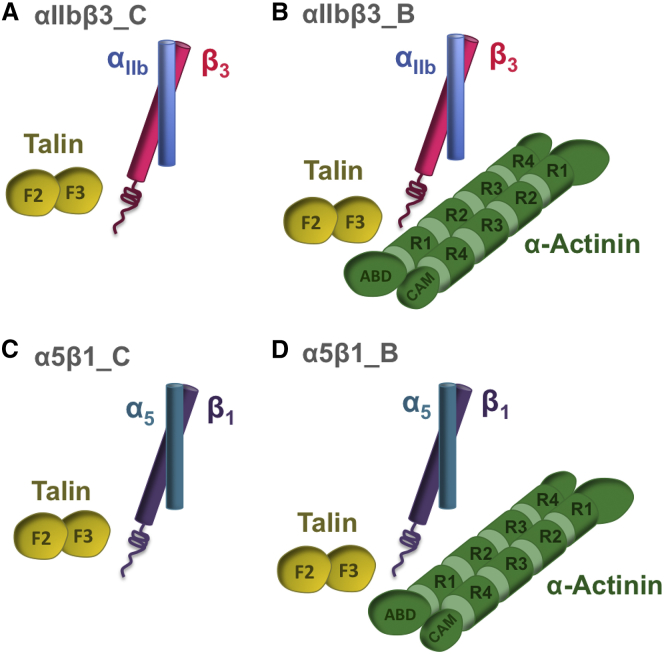
Figure 1.
A schematic overview of simulations carried out in this work. (A) The interaction between αIIbβ3 and talin was modeled in a set of control simulations marked as αIIbβ3_C. (B) α-Actinin, talin, and αIIbβ3 were all included in the binding simulations (αIIbβ3_B). (C) α-actinin was absent in the α5β1 control simulations, marked as α5β1_C, whereas (D) it was included along with talin and α5β1 in the binding simulations. For clarity, the lipid membrane is not shown. To see this figure in color, go online.
A triclinic box (size: 41.2 × 18.2 × 15.8 nm3) was generated to account for the elongated structure of α-actinin and also to ensure that molecules did not interact with their periodic image. The TIP3P water model was used for solvating the system but water molecules were deleted from the hydrophobic region of the lipid membrane (29). The excessive charge of the system was neutralized and the overall ion concentration was set to 150 mM of KCl, mimicking the physiological conditions. The number of atoms in the presence of α-actinin reached 1,918,344 and 1,918,175 for the αIIbβ3 and α5β1 simulations, respectively.
The SHAKE algorithm was applied to constrain bond lengths and geometry. The temperature was maintained at 310 K and the pressure was kept at 1 bar using Langevin dynamics and Langevin piston pressure control, respectively. Periodic boundary condition was applied in all three directions and the time-step of 2 fs was used in all simulations. The PME method was employed to account for the electrostatic interactions efficiently. The van der Waals interactions were modeled using a switching parameter for smoothly truncating the van der Waals potential at the cutoff distance. The initial configurations of simulations were minimized for 100,000 steps to relax the structures and remove all bad contacts. Minimized structures were then equilibrated for 1 ns after which energy, pressure, and the root mean square deviation were monitored to examine whether an equilibrium state had been reached. Fully equilibrated structures were then used in the final production simulations that each ran for 20 ns and was repeated three times to produce statistically significant results. Simulation parameters of the production runs were the same as those used in the equilibration simulations. The total simulation time performed for this work is 360 ns. From each simulation, 100 time frames were extracted for postprocessing the data, thereby, with three runs, 300 data points were available for each simulation set. The Visual MD package was used to visualize the results (26) and model mutations. The Bio3D package in R was utilized for analyzing the data (30). Normal mode analysis was done using the NOMAD-REF server (31).
Results
Inspired by the previous studies (15), here, we investigated the mechanisms by which α-actinin binding to the β3-tail suppresses integrin activation, whereas colocalization of α-actinin with talin promotes α5β1 activation. For that, we designed and performed four sets of simulations as shown in Fig. 1 to capture the effect of α-actinin binding on talin-induced activation of both αIIbβ3 and α5β1. The simulations are labeled in Fig. 1, and are referred to as such throughout the text. Two other sets of simulations were performed to test particular mutations speculated to be important in the first four sets of simulations described above.
The competition between talin and α-actinin at the cytoplasmic tail of αIIbβ3 integrin
The presence of α-actinin at the β3-tail resulted in significantly weaker interaction of talin with β3 in αIIbβ3_B simulations (Fig. 2 A). Conversely, talin engaged with the NPLY motif on the β3-tail in the absence of α-actinin in the control simulations (Fig. 2 B). This was in agreement with the previous studies that indicated a binding between the F3 subdomain of talin and the first NPxY motif of the β3-tail (Fig. 2 C) (8). In αIIbβ3_B simulations, the NPLY motif was drawn toward the integrin binding region between R1 and R2 repeats along the α-actinin rod domain (Fig. 2 D).
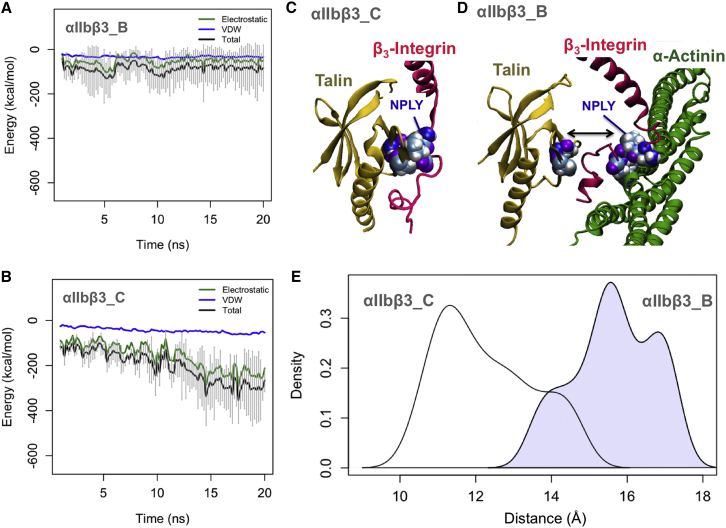
Figure 2.
The molecular mechanism of the competition between α-actinin and talin in regulating αIIbβ3 integrin. (A) The interaction energy between talin and the β3-tail in the presence of α-actinin averaged over three trials is relatively weak. (B) This interaction energy in αIIbβ3_C simulations drops to −300 kcal/mol after 15 ns. (C) Talin (yellow) is strongly associated with the NPLY motif of β3-integrin (mauve) in the last frame of the αIIbβ3_C simulations. (D) α-Actinin (green) pulls the NPLY motif away from talin toward its rod domain in αIIbβ3_B simulations. (E) The density plot of the distance between the NPLY motif and talin for αIIbβ3_C simulations (white) peaks at 11 Å, whereas for αIIbβ3_B simulations (blue), the major peak occurs at 16 Å. To see this figure in color, go online.
The density plot of the average distance between talin and β3 showed the effect of α-actinin in moving the β3-tail away from talin (Fig. 2 E). Specifically, the peak value of the talin-integrin distance in the αIIbβ3_C simulation was at 11 Å, whereas the peak was shifted to 16 Å in the αIIbβ3_B simulations showing that the β3-tail was pulled away from talin toward α-actinin. The common peak at 14 Å indicated that the initial distance between talin and integrin was the same in the αIIbβ3_C and αIIbβ3_B simulations. The peak at 17 Å in the αIIbβ3_B curve occurred as β3 moved further toward α-actinin in the last 5 ns of the simulations.
The formation of a kink in the transmembrane domain of β3-integrin
Upon α-actinin binding, a kink was formed at residue A711 on the β3-TMD, whereas such deformation was not observed in αIIbβ3_C simulations (Fig. 3). The resolved crystal structure of β3 with the A711P mutation (PDB: 2N9Y) shows a similar kink at P711. Moreover, we carried out a set of simulations in which residue A711 of the wild-type β3 was mutated to proline (referred to as αIIbβ3_IM simulations in Fig. 3). Our results confirmed the formation of a kink in both αIIbβ3_B and αIIbβ3_IM simulations, which most likely inhibit integrin signaling across the membrane as shown in Fig. 3.
Figure 3.
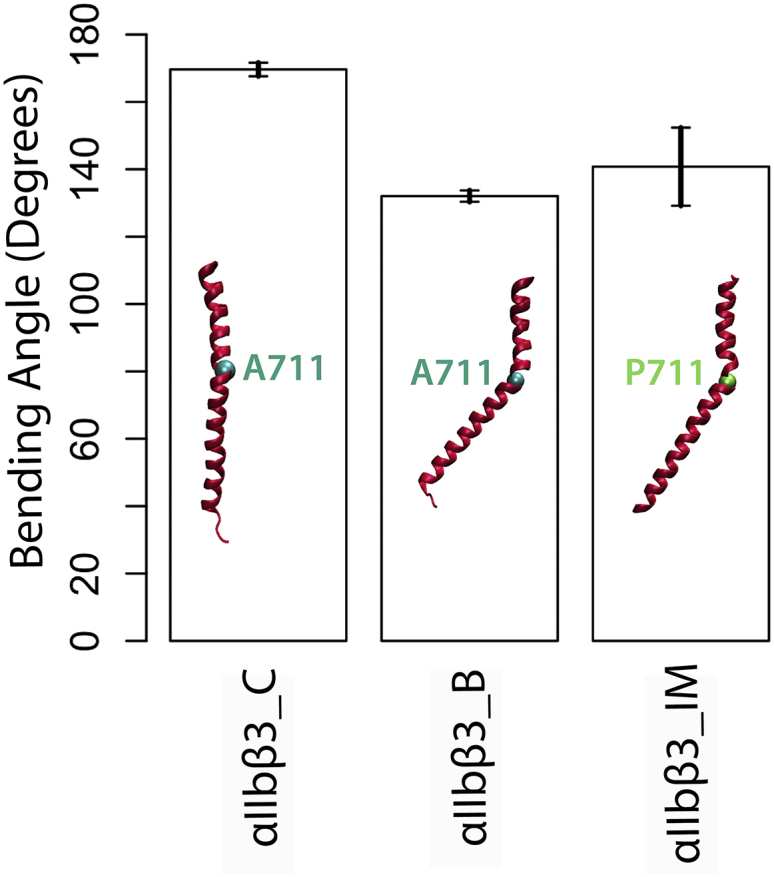
Formation of a kink in β3-TMD upon α-actinin binding. A kink centered at A711 formed in β3-TMD after α-actinin association with its cytoplasmic tail (middle) quantified by the bending angle of the TMD helix. Conversely, no such kink was observed in αIIbβ3_C simulations, indicating that talin alone cannot induce such deformation. Mutating residue A711 to proline created a similar kink. To see this figure in color, go online.
The continuum model of the TMD backbone structure could be a simple rod, thus its first natural mode is expected to be an in-plane bending. The normal mode analysis confirmed that the first nonrigid motion of the integrin TMD is bending as shown by the dashed arrows in Fig. S1 A. However, the motions of the N- and C termini of TMD were decoupled after the kink was established (Fig. S1 B). The fluctuations of residues were smoothly coupled in the straight TMD, and residue A711 acted as a nodal point. Also, the N-terminus (N-TMD) on the extracellular side moved in phase with the C terminus (C-TMD). In contrast, the root mean square fluctuations (RMSFs) of the kinked structure was highly jagged and motions of the N-TMD and C-TMD went out of phase (Fig. S1 B and C).
The cooperative role of α-actinin in the binding between talin and β1-integrin
To contrast the role of α-actinin in regulating talin association to αIIbβ3 and α5β1 integrins, the initial configurations of the two systems were designed to be similar. The interaction between talin and β1-integrin formed in both α5β1_C and α5β1_B simulations. In the first 15 ns of α5β1_B simulations, the interaction energy between talin and β1-integrin strengthened linearly, whereas this energy remained stable in α5β1_C simulations (Fig. 4, A and B). However, the talin-integrin interaction was slightly weakened in the second 15 ns of α5β1_B simulations and the average energy was comparable to that in α5β1_C simulations (Fig. 4, A and B). This demonstrated that the α-actinin molecule had a positive impact on the initial talin binding by aligning the β1-tail. As talin formed a stable interaction with β1, α-actinin binding remained intact. Thereby, talin and α-actinin molecules could simultaneously interact with opposite faces of the β1-tail.
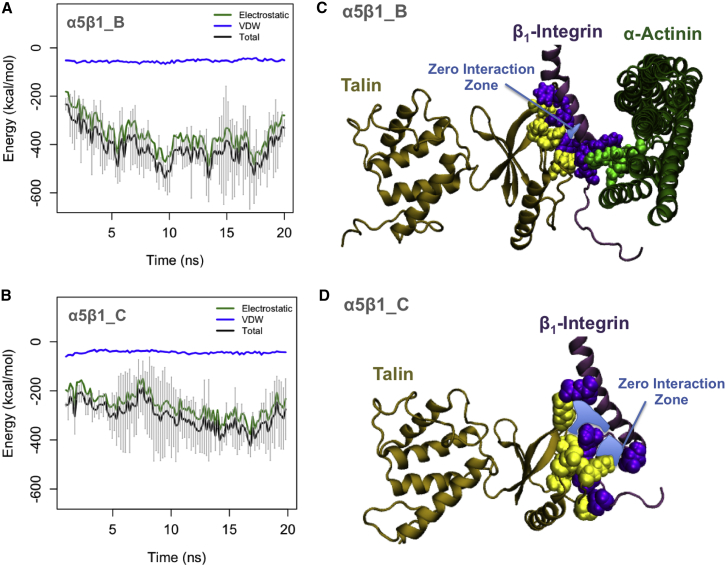
Figure 4.
The cooperative role of α-actinin in the binding between talin and β1-integrin. (A) The average interaction energy of talin and the β1-tail in the presence of α-actinin drops to around −400 kcal/mol within the first 15 ns and becomes stable in the last 10 ns of simulations. (B) In α5β1_C simulations, a large energy drop in the talin-β1 interaction is not observed and the enthalpy is reduced to ∼−250 kcal/mol. (C) α-Actinin (green) and talin (yellow) simultaneously interact with the β1-tail (purple), resulting in a closedly packed residues in the binding region. The zero interaction zone (the area shown in blue) is defined as a gap between atoms with no significant interaction between talin and the β1-tail. (D) In α5β1_C simulations, the zero-interaction zone is increased compared to α5β1_B simulations, indicating the cooperative role of α-actinin in the binding between talin and the β1-tail. To see this figure in color, go online.
The complex of talin, α-actinin, and β1-integrin formed several interactions at their binding interface (Fig. 4 C). To indicate the packing of the binding residues more clearly, the space in which no interaction occurred, also referred to as the zero interaction zone, was marked in blue. In the control simulation, talin and β1-integrin loosely interacted and the zero interaction zone was larger (Fig. 4 D). It should be noted that the zero interaction zone is not necessarily an empty space but it rather represents a region with no interaction between the molecules.
The effect of cytoplasmic interactions on activating integrins αIIbβ3 and α5β1
Integrin activation initiates as the TMDs of the integrin subunits dissociate in response to cytoplasmic interactions. We measured the association energy between αIIb-β3 and α5-β1 subunits in all simulations and observed that the presence of α-actinin markedly decreased the strength of α5–β1 interaction (Fig. 5 A), which makes it prone to activation.
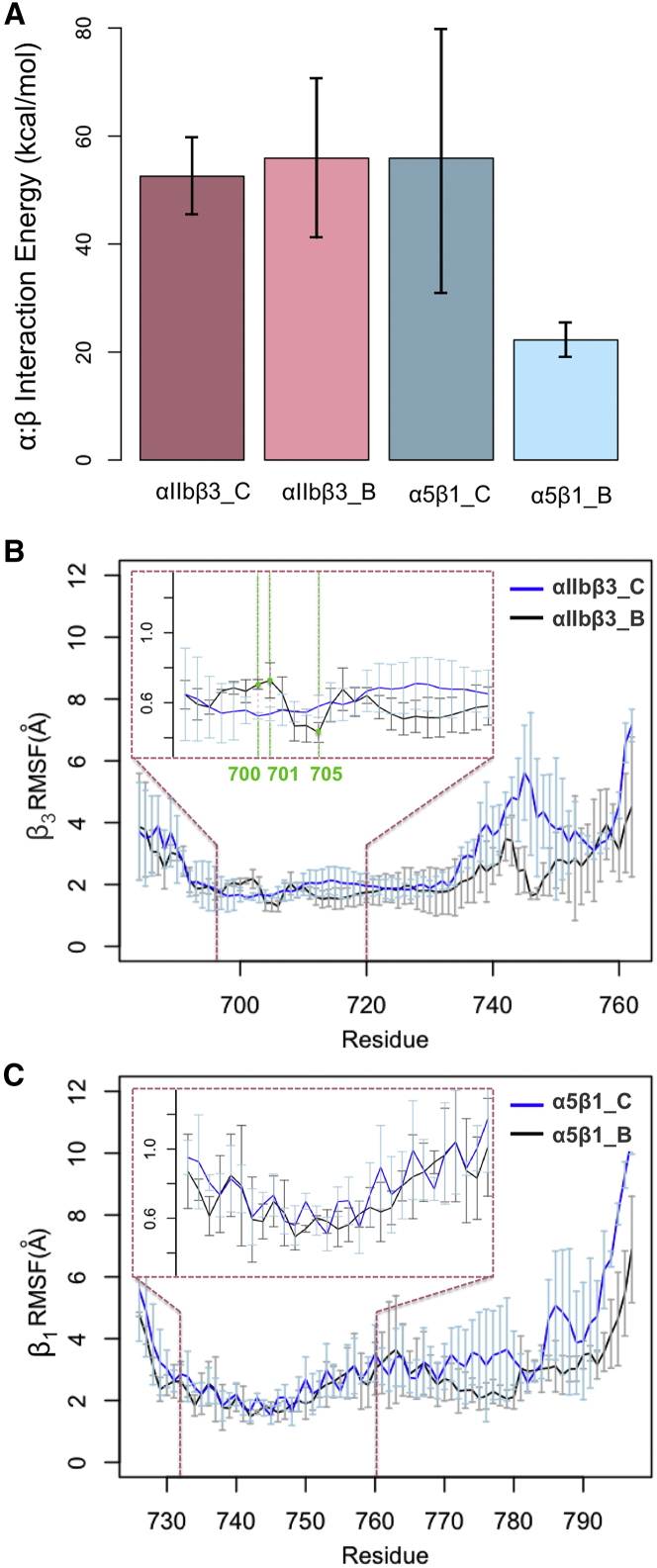
Figure 5.
The effect of cytoplasmic interactions on integrin activation. (A) The average energy between α5-β1 subunits in α5β1_B simulations is significantly decreased compared to that in α5β1_C simulations. Within the same simulation time, no change in the αIIb-β3 energy is observed upon α-actinin binding. (B) Atomic fluctuations are limited in the transmembrane domain relative to the cytoplasmic end of integrins in all simulations. The TMD regions are magnified in the insets. The RMSF of β3-tail is different between αIIbβ3_C and αIIbβ3_B simulations. Specifically, the error bars of atomic fluctuations do not overlap at residues 700, 701, and 705, which are all above residue 711 toward the extracellular side of TMD, showing a notable difference between the RMSF curves at those residues. (C) The RMSF of β1-TMD is similar between α5β1_C and αIIbβ3_B simulations. This most likely indicates that signal transmission through the β1-TMD is not significantly altered in the presence of α-actinin. To see this figure in color, go online.
Mechanical signal transmission along the β3-TMD strongly relies upon atomic motions. To find the correlation between high fluctuations of the interaction energy and actual physical movements of residues along TMDs, we calculated the RMSFs of β1- and β3-integrins in all simulations. The RMSF of every atom was obtained from averaging over 100 time frames, and the error bars were calculated by averaging over three runs for each simulation. The RMSFs of β3-TMD in αIIbβ3_C simulations as shown in Fig. 5 B was relatively smooth, indicating a tight coupling of residues connecting the key interactions between TMDs, i.e., inner and outer membrane clasps. Conversely, the RMSFs of β3-TMD showed a relatively sharp change in αIIbβ3_B, which may demonstrate local decoupling events along the β3-TMD (Fig. 5 B). Specifically, fluctuations of residues 700, 701, and 705 differed statistically more significantly between αIIbβ3_B and αIIbβ3_C as the error bars did not overlap. These residues are all located above A711 toward the N-terminus of β3-TMD, whereas RMSFs of TMD residues below A711 was relatively smooth. This may suggest that the kink at A711 allosterically affects fluctuations of the N-terminus of TMD, and breaks the continuity of the TMD domain.
The RMSF of β1-integrin in α5β1_B simulation closely followed that in α5β1_C along the extracellular and transmembrane sides of the β1-TMD, whereas the α5β1_B RMSF curve gets smoother toward the cytoplasmic side (Fig. 5 C). The RMSFs of the cytoplasmic domains of both β1- and β3-tails were higher in control simulations (Fig. 5 C), indicating that α-actinin binding constrained the motion of the cytoplasmic regions of both integrins, most likely decreasing the unfavorable entropic barrier for talin binding. Also, the RMSF of the cytoplasmic region was generally higher than the TMD domain, due to the confinement effect of the lipid molecules (Fig. 5 C). Differences in the RMSFs of β1 and β3 integrins suggested that signal transmission along integrin is strongly type-specific.
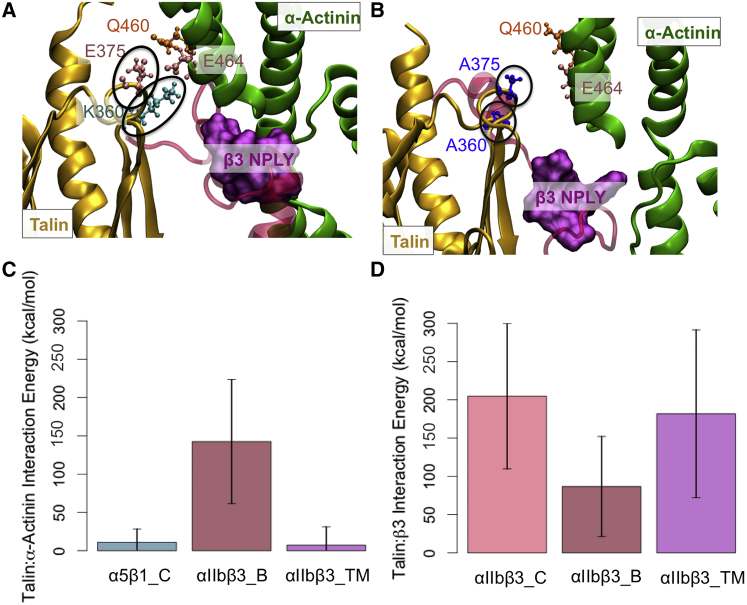
A direct interaction between talin and α-actinin
We observed a very stable interaction between talin and α-actinin at the β3-tail, which is most probably an important factor in regulating β3-integrin activation as these molecules compete for the same binding site on the integrin tail (Fig. 6). Two important interactions between talin and α-actinin were: 1) a salt bridge between residues K360 on talin and E464 on α-actinin, and 2) E375 on talin and Q460 on α-actinin as shown in Fig. 6 A. These residues were mutated to alanine (Fig. 6 B) and tested in a set of simulations referred to as αIIbβ3_TM simulations. It should be noted that the α-actinin-talin interaction was not observed at the β1 tail (Fig. 6 C). Also, the double mutation on talin completely disrupted its interaction with α-actinin (Fig. 6 C). The binding between talin and β3 was also correlated with the presence of α-actinin (Fig. 6 D); the less engaged talin became with α-actinin, the more strongly it associated with β3.
Figure 6.
The binding between talin and α-actinin is a possible mechanism for competition. (A) Strong interactions between α-actinin and talin are exclusively formed at the β3-tail in αIIbβ3_B simulations, mainly between residues E375 and K360 on talin, and residues Q460 and E464 on α-actinin. (B) Mutating both E375 and K360 to alanine changes the position of talin relative to both the NPLY motif and α-actinin. (C) The barplots indicate that the absolute value of interaction energy between talin and α-actinin is significantly reduced upon talin’s double mutation. It is also evident from the plot that α-actinin and talin do not associate at the β1-tail. (D) The interaction between talin and β3 is increased upon talin mutations. To see this figure in color, go online.
Discussion
Dynamic bidirectional signaling through integrins is required for regulating cell adhesion in both physiological and pathological conditions. Integrin-talin interaction provides the initial physical coupling essential for signal transmission to the extracellular matrix. As forces are increased at these nascent adhesions, other molecules are recruited to reinforce the extracellular matrix-cytoskeleton linkage and macromolecular complexes with distinct compositions are formed at integrin tails. Thereby, the interplay of cytoplasmic molecules with talin is essential for regulating integrin-mediated adhesions. Talin associates with the cytoplasmic tail of β-integrin and breaks the IMC, which leads to global extension of the integrin ectodomain (32). Other proteins such as filamin, α-actinin, kindlin, and FAK can also interact with the integrin tail and interfere with the activation process. The order in which these proteins bind to integrin and their potential impacts on talin's function can play an important regulatory role in adhesion formation and maturation. α-Actinin is an interesting molecule as it plays opposite roles in regulating αIIbβ3 versus α5β1 integrins and thus comparing its interplay with talin and integrin may reveal fundamental differences in type-specific regulatory mechanisms. Here, we investigated talin-mediated signal transmission from cytoplasm to the TMD domains of αIIbβ3 and α5β1 integrins in the presence and absence of α-actinin.
Our results showed that talin binding to β1-integrin was notably enhanced by including α-actinin in the simulations (Fig. 4). Specifically, α-actinin stabilized talin’s initial engagement by decreasing atomic motions, as indicated by lower RMSF of the β1-tail after α-actinin association, which most likely decreases the entropic barrier for talin binding (Fig. 5 C). Furthermore, the simultaneous interaction of α-actinin and talin lowered the binding energy between the α5β1 integrin subunits, elucidating the cooperative role of α-actinin.
Control simulations featuring β3-integrin and talin consistently showed successful association of talin with the NPLY motif of β3-tail, which was in agreement with previous studies (33, 34). However, this interaction was markedly lowered upon introducing α-actinin, as it pulled the β3-tail away from talin. As a result, α-actinin bound exclusively to β3-integrin and prevented talin from competing any further by directly associating with it. Although the mechanisms of integrin activation have been previously examined (33, 35), we report for the first time to our knowledge, the emergence of a striking kink along the β3-TMD upon α-actinin binding. The β3-TMD forms a ∼25° tilting angle with the αIIb-subunit, thus integrin monomers cross each other inside the lipid membrane. Talin binding increases the tilting angle of β3-integrin, resulting in the disruption of the IMC (24). The location of the kink observed in our simulations was near the crossing point of integrin subunits and occurred at the position of residue A711. We strongly hypothesize that the conformational change of the TMD domain is a critical factor in inhibiting integrin-mediated signaling. Previous studies also showed that the continuity of the integrin TMD is key to integrin activation (24). Interestingly, it was indicated that the A711P mutation also resulted in a flexible kink that broke the continuity of the β3-TMD, decoupling the tilting motion of integrin via talin (36). This supports our hypothesis on the mechanism by which α-actinin association suppresses integrin activation and signaling because the kink induced by α-actinin in the β3-TMD greatly resembles the effect of A711P mutation.
Normal mode analysis performed on both kinked and straight conformations of the β3-TMD indicated that the first bending mode of the TMD domain was significantly altered upon α-actinin binding (Fig. S1). Interestingly, the position of A711 after the appearance of the kink acted as a decoupling joint such that motions of the N-TMD and C-TMD went out-of-phase as illustrated by the gray arrows in Fig. S1, A and B. The RMSFs of Cα atoms in the first bending mode of the straight β3-TMD resembled a cosine wave function that most likely allowed for smooth transfer of mechanical signals along the TMD domain (Fig. S1 C). However, a jagged-looking RMSF plot of the first bending mode of the kinked structure suggests that random fluctuations of the TMD atoms most likely result in an inefficient transfer of mechanical signals across the membrane after α-actinin association. It should be noted the RMSF of β3-TMD atoms in the bending mode (Fig. S1 C) does not have to match that RMSF extracted from the MD simulation (Fig. 5, B and C) since this mode was not excited in the simulation. However, our analysis shows that the conformational changes in the TMD domain significantly affect affected the natural modes of the β3-TMD.
The observed interaction between talin and α-actinin, to our knowledge, has never been reported in the previous literature and could serve as a new target for future experimental studies. Our results suggested that one of the mechanisms by which α-actinin impairs β3-integrin-mediated signaling and competes with talin, is by forming simultaneous interactions with the β3-tail and talin head, which effectively prevents talin from associating with the integrin tail. Interestingly, α-actinin did not form any interaction with talin at the β1-tail, which confirmed our results on the exclusive occurrence of α-actinin-talin binding during competition.
Previous studies have discussed the effect of other cytoplasmic interactions at the integrin tail. For instance, it has been observed that the filamin competition with talin results in impaired integrin activation (11). On the contrary, kindlin binding promotes adhesion formation and reinforcement (24). Major approaches for examining the effect of these proteins are mostly limited to fluorescence imaging, cell-level assays (15, 36), which can only provide macroscopic information about the significance of cytoplasmic molecules in the integrin activation. This calls for new computational efforts to shed light on the mechanistic details and the factors involved in integrin activation (37, 38, 39). Recent crystallography studies provided a greatly valuable insight into the interhelical modifications within the αIIbβ3 heterodimer upon the A711P mutation, however such static snapshots provides no evidence on the dynamics of the system and potential allosteric regulations.
Taken together, our results suggest that α-actinin binding induces conformational changes in the β3-TMD, which can substantially modify mechanical signal transmission across the lipid membrane (Fig. 7). Furthermore, proline-induced kinks are shown to be evolutionarily important and critical for regulating interhelical packing of integrin receptors. However, to our knowledge, this is the first study that associates kink formation with the cytoplasmic interactions and explains its impact on the mechanism of signal transmission, which in turn cause macroscopic changes in cell adhesion and spreading.
Figure 7.
Possible molecular mechanism by which α-actinin suppresses integrin-mediated signaling. (A) In the inactive conformation of integrin, the TMD domains of integrin subunits directly interact. (B) As talin associates with the cytoplasmic tail of β3-integrin, the IMC interaction between integrin subunits is broken, changing the distance between αIIb and β3 subunits without altering the TMD conformation. (C) Talin and α-actinin compete for binding to the β3-tail. For clarity, only R1 and R2 repeats of α-actinin are shown. (D) α-Actinin directly engages with both the β3-tail and talin head and prevents talin from binding to integrin. It also induces a kink at A711 in the β3-TMD, which most likely impairs integrin signaling. For clarity, the full-length α-actinin is not shown but it was included in all simulations. To see this figure in color, go online.
Author Contributions
H.S. and M.R.K.M. designed the experiments, analyzed the data, and wrote the manuscript. H.S. performed the experiments. M.R.K.M. contributed materials, and analysis tools.
Acknowledgments
We thank Zainab Haydari and Zeinab Jahed and all other members of the Molecular Cell Biomechanics Laboratory for their fruitful discussions and feedbacks throughout this work.
This study was supported by National Science Foundation (NSF) grant CBET-0829205 and CAREER Award CBET-0955291 to M.R.K.M. In addition, this work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation (NSF) (grant ACI-1053575).
Editor: Markus Buehler.
Footnotes
One figure is available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(30751-8).
Supporting Material
References
- 1.Iskratsch T., Wolfenson H., Sheetz M.P. Appreciating force and shape the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 2014;15:825–833. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katta S., Krieg M., Goodman M.B. Feeling force: physical and physiological principles enabling sensory mechanotransduction. Annu. Rev. Cell Dev. Biol. 2015;31:347–371. doi: 10.1146/annurev-cellbio-100913-013426. [DOI] [PubMed] [Google Scholar]
- 3.Mofrad M.R.K., Kamm R.D. Cambridge University Press; New York, NY: 2014. Cellular Mechanotransduction: Diverse Perspectives From Molecules to Tissues. [Google Scholar]
- 4.Jahed Z., Shams H., Mofrad M.R.K. Mechanotransduction pathways linking the extracellular matrix to the nucleus. Int. Rev. Cell Mol. Biol. 2014;310:171–220. doi: 10.1016/B978-0-12-800180-6.00005-0. [DOI] [PubMed] [Google Scholar]
- 5.Galbraith C.G., Yamada K.M., Sheetz M.P. The relationship between force and focal complex development. J. Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carisey A., Tsang R., Ballestrem C. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr. Biol. 2013;23:271–281. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardel M.L., Sabass B., Waterman C.M. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J. Cell Biol. 2008;183:999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calderwood D.A., Yan B., Ginsberg M.H. The phosphotyrosine binding-like domain of talin activates integrins. J. Biol. Chem. 2002;277:21749–21758. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 9.Truong T., Shams H., Mofrad M.R.K. Mechanisms of integrin and filamin binding and their interplay with talin during early focal adhesion formation. Integr. Biol. 2015;7:1285–1296. doi: 10.1039/c5ib00133a. [DOI] [PubMed] [Google Scholar]
- 10.Calderwood D.A., Campbell I.D., Critchley D.R. Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 2013;14:503–517. doi: 10.1038/nrm3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiema T., Lad Y., Calderwood D.A. The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Chen H.S., Kolahi K.S., Mofrad M.R.K. Phosphorylation facilitates the integrin binding of filamin under force. Biophys. J. 2009;97:3095–3104. doi: 10.1016/j.bpj.2009.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderwood D.A., Huttenlocher A., Ginsberg M.H. Increased filamin binding to β-integrin cytoplasmic domains inhibits cell migration. Nat. Cell Biol. 2001;3:1060–1068. doi: 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- 14.Ye F., Lagarrigue F., Ginsberg M.H. SnapShot: talin and the modular nature of the integrin adhesome. Cell. 2014;156:1340. doi: 10.1016/j.cell.2014.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roca-Cusachs P., del Rio A., Sheetz M.P. Integrin-dependent force transmission to the extracellular matrix by α-actinin triggers adhesion maturation. Proc. Natl. Acad. Sci. USA. 2013;110:E1361–E1370. doi: 10.1073/pnas.1220723110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaidel-Bar R., Ballestrem C., Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 2003;116:4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 17.Hampton C.M., Taylor D.W., Taylor K.A. Novel structures for α-actinin:F-actin interactions and their implications for actin-membrane attachment and tension sensing in the cytoskeleton. J. Mol. Biol. 2007;368:92–104. doi: 10.1016/j.jmb.2007.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hotulainen P., Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciobanasu C., Faivre B., Le Clainche C. Reconstituting actomyosin-dependent mechanosensitive protein complexes in vitro. Nat. Protoc. 2015;10:75–89. doi: 10.1038/nprot.2014.200. [DOI] [PubMed] [Google Scholar]
- 20.Shams H., Golji J., Mofrad M.R.K. Dynamic regulation of α-actinin’s calponin homology domains on F-actin. Biophys. J. 2016;110:1444–1455. doi: 10.1016/j.bpj.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shams H., Golji J., Mofrad M.R.K. A molecular trajectory of α-actinin activation. Biophys. J. 2012;103:2050–2059. doi: 10.1016/j.bpj.2012.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tadokoro S., Nakazawa T., Tomiyama Y. A potential role for α-actinin in inside-out αIIbβ3 signaling. Blood. 2011;117:250–258. doi: 10.1182/blood-2009-10-246751. [DOI] [PubMed] [Google Scholar]
- 23.Edlund M., Lotano M.A., Otey C.A. Dynamics of α-actinin in focal adhesions and stress fibers visualized with α-actinin-green fluorescent protein. Cell Motil. Cytoskeleton. 2001;48:190–200. doi: 10.1002/1097-0169(200103)48:3<190::AID-CM1008>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 24.Ye F., Snider A.K., Ginsberg M.H. Talin and kindlin: the one-two punch in integrin activation. Front. Med. 2014;8:6–16. doi: 10.1007/s11684-014-0317-3. [DOI] [PubMed] [Google Scholar]
- 25.Kim C., Schmidt T., Ginsberg M.H. Basic amino-acid side chains regulate transmembrane integrin signalling. Nature. 2011;481:209–213. doi: 10.1038/nature10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 27.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 30.Grant B.J., Rodrigues A.P.C., Caves L.S.D. Bio3D: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 31.Lindahl E., Azuara C., Delarue M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. 2006;34:W52–W56. doi: 10.1093/nar/gkl082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim M., Carman C.V., Springer T.A. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 33.Provasi D., Negri A., Filizola M. Talin-driven inside-out activation mechanism of platelet αIIbβ3 integrin probed by multimicrosecond, all-atom molecular dynamics simulations. Proteins. 2014;82:3231–3240. doi: 10.1002/prot.24540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wegener K.L., Partridge A.W., Campbell I.D. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 35.Mehrbod M., Trisno S., Mofrad M.R.K. On the activation of integrin αIIbβ3: outside-in and inside-out pathways. Biophys. J. 2013;105:1304–1315. doi: 10.1016/j.bpj.2013.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim C., Ye F., Ginsberg M.H. Talin activates integrins by altering the topology of the β transmembrane domain. J. Cell Biol. 2012;197:605–611. doi: 10.1083/jcb.201112141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jamali Y., Jamali T., Mofrad M.R.K. An agent based model of integrin clustering: exploring the role of ligand clustering, integrin homo-oligomerization, integrin-ligand affinity, membrane crowdedness and ligand mobility. J. Comput. Phys. 2013;244:264–278. [Google Scholar]
- 38.Mehrbod M., Mofrad M.R.K. Localized lipid packing of transmembrane domains impedes integrin clustering. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1002948. 1:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shams H., Soheilypour M., Mofrad M.R.K. Looking “under the hood” of cellular mechanotransduction with multiscale computational tools: a systems biomechanics approach across multiple scales. ACS Biomat. Sci. Eng. 2017 doi: 10.1021/acsbiomaterials.7b00117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.